Multi-Omics Mining of Characteristic Quality Factors Boosts the Brand Enhancement of the Geographical Indication Product—Pingliang Red Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Sample Collection and Preparation
2.3. Metabolomic Profiling
2.4. RNA Library Construction and RNA-Seq
2.5. Statistical Analysis
3. Results and Discussion
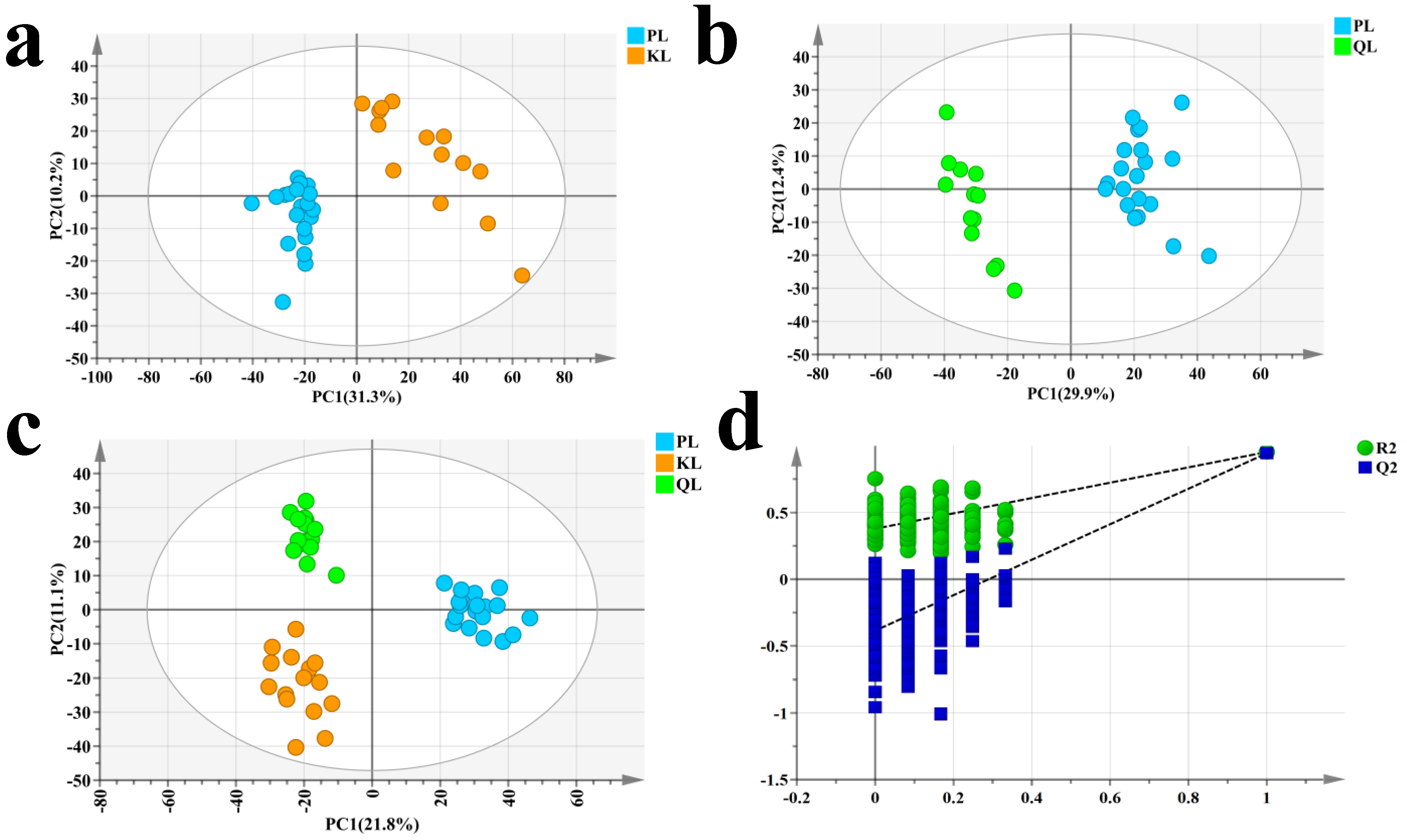
3.1. Identification of Differential Metabolites
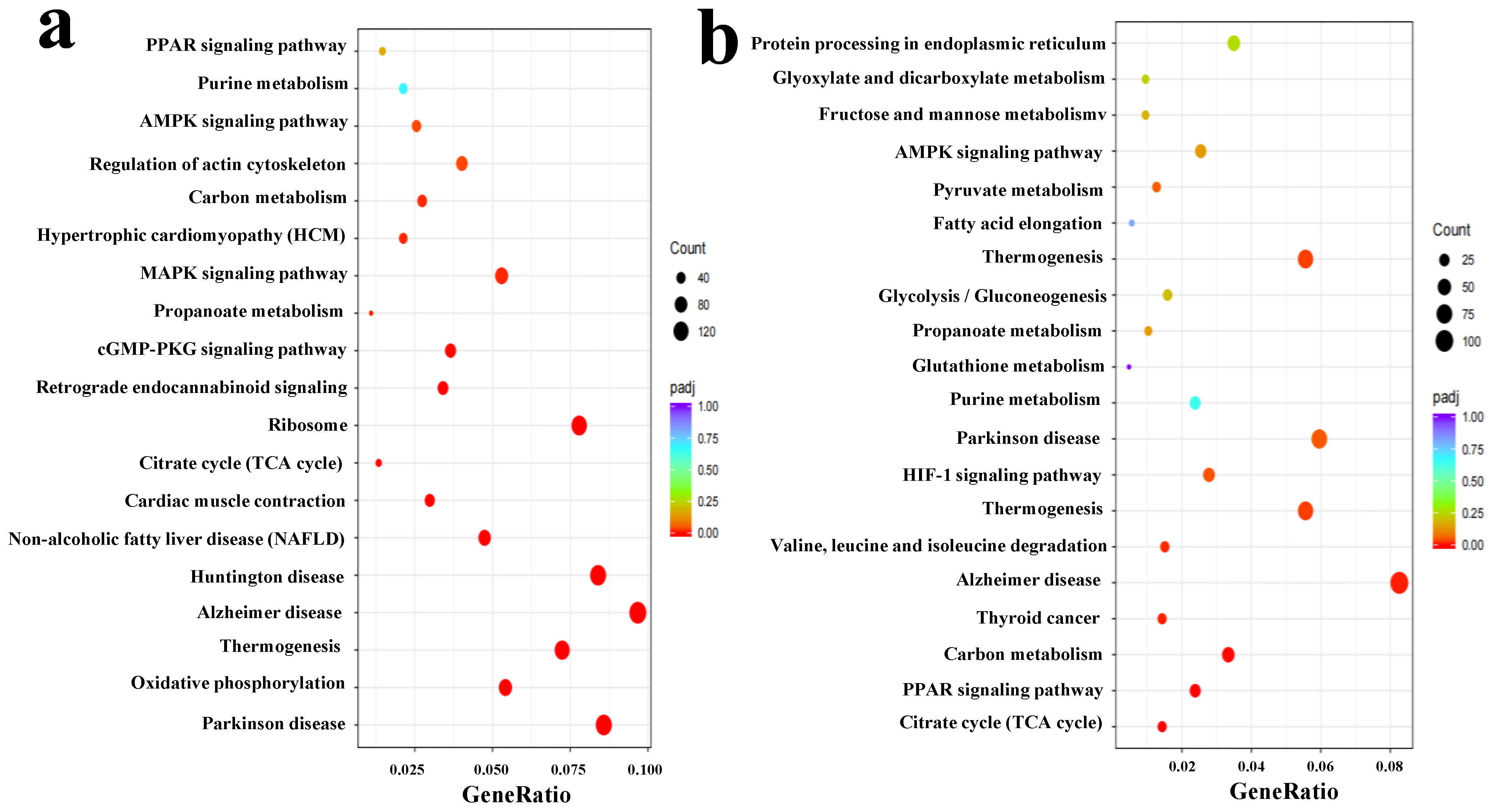
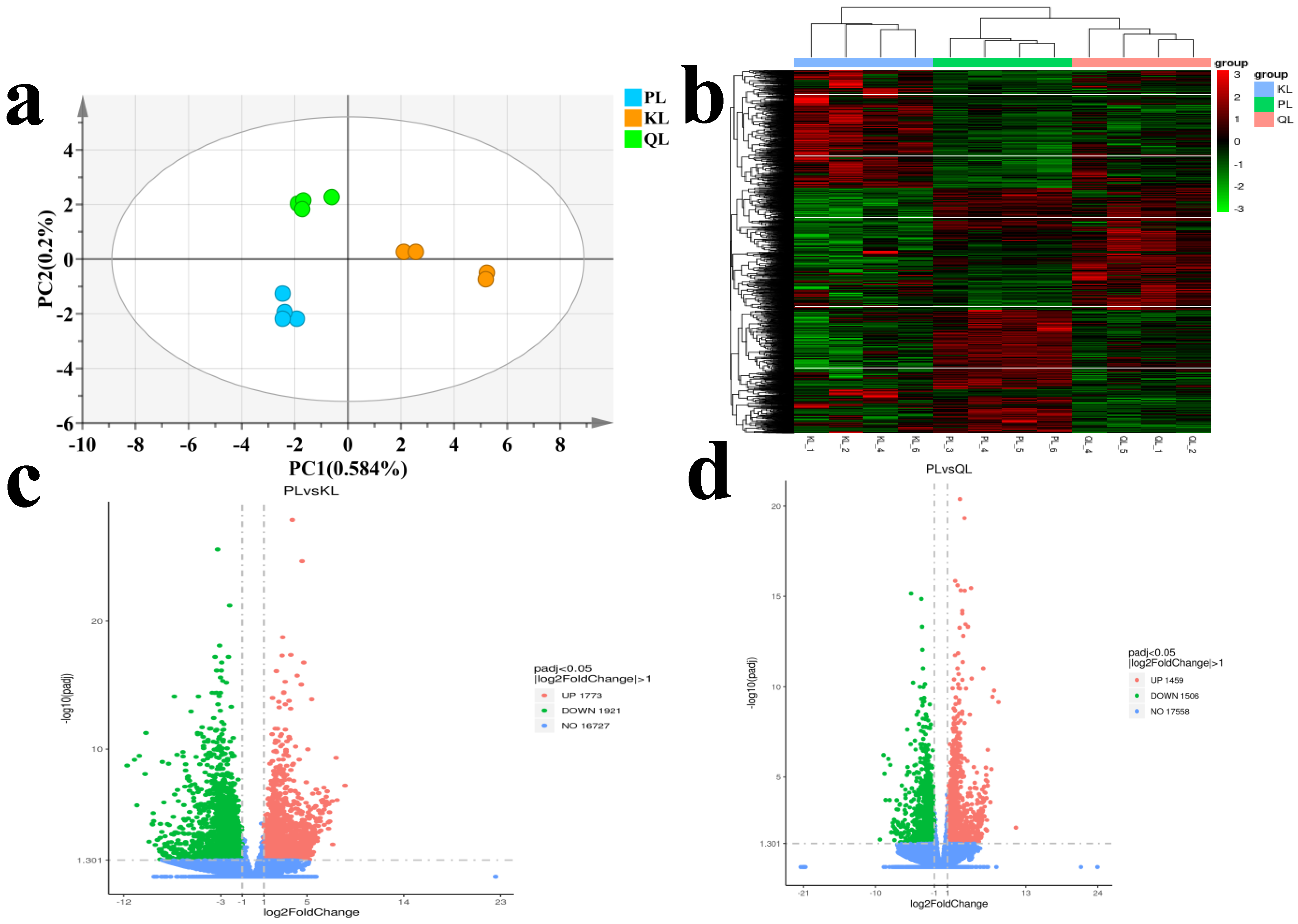
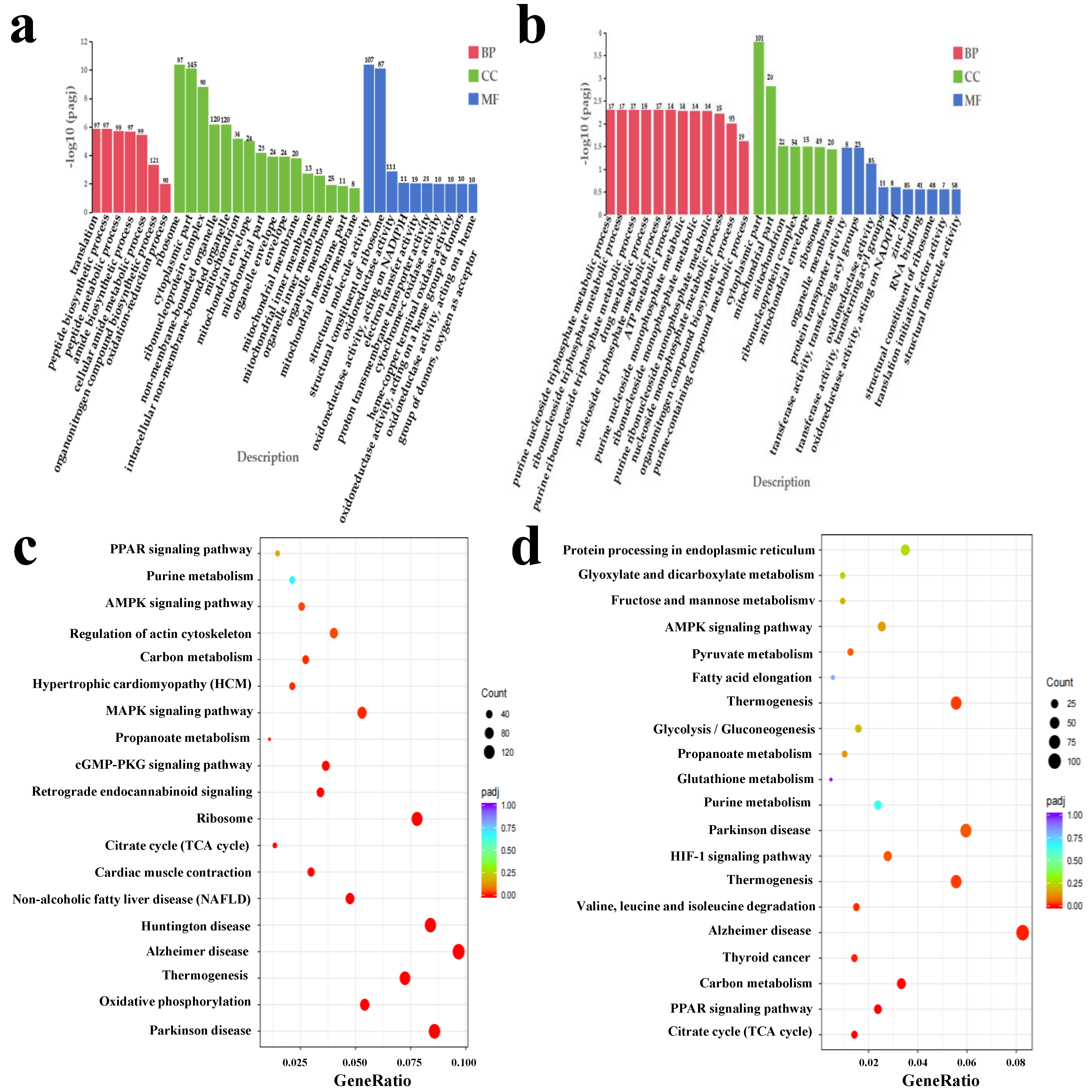
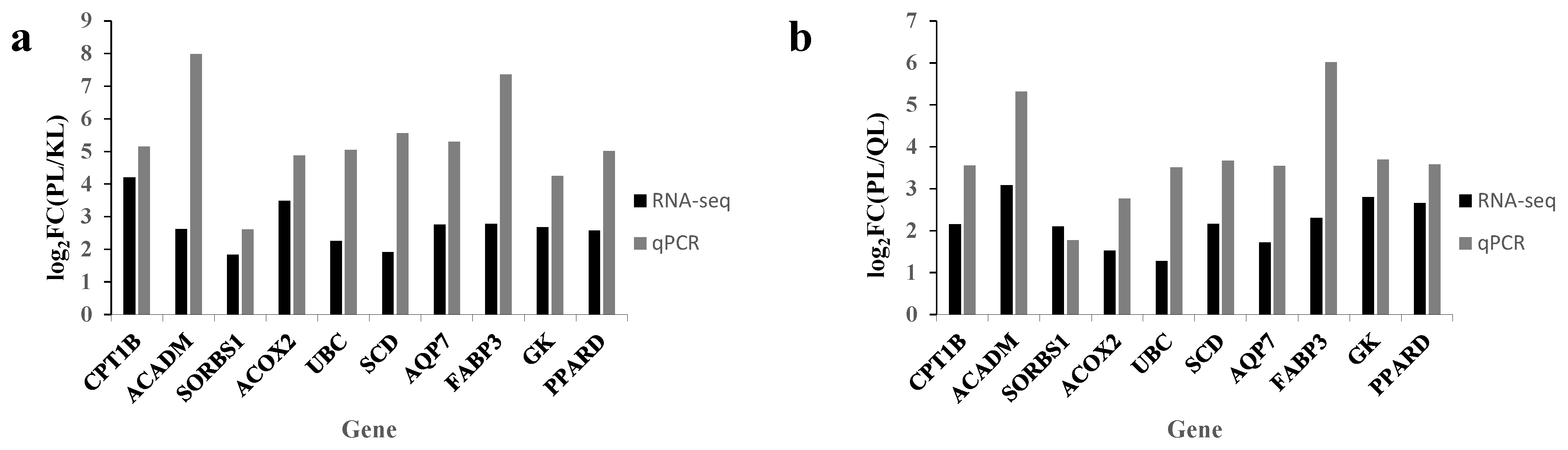
3.2. Identification of DEG and Verification of Transcriptomic Data
3.3. Unveil Pingliang Red Cattle’s Distinctive Quality Traits
3.3.1. IMP and Meat Flavor
3.3.2. GSH and Meat Color
3.3.3. DEGs and Meat Tenderness
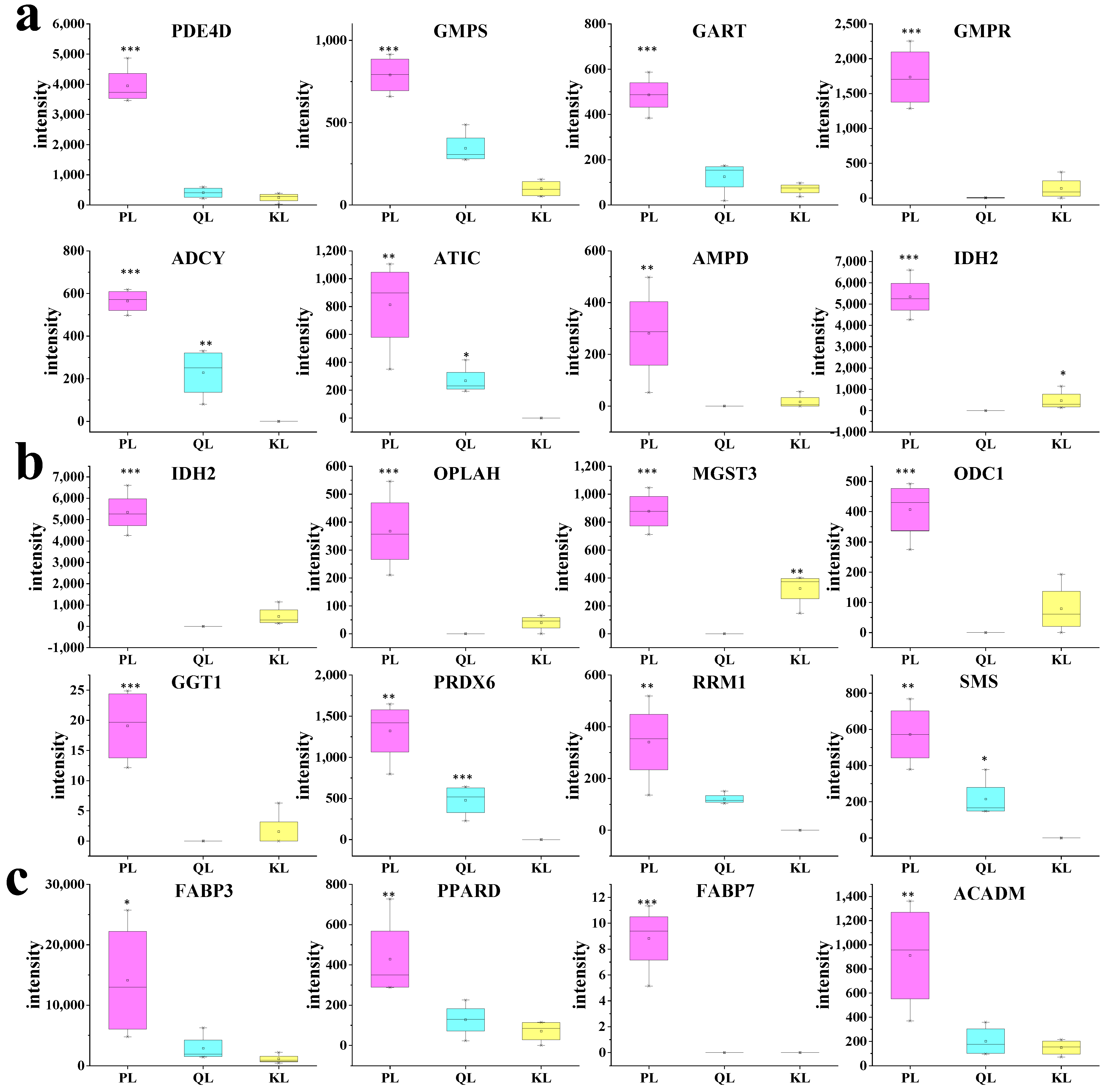
3.4. Bioactive Compounds
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Yang, X.T.; Li, Y.L.; Zhao, S.L.; Zhang, P.; Zhao, Y. Geographical origin authentication of agricultural products in the China-EU Geographical Indications Agreement: A comprehensive review of Chinese products. Trend Food Sci. Technol. 2024, 152, 104679. [Google Scholar] [CrossRef]
- Hu, X.Q.; Lu, L.; Li, S.M.; Zhang, W.X.; He, Y.T.; Chen, M.X. Comparison of appearance quality, cooking quality, and nutritional quality of geographical indication rice and their application in geographical indication discrimination. J. Food Compos. Anal. 2024, 135, 106668. [Google Scholar] [CrossRef]
- Li, W.B.; Xu, J.F.; Li, S.L. Study on meat index of Pingliang Red Cattle. China Cattle Sci. 2011, 37, 29–31. [Google Scholar]
