Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard?
Abstract
1. Introduction
2. Materials and Methods
2.1. Rationale for Isolate Selection
2.2. DNA Extraction and Whole-Genome Sequencing (WGS)
2.3. Sequence-Based Taxonomic Assignment and Sequence Type Definition
2.4. Phylogenetic Analysis by SNP Calling
2.5. Prediction of Resistance and Virulence Determinants and Mobilization
2.6. Antimicrobial Susceptibility Testing
2.7. Pathogenicity Assessment
3. Results
3.1. Citrobacter Prevalence and Taxonomic Assignment Across Food Processing Facilities
3.2. De Novo Assembly
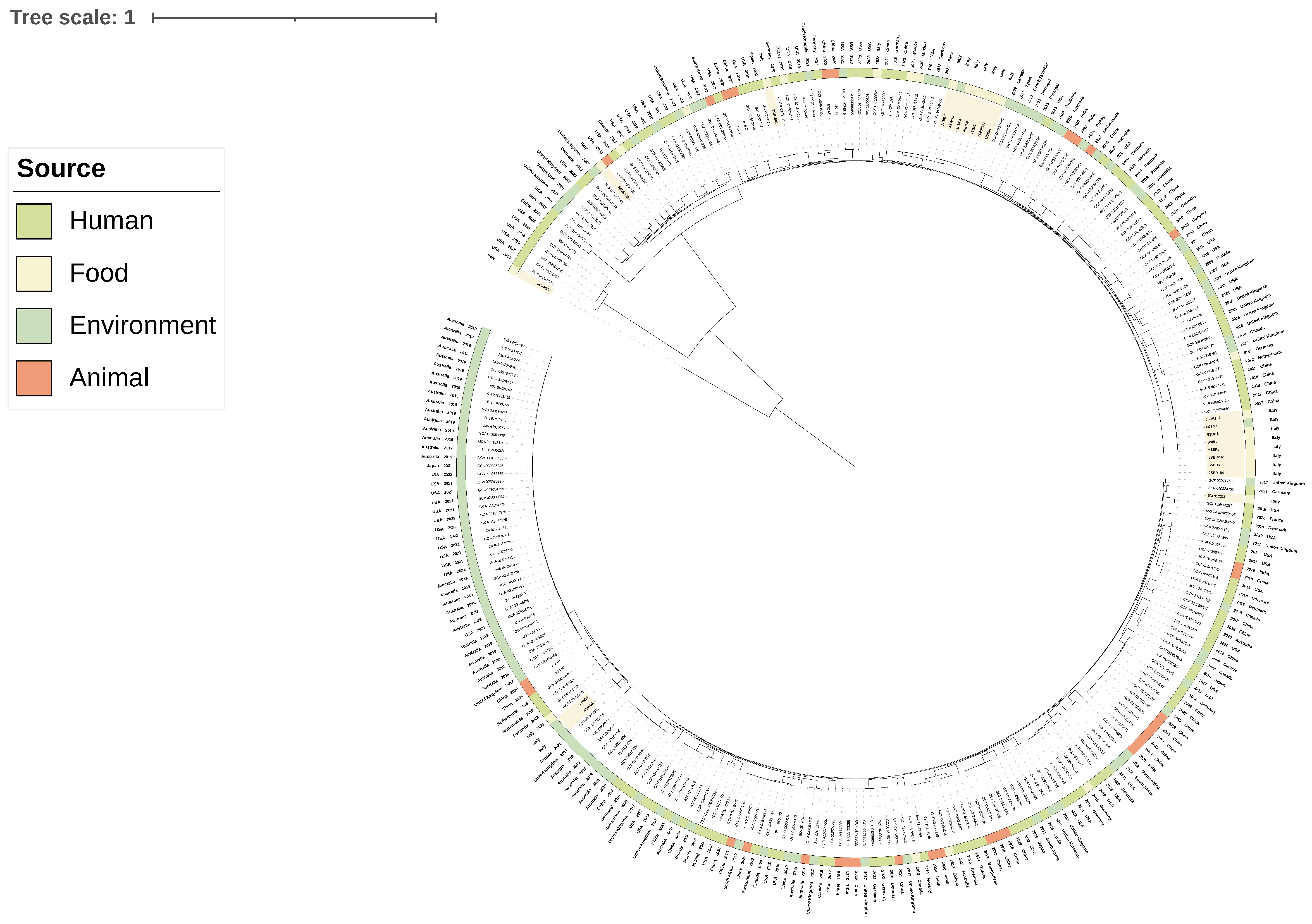
3.3. Phylogenetic Analyses and Sequence Type Distribution
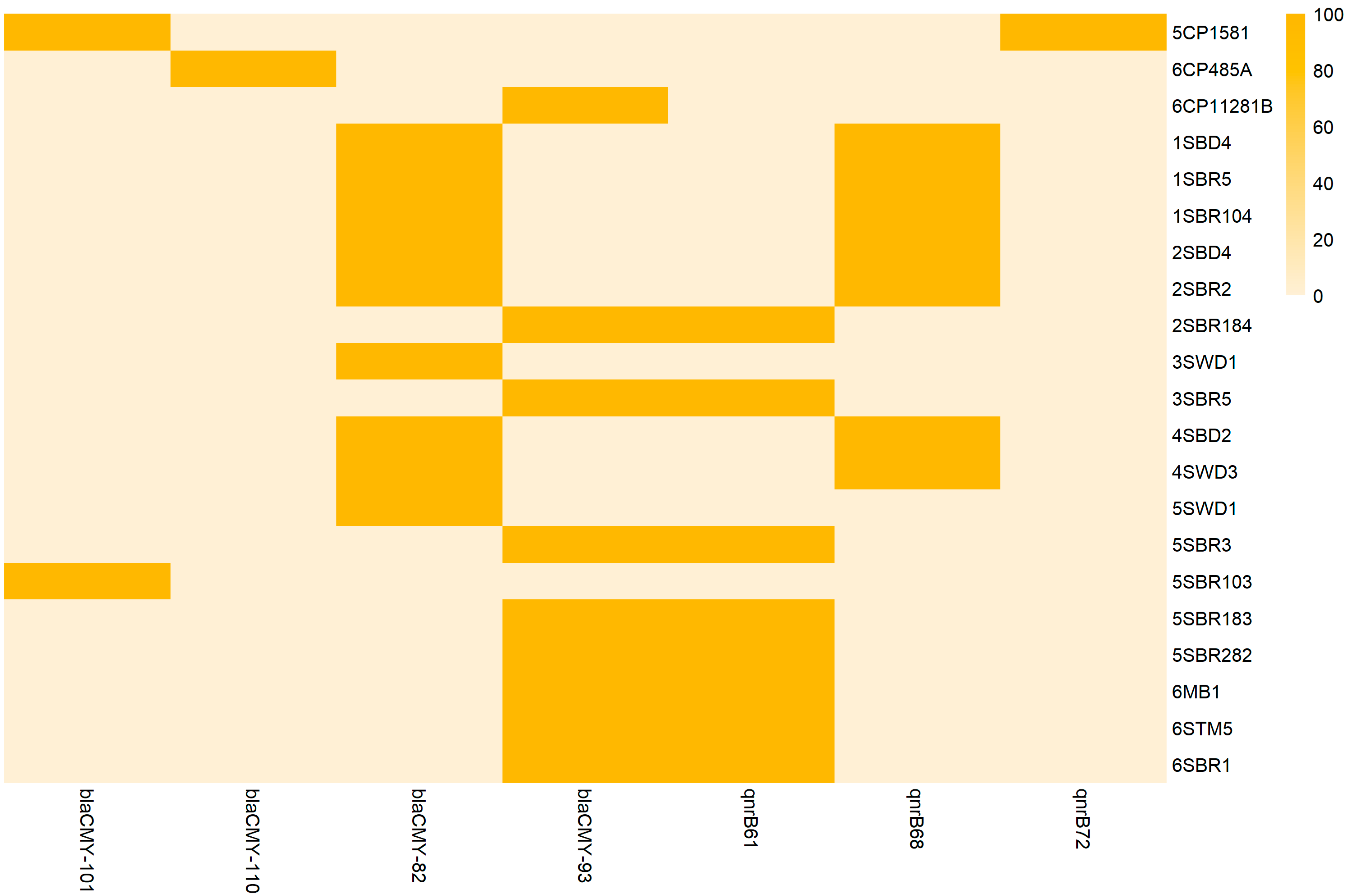
3.4. Resistome and Antimicrobial Susceptibility Testing
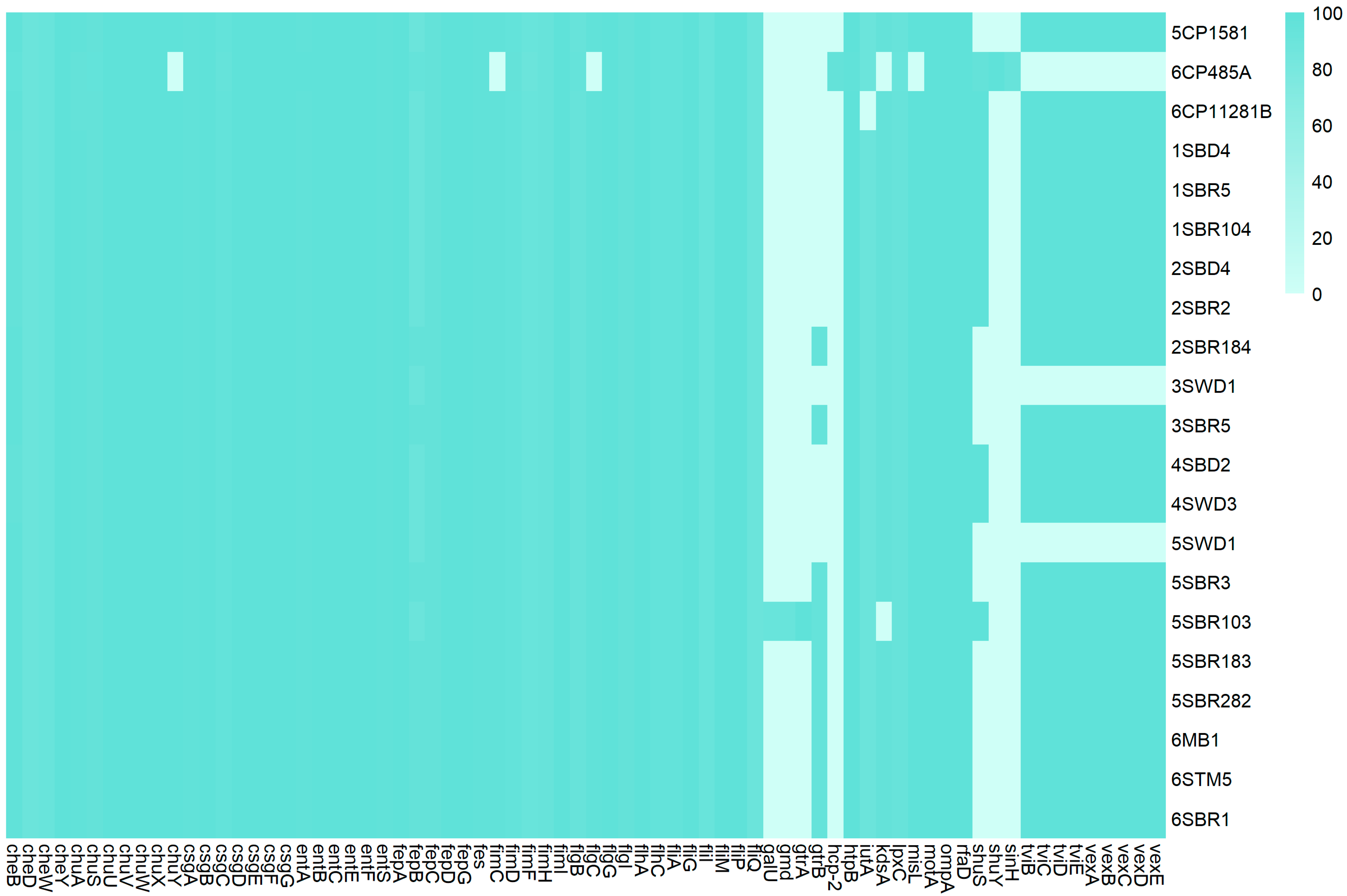
3.5. Virulome and Pathogenicity Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Fonton, P.; Hassoun-Kheir, N.; Harbarth, S. Epidemiology of Citrobacter spp. infections among hospitalized patients: A systematic review and meta-analysis. BMC Infect. Dis. 2024, 24, 662. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.; Choi, S.M.; Jo, S.J.; Lee, J.; Cho, S.Y.; Kim, S.H.; Lee, D.G.; Jeong, H.S. Clinical characteristics and antimicrobial susceptibility trends in Citrobacter bacteremia: An 11-year single-center experience. Infect. Chemother. 2019, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Parvez, S.; Khan, A.U. Hospital sewage water: A reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 2018, 51, 82–88. [Google Scholar] [CrossRef]
- Pletz, M.W.; Wollny, A.; Dobermann, U.H.; Rödel, J.; Neubauer, S.; Stein, C.; Brandt, C.; Hartung, A.; Mellmann, A.; Trommer, S.; et al. A nosocomial foodborne outbreak of a VIM carbapenemase-expressing Citrobacter freundii. Clin. Infect. Dis. 2018, 67, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Venditti, C.; Fortini, D.; Villa, L.; Vulcano, A.; D’Arezzo, S.; Capone, A.; Petrosillo, N.; Nisii, C.; Carattoli, A.; Di Caro, A. Circulation of blaKPC-3-carrying IncX3 plasmids among Citrobacter freundii isolates in an Italian hospital. Antimicrob. Agents Chemother. 2017, 61, e00505-17. [Google Scholar] [CrossRef]
