Exogenous L-Cysteine and Its Transport Through CtaP Play a Role in Biofilm Formation, Swimming Motility, and Swarming Motility of Listeria monocytogenes
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Determination of Planktonic Growth
2.3. Determination of Biofilm Formation
2.4. Cell Motility Assay: Defined Media Supplemented with Different Concentrations of L-Cysteine with Agar
2.5. RNA Sequencing (RNA-Seq) Sample Preparation and Analysis of L. monocytogenes 10403S WT and ΔctaP in DMs
2.6. Statistical Analysis
3. Results
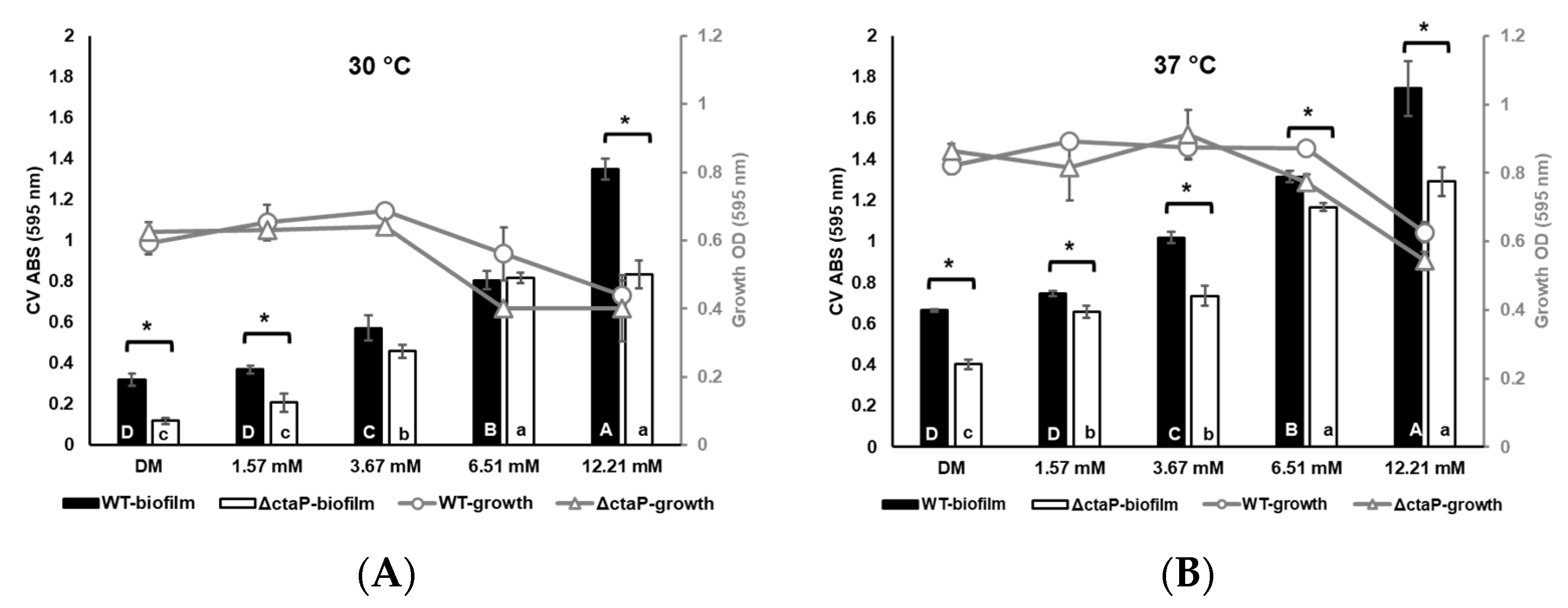
3.1. Planktonic Growth at the End of the 24 h Period
3.2. Biofilm Formation at the End of the 24 h Period
3.3. Relationship Between Growth and Biofilm Formation of L. monocytogenes
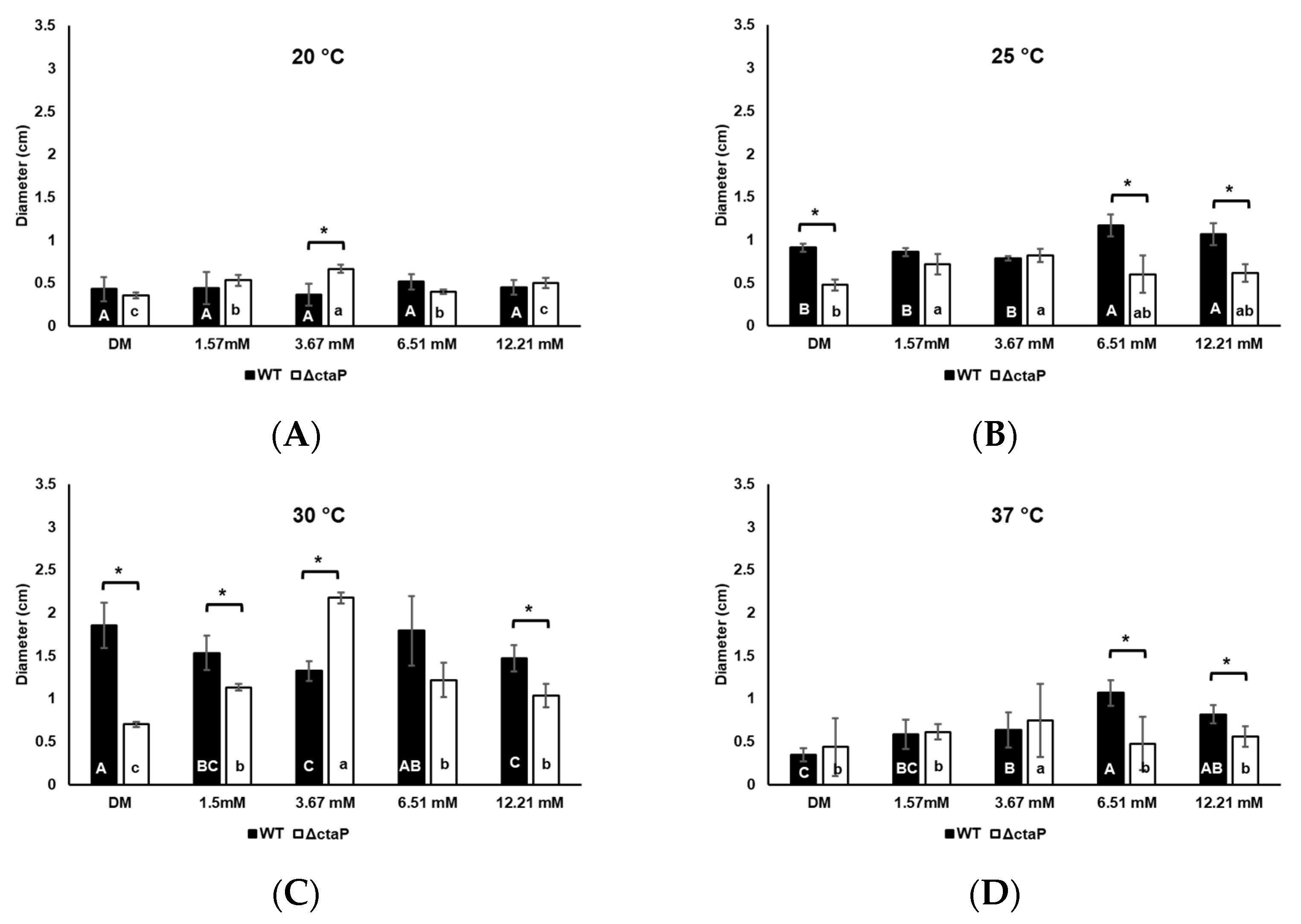
3.4. Effect of Cysteine on Swarming and Swimming Motility of L. monocytogenes 10403S WT and ΔctaP
3.5. Transcriptomic Analysis
- (i)
- Comparison of ΔctaP vs. WT in Basal DM
- (ii)
- Comparison of WT in Basal DM vs. DM with 1.57 mM L-cysteine
- (iii)
- Comparison of WT in Basal DM vs. DM with 3.67 mM L-cysteine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BHI | Brain heart infusion |
| CV | Crystal violet |
| c-di-GMP | Cyclic di-guanylate monophosphate |
| DMSO | Dimethyl sulfoxide |
| DM | Defined media |
| eDNA | Extracellular DNA |
| SAM | S-adenosylmethionine |
| QS | Quorum sensing |
References
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef] [PubMed]