- Liu, C.; Pan, D.; Ye, Y.; Cao, J. 1H NMR and multivariate data analysis of the relationship between the age and quality of duck meat. Food Chem. 2013, 141, 1281–1286. [Google Scholar] [CrossRef]
- Liu, Y.X.X.; Yang, X.Y.; Liu, Y.G.; Dong, P.C.; Liang, R.R.; Hopkins, D.L.; Holman, B.W.; Luo, X.; Zhu, L.X.; Yang, Z.G.; et al. Chinese consumer perception and purchasing behavior of beef—Mainly in North and East China. Meat Sci. 2025, 2020, 109696. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, B.L.; Mu, Y.M. Determination and analysis of nutritional components in meat of Pingliang Red Cattle. China Herbiv. Sci. 2015, 35, 73–75. [Google Scholar] [CrossRef]
- Stella, R.; Mastrorilli, E.; Pretto, T.; Tata, A.; Piro, R.; Arcangeli, G.; Biancotto, G. New strategies for the differentiation of fresh and frozen/thawed fish: Non-targeted metabolomics by LC-HRMS (part B). Food Control 2021, 132, 108461. [Google Scholar] [CrossRef]
- Su, G.Y.; Yu, C.; Liang, S.W.; Wang, W.; Wang, H.F. Multi-omics in food safety and authenticity in terms of food components. Food Chem. 2023, 437, 137943. [Google Scholar] [CrossRef]
- Liang, L.; Li, Y.H.; Mao, X.J.; Wang, Y.X. Metabolomics applications for plant-based foods origin tracing, cultivars identification and processing: Feasibility and future aspects. Food Chem. 2024, 449, 139227. [Google Scholar] [CrossRef]