- Hirai, J.; Uechi, K.; Hagihara, M.; Sakanashi, D.; Kinjo, T.; Haranaga, S.; Fujita, J. Bacteremia due to Citrobacter braakii: A case report and literature review. J. Infect. Chemother. 2016, 22, 819–821. [Google Scholar] [CrossRef]
- Khan, A.; Nandi, R.K.; Das, S.C.; Ramamurthy, T.; Khanam, J.; Shimizu, T.; Yamasaki, S.; Bhattacharya, S.K.; Chaicumpa, W.; Takeda, Y.; et al. Environmental isolates of Citrobacter braakii that agglutinate with Escherichia coli O157 antiserum but do not possess the genes responsible for the biosynthesis of O157 somatic antigen. Epidemiol. Infect. 2003, 130, 179–186. [Google Scholar] [CrossRef]
- Kim, H.-S.; Kim, Y.-J.; Chon, J.-W.; Kim, D.-H.; Kim, K.-Y.; Seo, K.-H. Citrobacter braakii: A major cause of false-positive results on MacConkey and Levine’s Eosin Methylene Blue selective agars used for the isolation of Escherichia coli from fresh vegetable samples. J. Food Saf. 2016, 36, 33–37. [Google Scholar] [CrossRef]
- Lai, C.C.; Tan, C.K.; Lin, S.H.; Liu, W.L.; Liao, C.H.; Huang, Y.T.; Hsueh, P.R. Bacteraemia caused by non-freundii, non-koseri Citrobacter species in Taiwan. J. Hosp. Infect. 2010, 76, 332–335. [Google Scholar] [CrossRef]
- Pławińska-Czarnak, J.; Wódz, K.; Kizerwetter-Świda, M.; Nowak, T.; Bogdan, J.; Kwieciński, P.; Kwieciński, A.; Anusz, K. Citrobacter braakii yield false-positive identification as Salmonella, a note of caution. Foods 2021, 10, 2177. [Google Scholar] [CrossRef]
- Pławińska-Czarnak, J.; Wódz, K.; Strzałkowska, Z.; Żychska, M.; Nowak, T.; Kwieciński, A.; Kwieciński, P.; Bielecki, W.; Rodo, A.; Rzewuska, M.; et al. Comparison of automatic methods MALDI-TOF, VITEK2 and manual methods for the identification of intestinal microbial communities on the example of samples from alpacas (Vicugna pacos). J. Vet. Res. 2023, 67, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Cortés-Sánchez, A.D.J.; Salgado-Cruz, M.d.l.P.; Diaz-Ramírez, M.; Torres-Ochoa, E.; Espinosa-Chaurand, L.D. A Review on Food Safety: The Case of Citrobacter sp., Fish and Fish Products. Appl. Sci. 2023, 12, 6907. [Google Scholar] [CrossRef]