- Ravindhiran, R.; Sivarajan, K.; Sekar, J.N.; Murugesan, R.; Dhandapani, K. Listeria monocytogenes an emerging pathogen: A comprehensive overview on listeriosis, virulence determinants, detection, and anti-listerial interventions. Microb. Ecol. 2023, 86, 2231–2251. [Google Scholar] [CrossRef] [PubMed]
- Matereke, L.T.; Okoh, A.I. Listeria monocytogenes virulence, antimicrobial resistance, and environmental persistence: A review. Pathogens 2020, 9, 528. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); European Centre for Disease Prevention and Control (ECDC). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-Borne Outbreaks in 2017; EFSA: Parma, Italy, 2018. [Google Scholar]
- Su, Y.; Liu, A.; Zhu, M.J. Mapping the landscape of listeriosis outbreaks (1998–2023): Trends, challenges, and regulatory responses in the United States. Trends Food Sci. Technol. 2024, 154, 104750. [Google Scholar] [CrossRef]
- Fernández-Martínez, N.F.; Ruiz-Montero, R.; Briones, E.; Baños, E.; Rodríguez-Alarcón, L.G.S.M.; Chaves, J.A.; Abad, R.; Varela, C.; Lorusso, N. Listeriosis outbreak caused by contaminated stuffed pork, Andalusia, Spain, July to October 2019. Eurosurveillance 2022, 27, 2200279. [Google Scholar] [CrossRef]
- Lachmann, R.; Halbedel, S.; Adler, M.; Becker, N.; Allerberger, F.; Holzer, A.; Boone, I.; Falkenhorst, G.; Kleta, S.; Al Dahouk, S.; et al. Nationwide outbreak of invasive listeriosis associated with consumption of meat products in health care facilities, Germany, 2014–2019. Clin. Microbiol. Infect. 2021, 27, 1035.e1–1035.e5. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Listeria (Listeriosis) Surveillance. CDC. Available online: https://www.cdc.gov/listeria/outbreaks/index.html (accessed on 20 April 2025).