- Bozza, D.; Barboni, D.; Spadafora, N.D.; Felletti, S.; De Luca, C.; Nosengo, C.; Compagnin, G.; Cavazzini, A.; Catani, M. Untargeted metabolomics approachesfor the characterization of cereals and their derived products by means of liquid chromatography coupled to high resolution mass spectrometry. J. Chromatogr. Open 2024, 6, 100168. [Google Scholar] [CrossRef]
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Viale, G.; Trapani, D.; Duso, B.A.; Curigliano, G. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit. Rev. Oncol. Hematol. 2019, 133, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Sunildutt, N.; Ahmed, F.; Salih, A.R.C.; Kim, H.C.; Choi, K.H. Unraveling new avenues in pancreatic cancer treatment: A comprehensive exploration of drug repurposing using transcriptomic data. Comput. Biol. Med. 2025, 185, 109481. [Google Scholar] [CrossRef] [PubMed]
- Mmbando, G.S. Omics: A new, promising technologies for boosting crop yield and stress resilience in African agriculture. Plant Stress 2024, 11, 100366. [Google Scholar] [CrossRef]
- Gao, J.; Rajib, M.M.R.; Dai, K.; Li, K.K.; Cui, Y.H.; Wang, W.X.; Li, J.Z.; Yin, H. Comparative transcriptomic analysis of the regulatory mechanisms underlying the resistance to Fusarium foetens mediated by konjac glucomannan oligosaccharides in tobacco. Physiol. Mol. Plant Pathol. 2025, 136, 102585. [Google Scholar] [CrossRef]
- Shi, S.; Shen, Y.; Zhao, Z.; Hou, Z.; Yang, Y.; Zhou, H.; Zou, J.; Guo, Y. Integrative analysis of transcriptomic and metabolomic profiling of ascites syndrome in broiler chickens induced by low temperature. Mol. Biosyst. 2014, 10, 2984–2993. [Google Scholar] [CrossRef]