- Mulita, F.; Tchabashvili, L.; Liolis, E.; Tasios, K.; Iliopoulos, F.; Kaplanis, C.; Parchas, N.; Plachouri, K.M. Green nail syndrome caused by Citrobacter braakii. Clin. Case Rep. 2021, 9, e04203. [Google Scholar] [CrossRef]
- Tollkuci, E.; Myers, R. Citrobacter braakii CLABSI in a hematopoietic stem cell transplant patient. J. Oncol. Pharm. Pract. 2021, 27, 1792–1794. [Google Scholar] [CrossRef]
- Yumoto, T.; Kono, Y.; Kawano, S.; Kamoi, C.; Iida, A.; Nose, M.; Sato, K.; Ugawa, T.; Okada, H.; Ujike, Y.; et al. Citrobacter braakii bacteremia-induced septic shock after colonoscopy preparation with polyethylene glycol in a critically ill patient: A case report. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 22. [Google Scholar] [CrossRef]
- Yu, M.; Xie, F.; Xu, C.; Yu, T.; Wang, Y.; Liang, S.; Dong, Q.; Wang, L. Characterization of cytotoxic Citrobacter braakii isolated from human stomach. FEBS Open Bio 2024, 14, 487–497. [Google Scholar] [CrossRef]
- Basra, P.; Koziol, A.; Wong, A.; Carrillo, C.D. Complete Genome Sequences of Citrobacter braakii Strains GTA-CB01 and GTA-CB04, isolated from ground beef. Genome Announc. 2015, 3, e01307–e01314. [Google Scholar] [CrossRef]
- Cole, S.D.; Healy, I.; Dietrich, J.M.; Redding, L.E. Evaluation of canine raw food products for the presence of extended-spectrum beta-lactamase- and carbapenemase-producing bacteria of the order Enterobacterales. Am. J. Vet. Res. 2022, 83, ajvr.21.12.0205. [Google Scholar] [CrossRef]
- Duceppe, M.O.; Phipps-Todd, B.; Carrillo, C.; Huang, H. Draft Genome Sequences of Eight Canadian Citrobacter braakii and Citrobacter freundii Strains. Microbiol. Resour. Announc. 2019, 8, e00273-19. [Google Scholar] [CrossRef]
- Jabeen, I.; Islam, S.; Hassan, A.K.M.I.; Tasnim, Z.; Shuvo, S.R. A brief insight into Citrobacter species-a growing threat to public health. Front. Antibiot. 2023, 2, 1276982. [Google Scholar] [CrossRef]