- Dos Reis, J.O.; Vieira, B.S.; Cunha Neto, A.; Castro, V.S.; de Souza Figueiredo, E.E. Antimicrobial Resistance of Listeria monocytogenes from Animal Foods to First- and Second-Line Drugs in the Treatment of Listeriosis from 2008 to 2021: A Systematic Review and Meta-Analysis. Can. J. Infect. Dis. Med. Microbiol. 2022, 2022, 1351983. [Google Scholar] [CrossRef]
- Demirkol, O.; Adams, C.; Ercal, N. Biologically Important Thiols in Various Vegetables and Fruits. J. Agric. Food Chem. 2004, 52, 8151–8154. [Google Scholar] [CrossRef] [PubMed]
- Larsson, S.C.; Håkansson, N.; Wolk, A. Dietary Cysteine and Other Amino Acids and Stroke Incidence in Women. Stroke 2015, 46, 922–926. [Google Scholar] [CrossRef]
- O’Toole, G.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Da Silva, E.P.; De Martinis, E.C.P. Current knowledge and perspectives on biofilm formation: The case of Listeria monocytogenes. Appl. Microbiol. Biotechnol. 2013, 97, 957–968. [Google Scholar] [CrossRef] [PubMed]
- Kamp, H.D.; Higgins, D.E. A protein thermometer controls temperature-dependent transcription of flagellar motility genes in Listeria monocytogenes. PLoS Pathog. 2011, 7, e1002153. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, C.; Heuner, K.; Brand, B.C.; Hacker, J.; Steinert, M. Flagellum of Legionella pneumophila positively affects the early phase of infection of eukaryotic host cells. Infect. Immun. 2001, 69, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- O’Neil, H.S.; Marquis, H. Listeria monocytogenes flagella are used for motility, not as adhesins, to increase host cell invasion. Infect. Immun. 2006, 74, 6675–6681. [Google Scholar] [CrossRef]
- Mattingly, A.E.; Weaver, A.A.; Dimkovikj, A.; Shrout, J.D. Assessing travel conditions: Environmental and host influences on bacterial surface motility. J. Bacteriol. 2018, 200, e00014-18. [Google Scholar] [CrossRef]
- Harshey, R.M. Bacterial motility on a surface: Many ways to a common goal. Annu. Rev. Microbiol. 2003, 57, 249–273. [Google Scholar] [CrossRef]
- Cherifi, T.; Jacques, M.; Quessy, S.; Fravalo, P. Impact of nutrient restriction on the structure of Listeria monocytogenes biofilm grown in a microfluidic system. Front. Microbiol. 2017, 8, 864. [Google Scholar] [CrossRef]
- Liu, Y.; Zhu, H.; Dou, X.; Jia, K.; Panagou, E.Z.; Zhang, H.; Dong, Q. The influence of nutrients on biofilm formation of an ST87 strain of Listeria monocytogenes. LWT 2024, 191, 115658. [Google Scholar] [CrossRef]
- Folsom, J.P.; Siragusa, G.R.; Frank, J.F. Formation of biofilm at different nutrient levels by various genotypes of Listeria monocytogenes. J. Food Sci. 2006, 69, 826–834. [Google Scholar] [CrossRef]
- Kohler, T.; Curty, L.K.; Barja, F.; Van Delden, C.; Pechère, J.C. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 2000, 182, 5990–5996. [Google Scholar] [CrossRef]
- Lee, Y.J.; Wang, C. Links between S-adenosylmethionine and Agr-based quorum sensing for biofilm development in Listeria monocytogenes EGD-e. MicrobiologyOpen 2020, 9, e1015. [Google Scholar] [CrossRef] [PubMed]
- Xayarath, B.; Marquis, H.; Port, G.C.; Freitag, N.E. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol. Microbiol. 2009, 74, 956–973. [Google Scholar] [CrossRef] [PubMed]
- Vaval Taylor, D.M.; Xayarath, B.; Freitag, N.E. Two permeases associated with the multifunctional CtaP cysteine transport system in Listeria monocytogenes play distinct roles in pathogenesis. Microbiol. Spectr. 2023, 11, e03317-22. [Google Scholar] [CrossRef] [PubMed]