- Cho, K.; Cho, K.S.; Sohn, H.B.; Ha, I.J.; Hong, S.Y.; Lee, H.; Kim, Y.M.; Nam, M.H. Network analysis of the metabolome and transcriptome reveals novel regulation of potato pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef]
- Zheng, N.N.; Wang, K.; He, J.J.; Qiu, Y.P.; Xie, G.X.; Su, M.M.; Li, H.K. Effects of ADMA on gene expression and metabolism in serum-starved LoVo cells. Sci. Rep. 2016, 6, 25892. [Google Scholar] [CrossRef]
- Masson, P.; Alves, A.C.; Ebbels, T.M.; Nicholson, J.K.; Want, E.J. Optimization and evaluation of metabolite extraction protocols for untargeted metabolic profiling of liver samples by UPLC-MS. Anal. Chem. 2010, 82, 7779–7786. [Google Scholar] [CrossRef]
- Want, E.J.; Wilson, I.D.; Gika, H.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Holmes, E.; Nicholson, J.K. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010, 5, 1005–1018. [Google Scholar] [CrossRef]
- Hammad, M.O. Simplified protocol modification of Trizol method for extraction of high-quality RNA yield from RNase-rich rat pancreas. Process Biochem. 2023, 130, 464–471. [Google Scholar] [CrossRef]
- Hew, B.E.; Gupta, S.; Sato, R.; Waller, D.F.; Stoytchev, I.; Short, J.E.; Sharek, L.; Tran, C.T.; Badran, A.H.; Owens, J.B. MetaboAnalyst 6.0: Towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 2024, 52, w398–w406. [Google Scholar] [CrossRef]
- Zhao, Q. A Review of Principal Component Analysis. Softw. Eng. 2016, 19, 1–3. [Google Scholar]
- Szeto, S.S.W.; Reinke, S.N.; Sykes, B.D.; Lemire, B.D. Mutations in the Saccharomyces cerevisiae succinate dehydrogenase result in distinct metabolic phenotypes revealed through (1)H-NMR-based metabolic footprinting. J. Proteome Res. 2010, 9, 672–6739. [Google Scholar] [CrossRef]