- Sennati, S.; Di Pilato, V.; Riccobono, E.; Di Maggio, T.; Villagran, A.L.; Pallecchi, L.; Bartoloni, A.; Rossolini, G.M.; Giani, T. Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J. Antimicrob. Chemother. 2017, 72, 2127–2129. [Google Scholar] [CrossRef] [PubMed]
- Zarzecka, U.; Chajęcka-Wierzchowska, W.; Zadernowska, A. Occurrence of antibiotic resistance among Enterobacterales isolated from raw and ready-to-eat food—Phenotypic and genotypic characteristics. Int. J. Environ. Health Res. 2022, 32, 1733–1744. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, F.; Valero, A.; Possas, A.; Lucchi, A.; Crippa, C.; Gambi, L.; Manfreda, G.; De Cesare, A. Occurrence of foodborne pathogens in Italian soft artisanal cheeses displaying different intra- and inter-batch variability of physicochemical and microbiological parameters. Front. Microbiol. 2022, 13, 959648. [Google Scholar] [CrossRef] [PubMed]
- Pasquali, F.; Valero, A.; Possas, A.; Lucchi, A.; Crippa, C.; Gambi, L.; Manfreda, G.; De Cesare, A. Variability in physicochemical parameters and its impact on microbiological quality and occurrence of foodborne pathogens in artisanal Italian organic salami. Foods 2023, 12, 4086. [Google Scholar] [CrossRef]
- Crippa, C.; Pasquali, F.; Rodrigues, C.; De Cesare, A.; Lucchi, A.; Gambi, L.; Manfreda, G.; Brisse, S.; Palma, F. Genomic features of Klebsiella isolates from artisanal ready-to-eat food production facilities. Sci. Rep. 2023, 131, 10957. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Contig_info—Estimating Standard Descriptive Statistics from Contig Sequences (Version 2.0.1). Available online: https://gitlab.pasteur.fr/GIPhy/contig_info (accessed on 8 March 2025).
- Schwengers, O.; Hain, T.; Chakraborty, T.; Goesmann, A. ReferenceSeeker: Rapid determination of appropriate reference genomes. J. Open Source Softw. 2020, 5, 1994. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- MLST—Scan Contig Files Against Traditional PubMLST Typing Schemes (Version 2.23.0). Available online: https://github.com/tseemann/mlst (accessed on 8 March 2025).
- NCBI—National Centre for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 8 March 2025).