- Huillet, E.; Gohar, M. Quorum sensing in Bacillus cereus in relation to cysteine metabolism and the oxidative stress response. In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria; Wiley: Hoboken, NJ, USA, 2016; pp. 1242–1251. [Google Scholar]
- Soutourina, O.; Poupel, O.; Coppée, J.Y.; Danchin, A.; Msadek, T.; Martin-Verstraete, I. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulfur source utilization and plays a role in biofilm formation. Mol. Microbiol. 2009, 73, 194–211. [Google Scholar] [CrossRef]
- Turnbull, A.L.; Surette, M.G. L-Cysteine is required for induced antibiotic resistance in actively swarming Salmonella enterica serovar Typhimurium. Microbiology 2008, 154, 3410–3419. [Google Scholar] [CrossRef]
- Anderson, M.T.; Mitchell, L.A.; Mobley, H.L.T. Cysteine biosynthesis controls Serratia marcescens phospholipase activity. J. Bacteriol. 2017, 199, e0077617. [Google Scholar] [CrossRef]
- Man, L.; Dale, A.L.; Klare, W.P.; Cain, J.A.; Sumer-Bayraktar, Z.; Niewold, P.; Solis, N.; Cordwell, S.J. Proteomics of Campylobacter jejuni growth in deoxycholate reveals Cj0025c as a cystine transport protein required for wild-type human infection phenotypes. Mol. Cell. Proteom. 2020, 19, 1263–1280. [Google Scholar] [CrossRef]
- Karatzas, K.A.G.; Brennan, O.; Heavin, S.; Morrissey, J.; O’Byrne, C.P. Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: Uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 2010, 76, 3529–3537. [Google Scholar] [CrossRef]
- Amezaga, M.R.; Davidson, I.; McLaggan, D.; Verheul, A.; Abee, T.; Booth, I.R. The role of peptide metabolism in the growth of Listeria monocytogenes ATCC 23074 at high osmolarity. Microbiology 1995, 141, 41–49. [Google Scholar] [CrossRef]
- Gou, H.; Cao, Q.; Wang, Z.; Liu, Y.; Sun, Y.; Wei, H.; Xue, H. Transcriptomic analysis of Listeria monocytogenes biofilm formation at different times. Can. J. Vet. Res. 2023, 87, 59–66. [Google Scholar]
- Alonso, A.N.; Perry, K.J.; Regeimbal, J.M.; Regan, P.M.; Higgins, D.E. Identification of Listeria monocytogenes determinants required for biofilm formation. PLoS ONE. 2014, 9, e0113696. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, Q.; Tian, S.; Zhang, Y.; Liu, S.; Ou, Q.; Chen, Z.; Wang, C. Hemolysin function of Listeria is related to biofilm formation: Transcriptomics analysis. Vet. Res. 2022, 53, 113. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, A.L.; Surette, M.G. Cysteine biosynthesis, oxidative stress, and antibiotic resistance in Salmonella typhimurium. Res. Microbiol. 2010, 161, 643–650. [Google Scholar] [CrossRef]
- Fan, Y.; Qiao, J.; Lu, Z.; Fen, Z.; Tao, Y.; Lv, F.; Zhao, H.; Zhang, C.; Bie, X. Influence of different factors on biofilm formation of Listeria monocytogenes and the regulation of cheY gene. Food Res. Int. 2020, 137, 109405. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, M.A.; Pedersen, S. Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 1991, 173, 5244–5246. [Google Scholar] [CrossRef]
- Kadam, S.R.; den Besten, H.M.W.; van der Veen, S.; Zwietering, M.H.; Moezelaar, R.; Abee, T. Diversity assessment of Listeria monocytogenes biofilm formation: Impact of growth condition, serotype, and strain origin. Int. J. Food Microbiol. 2013, 165, 259–264. [Google Scholar] [CrossRef]
- Zetzmann, M.; Okshevsky, M.; Endres, J.; Sedlag, A.; Caccia, N.; Auchter, M.; Waidmann, M.S.; Desvaux, M.; Meyer, R.L.; Riedel, C.U. DNase-sensitive and -resistant modes of biofilm formation by Listeria monocytogenes. Front. Microbiol. 2015, 6, 1428. [Google Scholar] [CrossRef]