- Masic, U.; Yeomans, M.R. Umami flavor enhances appetite but also increases satiety. Am. J. Clin. Nutr. 2014, 100, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Bonagurio, L.P.; Murakami, A.E.; Pereira-Maróstica, H.V.; Almeida, F.L.A.; Santos, T.C.; Pozza, P.C. Effects of different levels of inosine-5′-monophosphate (5′-IMP) supplementation on the growth performance and meat quality of finishing pigs (75 to 100 kg). Meat Sci. 2023, 196, 109016. [Google Scholar] [CrossRef]
- Riedinger, C.; Mendler, M.; Schlotterer, A.; Fleming, T.; Okun, J.; Hammes, H.P.; Herzig, S.; Nawroth, P.P. High-glucose toxicity is mediated by AICAR-transformylase/IMP cyclohydrolase and mitigated by AMP-activated protein kinase in Caenorhabditis elegans. J. Biol. Chem. 2018, 293, 4845–4859. [Google Scholar] [CrossRef]
- Cao, Q.; Wan, X.; Shu, X.B.; Xie, L.Q. Bioaccumulation and detoxication of microcystin-LR in three submerged macrophytes: The important role of glutathione biosynthesis. Chemosphere 2019, 225, 935–942. [Google Scholar] [CrossRef]
- Su, Y.H.; Pan, H.Y.; Guo, Z.W.; Zhou, W.Y.; Zhang, B.M. Bacterial translocation and endotoxemia after Pringle maneuver in cirrhotic rats. Dig. Dis. Sci. 2015, 60, 414–419. [Google Scholar] [CrossRef]
- Shen, K.J. Studies on Preservation and Quality Control for Chilled Beef. Master’s Thesis, Zhejiang University of Technology, Hangzhou, China, 2015. [Google Scholar]
- Waerp, H.K.; Waters, S.M.; McCabe, M.S.; Cormican, P.; Salte, R. RNA-seq analysis of bovine adipose tissue in heifers fed diets differing in energy and protein content. PLoS ONE 2018, 13, e0201284. [Google Scholar] [CrossRef]