- PubMLST—Public Databases for Molecular Typing and Microbial Genome Diversity. Available online: https://pubmlst.org/ (accessed on 8 March 2025).
- Eaton, O.C. NCBImeta: Efficient and comprehensive metadata retrieval from NCBI databases. J. Open Source Softw. 2020, 5, 1990. [Google Scholar] [CrossRef]
- Hall, B.G.; Nisbet, J. Building phylogenetic trees from genome sequences with kSNP4. Mol. Biol. Evol. 2023, 40, msad235. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Taouk, M.L.; Featherstone, L.A.; Taiaroa, G.; Seemann, T.; Ingle, D.J.; Stinear, T.P.; Wick, R.R. Exploring SNP Filtering Strategies: The Influence of Strict vs. Soft Core. Microb. Genom. 2025, 11, e001346. [Google Scholar] [CrossRef] [PubMed]
- Abricate—Mass Screening of Contigs for Antimicrobial Resistance or Virulence Genes (Version 1.0.1). Available online: https://github.com/tseemann/abricate (accessed on 8 March 2025).
- SnapGene Viewer (Version 7.1.1). Available online: https://www.snapgene.com/snapgene-viewer (accessed on 17 April 2025).
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software Tools for Clustering, Reconstruction and Typing of Plasmids from Draft Assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef]
- ISO Norm 20776-1:2019; Susceptibility Testing of Infectious Agents and Evaluation of Performance of Antimicrobial Susceptibility Test Devices. Part 1: Broth Micro-Dilution Reference Method for Testing the In Vitro Activity of Antimicrobial Agents Against Rapidly Growing Aerobic Bacteria Involved in Infectious Diseases. International Organization for Standardization: Geneva, Switzerland, 2019.
- EUCAST—European Committee of Susceptibility Testing. Clinical Breakpoints (Version 15.0). Available online: https://www.eucast.org/clinical_breakpoints (accessed on 8 March 2025).
- Dindo, M.L.; Francati, S. Soy flour versus skimmed milk powder in artificial diets for Galleria mellonella, a factitious host for Exorista larvarum. Bull. Insectology 2022, 75, 273–280. [Google Scholar]
- Gallorini, M.; Marinacci, B.; Pellegrini, B.; Cataldi, A.; Dindo, M.L.; Carradori, S.; Grande, R. Immunophenotyping of hemocytes from infected Galleria mellonella larvae as an innovative tool for immune profiling, infection studies and drug screening. Sci. Rep. 2024, 14, 759. [Google Scholar] [CrossRef]
- Zhang, G.; Zhao, Q.; Ye, K.; Ye, L.; Ma, Y.; Yang, J. Molecular analysis of clinical Citrobacter spp. isolates: Acquisition of the Yersinia high-pathogenicity island mediated by ICEkp in C. freundii. Front. Microbiol. 2023, 14, 1056790. [Google Scholar] [CrossRef]
- Barlow, M.; Hall, B.G. Origin and evolution of the AmpC beta-lactamases of Citrobacter freundii. Antimicrob. Agents Chemother. 2002, 46, 1190–1198. [Google Scholar] [CrossRef]
- Bartowsky, E.; Normark, S. Purification and mutant analysis of Citrobacter freundii AmpR, the regulator for chromosomal AmpC beta-lactamase. Mol. Microbiol. 1991, 5, 1715–1725. [Google Scholar] [CrossRef]
- Kong, K.F.; Jayawardena, S.R.; Indulkar, S.D.; Del Puerto, A.; Koh, C.L.; Høiby, N.; Mathee, K. Pseudomonas aeruginosa AmpR is a global transcriptional factor that regulates expression of AmpC and PoxB beta-lactamases, proteases, quorum sensing, and other virulence factors. Antimicrob. Agents Chemother. 2005, 49, 4567–4575. [Google Scholar] [CrossRef]