- Okshevsky, M.; Meyer, R.L. The role of extracellular DNA in the establishment, maintenance, and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015, 41, 341–352. [Google Scholar] [CrossRef]
- Harmsen, M.; Lappann, M.; Knøchel, S.; Molin, S. Role of extracellular DNA during biofilm formation by Listeria monocytogenes. Appl. Environ. Microbiol. 2010, 76, 2271–2279. [Google Scholar] [CrossRef]
- Ivanova, L.A.; Egorov, V.V.; Zabrodskaya, Y.A.; Shaldzhyan, A.A.; Baranchikov, A.Y.; Tsvigun, N.V.; Lykholay, A.N.; Yapryntsev, A.D.; Lebedev, D.V.; Kulminskaya, A.A. Matrix is everywhere: Extracellular DNA is a link between biofilm and mineralization in Bacillus cereus planktonic lifestyle. NPJ Biofilms Microbiomes 2023, 9, 9. [Google Scholar] [CrossRef]
- Wen, H.Q.; Xing, D.F.; Xie, G.J.; Yin, T.M.; Ren, N.Q.; Liu, B.F. Enhanced photo-fermentative hydrogen production by synergistic effects of formed biofilm and added L-cysteine. Renew. Energy 2019, 139, 643–650. [Google Scholar] [CrossRef]
- Nyenje, M.E.; Green, E.; Ndip, R.N. Evaluation of the effect of different growth media and temperature on the suitability of biofilm formation by Enterobacter cloacae strains isolated from food samples in South Africa. Molecules 2013, 18, 9582–9593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. Royal Soc. Chem. 2017, 58, 36670–36683. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial ‘protective clothing’ in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef]
- Parrilli, E.; Tutino, M.L.; Marino, G. Biofilm as an adaptation strategy to extreme conditions. Springer Sci. Bus. Media Dtschl. GmbH. 2022, 33, 527–536. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef]
- Park, M.; Kim, J.; Horn, L.; Haan, J.; Strickland, A.; Lappi, V.; Boxrud, D.; Hedberg, C.; Ryu, S.; Jeon, B. Sugar modification of wall teichoic acids determines serotype-dependent strong biofilm production in Listeria monocytogenes. Microbiol. Spectr. 2022, 10, e0276922. [Google Scholar] [CrossRef]
- Di Bonaventura, G.; Piccolomini, R.; Paludi, D.; D’Orio, V.; Vergara, A.; Conter, M.; Ianieri, A. Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. J. Appl. Microbiol. 2008, 104, 1552–1561. [Google Scholar] [CrossRef]
- Doijad, S.P.; Barbuddhe, S.B.; Garg, S.; Poharkar, K.V.; Kalorey, D.R.; Kurkure, N.V.; Rawool, D.B.; Chakraborty, T. Biofilm-forming abilities of Listeria monocytogenes serotypes isolated from different sources. PLoS ONE 2015, 10, e0137046. [Google Scholar] [CrossRef]
- Verstraeten, N.; Braeken, K.; Debkumari, B.; Fauvart, M.; Fransaer, J.; Vermant, J.; Michiels, J. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008, 16, 496–506. [Google Scholar] [CrossRef]
- Morgan, R.; Kohn, S.; Hwang, S.H.; Hassett, D.J.; Sauer, K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7335–7343. [Google Scholar] [CrossRef] [PubMed]
- Köseoğlu, V.K.; Heiss, C.; Azadi, P.; Topchiy, E.; Güvener, Z.T.; Lehmann, T.E.; Miller, K.W.; Gomelsky, M. Listeria monocytogenes exopolysaccharide: Origin, structure, biosynthetic machinery and c-di-GMP-dependent regulation. Mol. Microbiol. 2015, 96, 728–743. [Google Scholar] [CrossRef]
- Valentini, M.; Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: Lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 2016, 291, 12547–12555. [Google Scholar] [CrossRef] [PubMed]