- Musatti, A.; Manzoni, M.; Rollini, M. Post-fermentative production of glutathione by baker’s yeast (S. cerevisiae) in compressed and dried forms. New Biotechnol. 2013, 30, 219–226. [Google Scholar] [CrossRef]
- Wang, X.J.; Gao, H.; Li, H.P.; Gao, X.; Sun, B.Z.; Cheng, Q.; Xu, L.; Zhang, Y.P.; Lei, Y.H.; Wei, M.; et al. Analysis of growth performance as well as carcass and meat. Sci. Agric. Sin. 2023, 56, 559–571. [Google Scholar] [CrossRef]
- Wang, J.J.; Deng, Y.X.; Hu, Y.Y.; Yang, N.N.; Wang, D.D.; Ding, H.; Yao, B.Q. Nutritional value of beef and research progress of beef tendering technology. Anhui Agric. Sci. Bull. 2019, 25, 140–144. [Google Scholar] [CrossRef]
- Gandolfi, G.; Pomponio, L.; Ertbjerg, P.; Karlsson, A.H.; Costa, L.N.; Lametsch, R.; Russo, V.; Davoli, R. Investigation on CAST, CAPN1 and CAPN3 porcine gene polymorphisms and expression in relation to post-mortem calpain activity in muscle and meat quality. Meat Sci. 2011, 88, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kim, S.W.; Choi, J.S.; Lee, Y.M.; Lee, C.K.; Choi, B.H.; Kim, T.H.; Choi, Y.I.; Kim, J.J.; Kim, K.S. Investigation of porcine FABP3 and LEPR gene polymorphisms and mRNA expression for variation in intramuscular fat content. Mol. Biol. Rep. 2010, 37, 3931–3939. [Google Scholar] [CrossRef]
- Picard, B.; Gagaoua, M. Meta-proteomics for the discovery of protein biomarkers of beef tenderness: An overview of integrated studies. Food Res. Int. 2020, 127, 108739. [Google Scholar] [CrossRef]