- Ribeiro, T.G.; Novais, Â.; Branquinho, R.; Machado, E.; Peixe, L. Phylogeny and comparative genomics unveil independent diversification trajectories of qnrB and genetic platforms within particular Citrobacter species. Antimicrob. Agents Chemother. 2015, 59, 5951–5958. [Google Scholar] [CrossRef] [PubMed]
- Tariq, F.N.; Shafiq, M.; Khawar, N.; Habib, G.; Gul, H.; Hayat, A.; Rehman, M.U.; Moussa, I.M.; Mahmoud, E.A.; Elansary, H.O. The functional repertoire of AmpR in the AmpC β-lactamase high expression and decreasing β-lactam and aminoglycosides resistance in ESBL Citrobacter freundii. Heliyon 2023, 9, e19486. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E.F.; Porwollik, S.; Blackmer, F.; McClelland, M. Differences in gene content among Salmonella enterica serovar typhi isolates. J. Clin. Microbiol. 2003, 41, 3823–3828. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stessl, B.; Szakmary-Brändle, K.; Vorberg, U.; Schoder, D.; Wagner, M. Temporal analysis of the Listeria monocytogenes population structure in floor drains during reconstruction and expansion of a meat processing plant. Int. J. Food Microbiol. 2020, 314, 108360. [Google Scholar] [CrossRef]
- Dzieciol, M.; Schornsteiner, E.; Muhterem-Uyar, M.; Stessl, B.; Wagner, M.; Schmitz-Esser, S. Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Int. J. Food Microbiol. 2016, 223, 33–40. [Google Scholar] [CrossRef]
- Paradis, A.; Beaudet, M.F.; Boisvert Moreau, M.; Huot, C. Investigation of a Salmonella Montevideo outbreak related to the environmental contamination of a restaurant kitchen drainage system, Québec, Canada, 2020–2021. J. Food Prot. 2023, 86, 100131. [Google Scholar] [CrossRef]
- Hamerlinck, H.; Aerssens, A.; Boelens, J.; Dehaene, A.; McMahon, M.; Messiaen, A.S.; Vandendriessche, S.; Velghe, A.; Leroux-Roels, I.; Verhasselt, B. Sanitary installations and wastewater plumbing as reservoir for the long-term circulation and transmission of carbapenemase-producing Citrobacter freundii clones in a hospital setting. Antimicrob. Resist. Infect. Control 2023, 12, 58. [Google Scholar] [CrossRef]
- Zhou, G.; Wang, Y.S.; Peng, H.; Li, S.J.; Sun, T.L.; Li, C.L.; Shi, Q.S.; Xie, X.B. ompX contribute to biofilm formation, osmotic response and swimming motility in Citrobacter werkmanii. Gene 2023, 851, 147019. [Google Scholar] [CrossRef]
- Muhterem-Uyar, M.; Dalmasso, M.; Bolocan, A.S.; Hernandez, M.; Kapetanakou, A.E.; Kuchta, T.; Manios, S.G.; Melero, B.; Minarovičová, J.; Nicolau, A.I.; et al. Environmental sampling for Listeria monocytogenes control in food processing facilities reveals three contamination scenarios. Food Control 2015, 51, 94–107. [Google Scholar] [CrossRef]
- Pakdel, M.; Olsen, A.; Bar, E.M.S. A review of food contaminants and their pathways within food processing facilities using open food processing equipment. J. Food Prot. 2023, 86, 100184. [Google Scholar] [CrossRef]
- Bonardi, S.; Cabassi, C.S.; Manfreda, G.; Parisi, A.; Fiaccadori, E.; Sabatino, A.; Cavirani, S.; Bacci, C.; Rega, M.; Spadini, C.; et al. Survey on Carbapenem-resistant bacteria in pigs at slaughter and comparison with human clinical isolates in Italy. Antibiotics 2022, 11, 777. [Google Scholar] [CrossRef] [PubMed]