- Purcell, E.B.; McKee, R.W.; Courson, D.S.; Garrett, E.M.; McBride, S.M.; Cheney, R.E.; Tamayo, R. A nutrient-regulated cyclic diguanylate phosphodiesterase controls Clostridium difficile biofilm and toxin production during the stationary phase. Infect. Immun. 2017, 85, e01012-17. [Google Scholar] [CrossRef]
- Krypotou, E.; Scortti, M.; Grundström, C.; Oelker, M.; Luisi, B.F.; Sauer-Eriksson, A.E.; Vazquez-Boland, J. Control of bacterial virulence through the peptide signature of the habitat. Cell Rep. 2019, 26, 1815–1827. [Google Scholar] [CrossRef]
- Xayarath, B.; Alonzo, F.; Freitag, N.E. Identification of a peptide-pheromone that enhances Listeria monocytogenes escape from host cell vacuoles. PLoS Pathog. 2015, 11, e1004707. [Google Scholar] [CrossRef] [PubMed]
- Brenner, M.; Friedman, S.; Haber, A.; Livnat-Levanon, N.; Borovok, I.; Sigal, N.; Lewinson, O.; Herskovits, A.A. Listeria monocytogenes TcyKLMN cystine/cysteine transporter facilitates glutathione synthesis and virulence gene expression. mBio 2022, 13, e0044822. [Google Scholar] [CrossRef]
- Berude, J.C.; Kennouche, P.; Reniere, M.L.; Portnoy, D.A. Listeria monocytogenes utilizes glutathione and limited inorganic sulfur compounds as sources of essential cysteine. Infect. Immun. 2024, 92, e00422-23. [Google Scholar] [CrossRef] [PubMed]
- Sela, S.; Frank, S.; Belausov, E.; Pinto, R. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl. Environ. Microbiol. 2006, 72, 5653–5658. [Google Scholar] [CrossRef]
- Challan Belval, S.; Gal, L.; Margiewes, S.; Garmyn, D.; Piveteau, P.; Guzzo, J. Assessment of the roles of LuxS, S-ribosyl homocysteine, and autoinducer 2 in cell attachment during biofilm formation by Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 2006, 72, 2644–2650. [Google Scholar] [CrossRef]
- Ju, X.; Li, J.; Zhu, M.; Lu, Z.; Lv, F.; Zhu, X.; Bie, X. Effect of the luxS gene on biofilm formation and antibiotic resistance by Salmonella serovar Dublin. Food Res. Int. 2018, 107, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Cai, H.; Xu, B.; Yang, F.; Dou, X.; Dong, Q.; Yan, H.; Bu, X.; Li, Z. Growth, biofilm formation, and motility of Listeria monocytogenes strains isolated from food and clinical samples located in Shanghai (China). Food Res. Int. 2024, 184, 114232. [Google Scholar] [CrossRef] [PubMed]
- Bonsaglia, E.C.R.; Silva, N.C.C.; Fernades Júnior, A.; Araújo Júnior, J.P.; Tsunemi, M.H.; Rall, V.L.M. Production of biofilm by Listeria monocytogenes in different materials and temperatures. Food Control. 2014, 35, 386–391. [Google Scholar] [CrossRef]
- Jiang, X.; Jiang, C.; Yu, T.; Jiang, X.; Ren, S.; Kang, R.; Qiu, S. Benzalkonium chloride adaptation increases expression of the Agr system, biofilm formation, and virulence in Listeria monocytogenes. Front. Microbiol. 2022, 13, 856274. [Google Scholar] [CrossRef] [PubMed]
- Ondrusch, N.; Kreft, J. Blue and Red Light Modulates SigB-Dependent Gene Transcription, Swimming Motility and Invasiveness in Listeria monocytogenes. PLoS ONE 2011, 6, e16151. [Google Scholar] [CrossRef]
- Gorski, L.; Palumbo, J.D.; Mandrell, R.E. Attachment of Listeria monocytogenes to radish tissue is dependent upon temperature and flagellar motility. Appl. Environ. Microbiol. 2003, 69, 258–266. [Google Scholar] [CrossRef]
- Peel, M.; Donachie, W.; Shaw, A. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE, and Western blotting. Microbiology 1988, 134, 2171–2178. [Google Scholar] [CrossRef]