- Hondal, R.J. Selenium vitaminology: The connection between selenium, vitamin C, vitamin E, and ergothioneine. Curr. Opin. Chem. Biol. 2023, 75, 102328. [Google Scholar] [CrossRef]
- Turck, D.; Bresson, J.L.; Burlingame, B.; Dean, T.; Fairweather-Tait, S.; Heinonen, M.; Hirsch-Ernst, K.I.; Mangelsdorf, I.; McArdle, H.J.; Naska, A.; et al. Statement on the safety of synthetic L-ergothioneine as a novel food–supplementary dietary exposure and safety assessment for infants and young children, pregnant and breastfeeding women. Efsa J. 2017, 15, 5060. [Google Scholar] [CrossRef]


| Gene Name (Symbol) | Primer Sequences (5′–3′) | Product Length (bp) |
|---|---|---|
| Carnitine palmitoyltransferase 1B (CPT1B) | CCCAGGGAAGGACACAGATGT | 149 |
| GGATCCTCTGGAACTGCATCTC | ||
| Acyl-CoA dehydrogenase medium chain (ACADM) | GCTTGGGAAGTTGATTCTGGTC | 224 |
| TGTTCACGGGCTATAATAAGCC | ||
| Peroxisome proliferator-activated receptor delta (PPARD) | TGCAAAATCCAGAAGAAGAACC | 171 |
| CTGGGGGTTGTGCTGACTC | ||
| Acyl-CoA oxidase 2 (ACOX2) | TTCCTGTCTGGTGCCCAAATA | 160 |
| GACGTTCATAGGCATGTCCATC | ||
| Ubiquitin C (UBC) | CCGGACCGGGAGTTCAGT | 172 |
| GGGATGCCTTCTTTTTCTTGTAT | ||
| Stearoyl-CoA desaturase (SCD) | CCAGGGCACCCATCAGATAG | 162 |
| TCCAAGGTGGTCTCGACA | ||
| Sorbin and SH3 domain containing 1 (SORBS1) | TGTCCTGGAAGGAGGAGACATC | 110 |
| CAGCTGGTATAAAATGCCTTGG | ||
| Aquaporin 7 (AQP7) | TCCAAGGTGGTCTCGACA | 204 |
| ACCTATGGTGACTCCGAAGC | ||
| Fatty acid binding protein 3 (FABP3) | GCGTTCTCTGTCGTCTTTCC | 169 |
| GATGATTGTGGTAGGCTTGGT | ||
| Glycerol kinase (GK) | TAAGGAAATTCTGCAGTCTGTCT | 139 |