- Hidayatullah, A.R.; Effendi, M.H.; Plumeriastuti, H.; Wibisono, F.M.; Hartadi, E.B.; Sofiana, E.D. A Review of the opportunistic pathogen Citrobacter freundii in piglets post weaning: Public health importance. Sys. Rev. Pharm. 2020, 11, 767–773. [Google Scholar]
- Hem, S.; Cummins, M.L.; Wyrsch, E.R.; Drigo, B.; Hoye, B.J.; Maute, K.; Sanderson-Smith, M.; Gorman, J.; Bogema, D.R.; Jenkins, C.; et al. Genomic analysis of Citrobacter from Australian wastewater and silver gulls reveals novel sequence types carrying critically important antibiotic resistance genes. Sci. Total Environ. 2024, 909, 168608. [Google Scholar] [CrossRef] [PubMed]
- Hone, D.M.; Attridge, S.R.; Forrest, B.; Morona, R.; Daniels, D.; LaBrooy, J.T.; Bartholomeusz, R.C.; Shearman, D.J.; Hackett, J. A galE via (Vi antigen-negative) mutant of Salmonella typhi Ty2 retains virulence in humans. Infect. Immun. 1988, 56, 1326–1333. [Google Scholar] [CrossRef]
- Karlinsey, J.E.; Stepien, T.A.; Mayho, M.; Singletary, L.A.; Bingham-Ramos, L.K.; Brehm, M.A.; Greiner, D.L.; Shultz, L.D.; Gallagher, L.A.; Bawn, M.; et al. Genome-wide analysis of Salmonella enterica serovar Typhi in humanized mice reveals key virulence features. Cell Host Microbe 2019, 26, 426–434.e6. [Google Scholar] [CrossRef]
- Chen, L.; Zou, Y.; She, P.; Wu, Y. Composition, function, and regulation of T6SS in Pseudomonas aeruginosa. Microbiol. Res. 2015, 172, 19–25. [Google Scholar] [CrossRef]
- Durand, E.; Cambillau, C.; Cascales, E.; Journet, L. VgrG, Tae, Tle, and beyond: The versatile arsenal of Type VI secretion effectors. Trends Microbiol. 2014, 22, 498–507. [Google Scholar] [CrossRef]
- Merciecca, T.; Bornes, S.; Nakusi, L.; Theil, S.; Rendueles, O.; Forestier, C.; Miquel, S. Role of Klebsiella pneumoniae Type VI secretion system (T6SS) in long-term gastrointestinal colonization. Sci. Rep. 2022, 12, 16968. [Google Scholar] [CrossRef]
- Russell, A.B.; Peterson, S.B.; Mougous, J.D. Type VI secretion system effectors: Poisons with a purpose. Nat. Rev. Microbiol. 2014, 12, 137–148. [Google Scholar] [CrossRef]
- Champion, O.L.; Cooper, I.A.M.; James, S.L.; Ford, D.; Karlyshev, A.; Wren, B.W.; Duffield, M.; Oyston, P.C.F.; Titball, R.W. Galleria mellonella as an alternative infection model for Yersinia pseudotuberculosis. Microbiology 2009, 155, 1516–1522. [Google Scholar] [CrossRef]
- Chen, R.Y.; Keddie, B.A. The Galleria mellonella-Enteropathogenic Escherichia coli model system: Characterization of pathogen virulence and insect Immune Responses. J. Insect Sci. 2021, 21, 7. [Google Scholar] [CrossRef] [PubMed]
- Jander, G.; Rahme, L.G.; Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000, 182, 3843–3845. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Altincicek, B.; Hain, T.; Domann, E.; Vilcinskas, A.; Chakraborty, T. Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 2010, 76, 310–317. [Google Scholar] [CrossRef]
- Seed, K.D.; Dennis, J.J. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 2008, 76, 1267–1275. [Google Scholar] [CrossRef]