- Abdulkadieva, M.M.; Sysolyatina, E.V.; Vasilieva, E.V.; Litvinenko, V.V.; Kalinin, E.V.; Zhukhovitsky, V.G.; Shevlyagina, N.V.; Andreevskaya, S.G.; Stanishevskyi, Y.M.; Vasiliev, M.M.; et al. Motility provides specific adhesion patterns and improves Listeria monocytogenes invasion into human HEp-2 cells. PLoS ONE 2023, 18, e0290842. [Google Scholar] [CrossRef]
- Minamino, T. Hierarchical protein export mechanism of the bacterial flagellar type III protein export apparatus. FEMS Lett. 2018, 365, fny117. [Google Scholar] [CrossRef]
- Dons, L.; Olsen, J.E.; Rasmussen, O.F. Characterization of two putative Listeria monocytogenes genes encoding polypeptides homologous to the sensor protein CheA and the response regulator CheY of chemotaxis. DNA Seq. 1994, 4, 301–311. [Google Scholar] [CrossRef]
- Bischoff, D.S.; Bourret, R.B.; Kirsch, M.L.; Ordal, G.W. Purification and characterization of Bacillus subtilis CheY. Biochemistry 1993, 32, 9256–9261. [Google Scholar] [CrossRef] [PubMed]
- Croze, O.A.; Ferguson, G.P.; Cates, M.E.; Poon, W.C.K. Migration of chemotactic bacteria in soft agar: Role of gel concentration. Biophys. J. 2011, 101, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Dons, L.; Eriksson, E.; Jin, Y.; Rottenberg, M.E.; Kristensson, K.; Larsen, C.N.; Bresciani, J.; Olsen, J.E. Role of flagellin and the two-component CheA/CheY system of Listeria monocytogenes in host cell invasion and virulence. Infect. Immun. 2004, 72, 3237–3244. [Google Scholar] [CrossRef]
- Sánchez-Clemente, R.; Igeño, M.I.; Población, A.G.; Guijo, M.I.; Merchán, F.; Blasco, R. Study of pH changes in media during bacterial growth of several environmental strains. Proceedings 2018, 2, 1297. [Google Scholar] [CrossRef]

| Strain | Relevant Properties | Reference Source |
|---|---|---|
| 10403S | Serotype ½ a, wild type | [31] |
| 10403S Δlmo0135 | 10403S with Δlmo0135::erm (ΔctaP::erm) deletion | [24] |
| Temperature (30 °C) | Temperature (37 °C) | |||
|---|---|---|---|---|
| Condition | WT Correlation (r) | ΔctaP Correlation (r) | WT Correlation (r) | ΔctaP Correlation (r) |
| Overall | −0.72698 * | −0.86986 * | −0.68798 * | −0.70016 * |
| DM | 0.182 | 0.87 | 0.857 | −0.207 |
| 1.57 mM | 0.424 | −0.721 | 0.96 | −0.519 |
| 3.67 mM | −0.998 * | −0.84 | −0.703 | 0.591 |
| 6.51 mM | −0.41 | −0.989 | 0.942 | 0.521 |
| 12.21 mM | 0.714 | −0.998 * | 0.784 | 0.997 * |
| Gene Symbol | Locus Tag | Gene ID for 10403S | Log2 Fold-Change WT in 1.57 mM vs. DM | Log2 Fold-Change WT in 3.67 mM vs. DM | Log2 Fold-Change ΔctaP vs. WT in DM | Gene Description |
|---|---|---|---|---|---|---|
| bdlA | lmo1699 | LMRG_RS08590 | −3.11 * | −1.48 * | −1.17 | Methyl-accepting chemotaxis protein |
| dltA | LMRG_RS04905 | −2.19 * | +0.62 * | −1.50 * | D-alanine--poly (phosphoribitol) ligase subunit DltA | |
| dltB | lmo0973 | LMRG_RS04900 | −2.30 * | −0.03 | −1.78 * | D-alanyl-lipoteichoic acid biosynthesis protein DltB |
| NI | NI | Novel | +0.11 | +4.21 * | +0.56 | Biofilm formation stimulator VEG |
| lmo0189 | LMRG_RS00910 | −0.56 | +4.15 * | +0.63 | Veg family protein | |
| hly | lmo0202 | LMRG_RS00975 | −0.45 | +7.38 * | −1.34 * | Cholesterol-dependent cytolysin listeriolysin O |
| phoR | lmo2500 | LMRG_RS12690 | −0.81* | −0.56 * | −0.73 * | Alkaline phosphatase synthesis sensor protein PhoR |
| Gene Symbol | Locus Tag | Gene ID for 10403S | Log2 Fold-Change WT in 1.57 mM vs. DM | Log2 Fold-Change WT in 3.67 mM vs. DM | Log2 Fold-Change ΔctaP vs. WT in DM | Gene Description |