| TAACTTGTCCCAGACTACAGTGG | ||
| 18S rRNA | CGGAACTGAGGCCATGATT | 145 |
| CCTCCGACTTTCGTTCTTGAT |
| Procedure | Temperature (°C) | Time |
|---|---|---|
| Pre-incubation | 95 | 5 min |
| Amplification (40 cycles) | 95 | 3 s |
| 63 | 20 s | |
| Melting curves | 95 | 15 s |
| 60 | 15 s | |
| 95 | 15 s |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Zhu, Y.; Liu, X.; Zhang, J.; Liu, C.; Zhao, Y.; Yang, S.; Chen, A.; Zhao, J. Multi-Omics Mining of Characteristic Quality Factors Boosts the Brand Enhancement of the Geographical Indication Product—Pingliang Red Cattle. Foods 2025, 14, 1770. https://doi.org/10.3390/foods14101770
Liu J, Zhu Y, Liu X, Zhang J, Liu C, Zhao Y, Yang S, Chen A, Zhao J. Multi-Omics Mining of Characteristic Quality Factors Boosts the Brand Enhancement of the Geographical Indication Product—Pingliang Red Cattle. Foods. 2025; 14(10):1770. https://doi.org/10.3390/foods14101770
Chicago/Turabian StyleLiu, Jing, Yu Zhu, Xiaoxia Liu, Juan Zhang, Chuan Liu, Yan Zhao, Shuming Yang, Ailiang Chen, and Jie Zhao. 2025. "Multi-Omics Mining of Characteristic Quality Factors Boosts the Brand Enhancement of the Geographical Indication Product—Pingliang Red Cattle" Foods 14, no. 10: 1770. https://doi.org/10.3390/foods14101770
APA StyleLiu, J., Zhu, Y., Liu, X., Zhang, J., Liu, C., Zhao, Y., Yang, S., Chen, A., & Zhao, J. (2025). Multi-Omics Mining of Characteristic Quality Factors Boosts the Brand Enhancement of the Geographical Indication Product—Pingliang Red Cattle. Foods, 14(10), 1770. https://doi.org/10.3390/foods14101770