- Yang, H.F.; Pan, A.J.; Hu, L.F.; Liu, Y.Y.; Cheng, J.; Ye, Y.; Li, J.B. Galleria mellonella as an in vivo model for assessing the efficacy of antimicrobial agents against Enterobacter cloacae infection. J. Microbiol. Immunol. Infect. 2017, 50, 55–61. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Zaragoza, O. Immune Response of Galleria mellonella against Human Fungal Pathogens. J. Fungi 2019, 5, 3. [Google Scholar] [CrossRef]
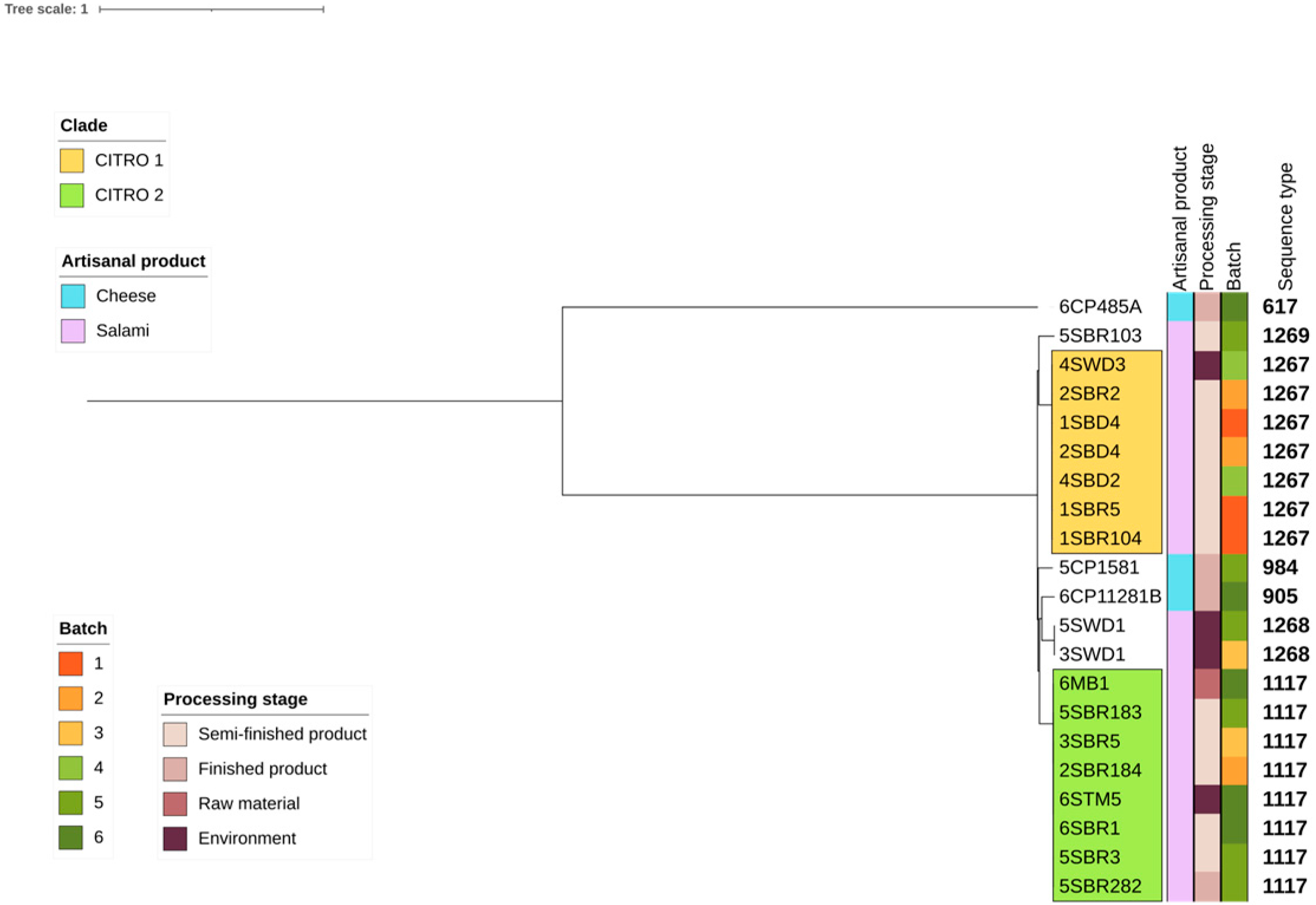
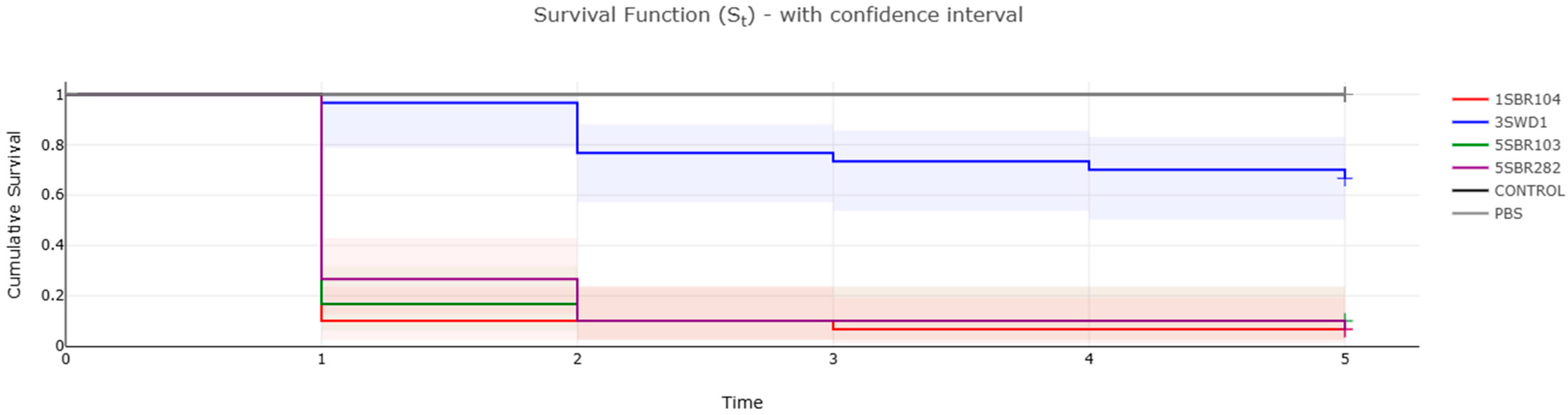
| Sample | Biotyping | WGS a Refseq | ANI b (%) | Food Production | Sample Origin | Batch | ST c |
|---|---|---|---|---|---|---|---|
| 5CP1581 | C. freundii | C. braakii | 99.08 | Cheese | Finished product | 5 | 984 |
| 6CP485A | K. pneumoniae | C. freundii | 99.75 | Cheese | Finished product | 6 | 617 |
| 6CP11281B | C. freundii | C. braakii | 99.10 | Cheese | Finished product | 6 | 905 |
| 1SBD4 | C. freundii | C. braakii | 99.07 | Salami | Semi-finished product | 1 | 1267 |
| 1SBR5 | C. freundii | C. braakii | 99.06 | Salami | Semi-finished product | 1 | 1267 |
| 1SBR104 | C. freundii | C. braakii | 99.05 | Salami | Semi-finished product | 1 | 1267 |
| 2SBD4 | C. freundii | C. braakii | 99.05 | Salami | Semi-finished product | 2 | 1267 |
| 2SBR2 | C. freundii | C. braakii | 99.07 | Salami | Semi-finished product | 2 | 1267 |
| 2SBR184 | C. freundii | C. braakii | 99.20 | Salami | Semi-finished product | 2 | 1117 |
| 3SWD1 | C. freundii | C. braakii | 99.22 | Salami | Environment | 3 | 1268 |
| 3SBR5 | C. freundii | C. braakii | 99.20 | Salami | Semi-finished product | 3 | 1117 |
| 4SBD2 | C. freundii | C. braakii | 99.04 | Salami | Semi-finished product | 4 | 1267 |
| 4SWD3 | C. freundii | C. braakii | 99.04 | Salami | Environment | 4 | 1267 |
| 5SWD1 | C. freundii | C. braakii | 99.22 | Salami | Environment | 5 | 1268 |
| 5SBR3 | C. freundii | C. braakii | 99.20 | Salami | Semi-finished product | 5 | 1117 |
| 5SBR103 | C. freundii | C. braakii | 99.22 | Salami | Semi-finished product | 5 | 1269 |
| 5SBR183 | C. freundii | C. braakii | 99.20 | Salami | Semi-finished product | 5 | 1117 |
| 5SBR282 | C. freundii | C. braakii | 99.40 | Salami | Semi-finished product | 5 | 1117 |
| 6MB1 | C. freundii | C. braakii | 99.20 | Salami | Raw material | 6 | 1117 |
| 6STM5 | K. oxytoca | C. braakii | 99.20 | Salami | Environment | 6 | 1117 |
| 6SBR1 | C. freundii | C. braakii | 99.22 | Salami | Semi-finished product | 6 | 1117 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pasquali, F.; Crippa, C.; Lucchi, A.; Francati, S.; Dindo, M.L.; Manfreda, G. Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard? Foods 2025, 14, 1887. https://doi.org/10.3390/foods14111887
Pasquali F, Crippa C, Lucchi A, Francati S, Dindo ML, Manfreda G. Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard? Foods. 2025; 14(11):1887. https://doi.org/10.3390/foods14111887
Chicago/Turabian StylePasquali, Frédérique, Cecilia Crippa, Alex Lucchi, Santolo Francati, Maria Luisa Dindo, and Gerardo Manfreda. 2025. "Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard?" Foods 14, no. 11: 1887. https://doi.org/10.3390/foods14111887
APA StylePasquali, F., Crippa, C., Lucchi, A., Francati, S., Dindo, M. L., & Manfreda, G. (2025). Citrobacter braakii Isolated from Salami and Soft Cheese: An Emerging Food Safety Hazard? Foods, 14(11), 1887. https://doi.org/10.3390/foods14111887