|---|---|---|---|---|---|---|
| tcyK | lmo2349 | LMRG_RS11840 | +2.02 * | −1.88 * | +2.55 * | Amino acid ABC transporter substrate-binding protein |
| tcyL | lmo2348 | LMRG_RS11835 | +2.71 * | −2.23 * | +1.54 * | Amino acid ABC transporter permease |
| tcyM | lmo2347 | LMRG_RS11830 | +1.69 * | −6.05 | +1.46 * | Amino acid ABC transporter permease |
| tcyN | lmo2346 | LMRG_RS11825 | +2.02 * | −2.87 * | +1.60 * | Amino acid ABC transporter ATP-binding protein |
| CymR | lmo1515 | LMRG_RS07540 | −0.77 * | +0.86 * | +0.58 * | Rrf2 family transcriptional regulator |
| oppA | lmo0152 | LMRG_RS00730 | −3.69 * | −2.42 * | +3.15 | Peptide ABC transporter substrate-binding protein PF00496: bacterial extracellular solute-binding proteins, family 5 middle |
| oppB | lmo2195 | LMRG_RS11135 | −3.01 * | −1.82 * | −0.74 * | ABC transporter permease |
| oppC | LMRG_RS11130 | −2.23 * | −1.68 * | −0.45 | ABC transporter permease | |
| oppD | LMRG_RS11125 | −2.85 * | −0.94 * | −0.08 | ABC transporter ATP-binding protein | |
| oppF | lmo2192 | LMRG_RS11120 | −2.64 * | −1.21 * | +0.04 | ATP-binding cassette domain-containing protein |
| luxS | lmo1288 | LMRG_RS06405 | −3.45 * | −0.47 * | +0.08 | S-ribosylhomocysteine lyase |
| Gene Symbol | Locus Tag | Gene ID for 10403S | Log2 Fold-Change WT in 1.57 mM vs. DM | Log2 Fold-Change WT in 3.67 mM vs. DM | Log2 Fold-Change ΔctaP vs. WT in DM | Gene Description |
|---|---|---|---|---|---|---|
| flhA | lmo0680 | LMRG_RS03415 | +2.34 * | +1.10 * | +0.77 | Flagellar biosynthesis protein FlhA |
| flhB | lmo0679 | LMRG_RS03410 | +1.26 * | −2.99 * | +0.04 | Flagellar biosynthesis protein FlhB |
| cheA | lmo0692 | LMRG_RS03475 | −3.51 * | −0.72 * | −1.51 | Chemotaxis protein CheA |
| cheY | lmo0691 | LMRG_RS03470 | −2.89 * | −0.36 | −1.35 | Chemotaxis protein CheY |
| motA | lmo0685 | LMRG_RS03440 | +2.75 * | −1.89 * | +1.27 | Flagellar motor stator protein MotA |
| motB | lmo0686 | LMRG_RS03445 | +3.39 * | −1.60 * | +1.47 | Flagellar motor protein MotB |
| plcA | lmo0201 | LMRG_RS00970 | −1.42 * | +5.40 * | −1.15 * | Phosphatidylinositol-specific phospholipase C |
| plcB | lmo0205 | LMRG_RS00990 | −0.18 | +4.65 * | −0.74 | Phosphatidylcholine phospholipase C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yilmaz Topcam, M.M.; Prayoonwiwat, N.; Bruschi, C.; Karatzas, K.A.G. Exogenous L-Cysteine and Its Transport Through CtaP Play a Role in Biofilm Formation, Swimming Motility, and Swarming Motility of Listeria monocytogenes. Foods 2025, 14, 1845. https://doi.org/10.3390/foods14111845
Yilmaz Topcam MM, Prayoonwiwat N, Bruschi C, Karatzas KAG. Exogenous L-Cysteine and Its Transport Through CtaP Play a Role in Biofilm Formation, Swimming Motility, and Swarming Motility of Listeria monocytogenes. Foods. 2025; 14(11):1845. https://doi.org/10.3390/foods14111845
Chicago/Turabian StyleYilmaz Topcam, Mahide Muge, Nattanicha Prayoonwiwat, Carolina Bruschi, and Kimon Andreas G. Karatzas. 2025. "Exogenous L-Cysteine and Its Transport Through CtaP Play a Role in Biofilm Formation, Swimming Motility, and Swarming Motility of Listeria monocytogenes" Foods 14, no. 11: 1845. https://doi.org/10.3390/foods14111845
APA StyleYilmaz Topcam, M. M., Prayoonwiwat, N., Bruschi, C., & Karatzas, K. A. G. (2025). Exogenous L-Cysteine and Its Transport Through CtaP Play a Role in Biofilm Formation, Swimming Motility, and Swarming Motility of Listeria monocytogenes. Foods, 14(11), 1845. https://doi.org/10.3390/foods14111845






