Surveillance of Antimicrobial Resistance in the Asian Seabass (Lates calcarifer) Supply Chain Using Nanopore Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Library Preparation and Sequencing
2.3. Bioinformatic Analysis
3. Results
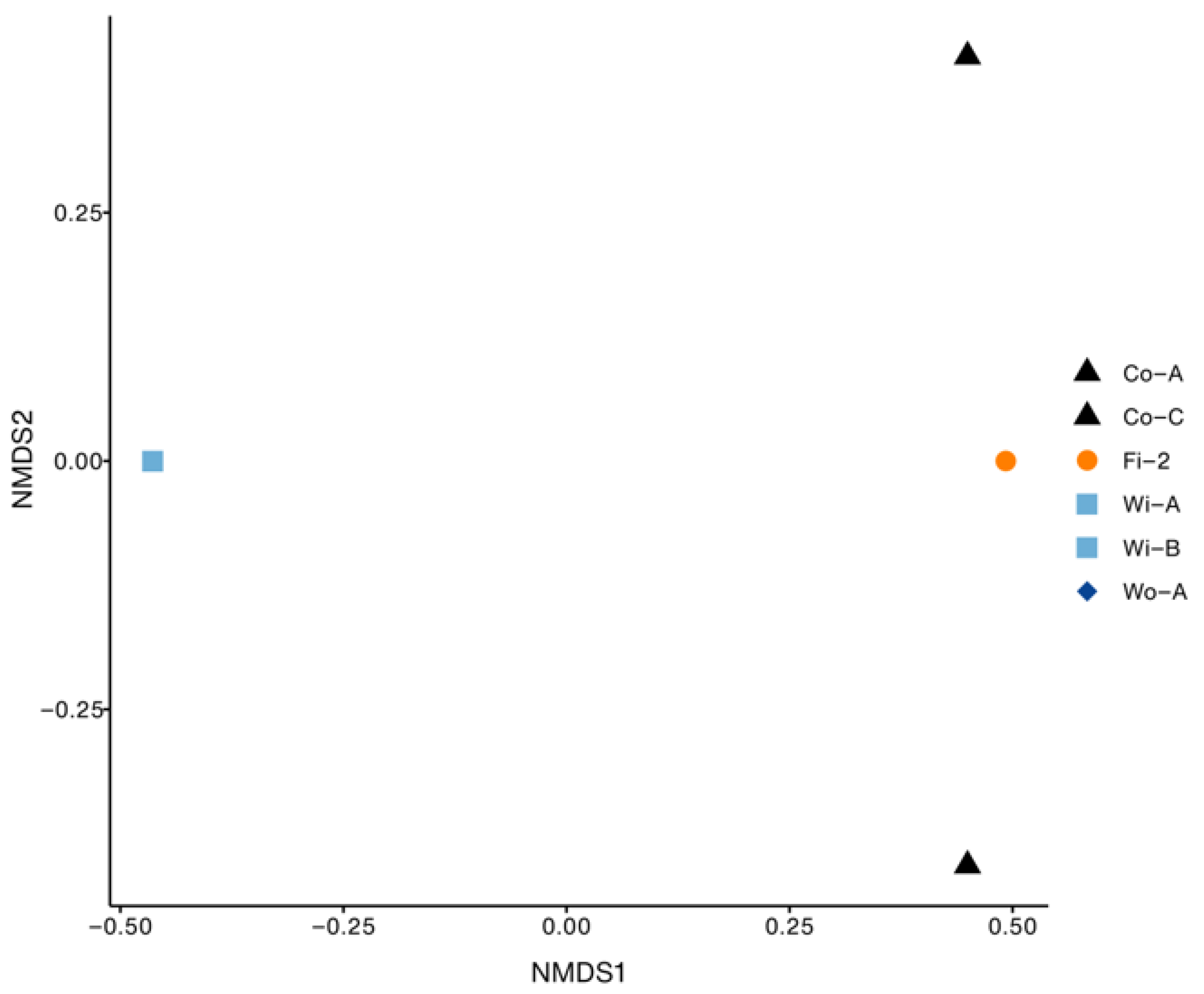
3.1. Bacterial Diversity
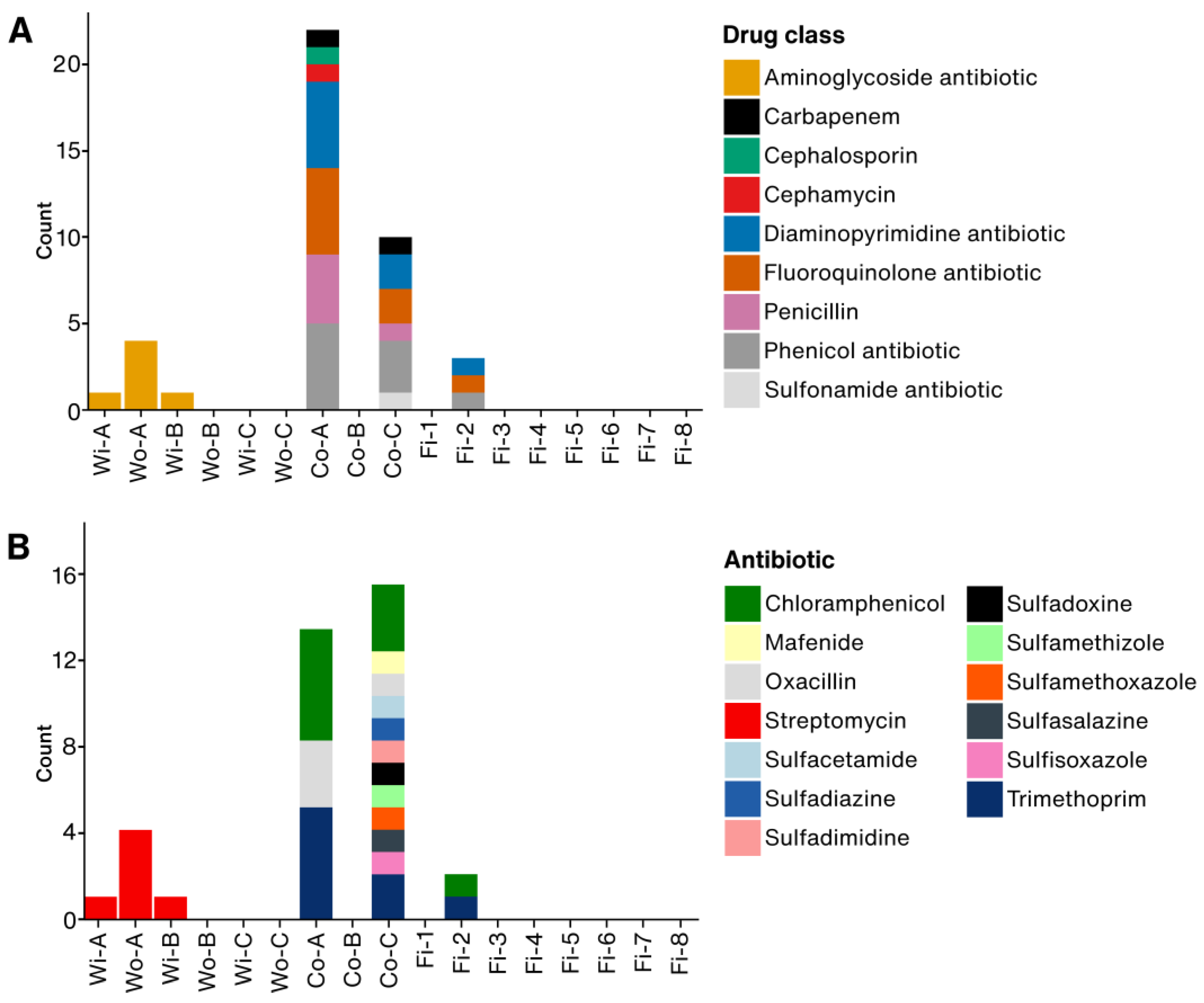
3.2. Antimicrobial Resistance Genes
3.3. Gene Family and Resistant Drug Class of ARGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samtiya, M.; Matthews, K.R.; Dhewa, T.; Puniya, A.K. Antimicrobial Resistance in the Food Chain: Trends, Mechanisms, Pathways, and Possible Regulation Strategies. Foods 2022, 11, 2966. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Maciel-Guerra, A.; Baker, M.; Zhang, X.; Hu, Y.; Wang, W.; Rong, J.; Zhang, J.; Xue, N.; Barrow, P.; et al. Whole-Genome Sequencing and Gene Sharing Network Analysis Powered by Machine Learning Identifies Antibiotic Resistance Sharing between Animals, Humans and Environment in Livestock Farming. PLoS Comput. Biol. 2022, 18, e1010018. [Google Scholar] [CrossRef] [PubMed]
- Ardakani, Z.; Canali, M.; Aragrande, M.; Tomassone, L.; Simoes, M.; Balzani, A.; Beber, C.L. Evaluating the Contribution of Antimicrobial Use in Farmed Animals to Global Antimicrobial Resistance in Humans. One Health 2023, 17, 100647. [Google Scholar] [CrossRef]
- Rahman, S.; Hollis, A. The Effect of Antibiotic Usage on Resistance in Humans and Food-Producing Animals: A Longitudinal, One Health Analysis Using European Data. Front. Public Health 2023, 11, 1170426. [Google Scholar] [CrossRef] [PubMed]
- Raza, M.F.; Wang, Y.; Cai, Z.; Bai, S.; Yao, Z.; Awan, U.A.; Zhang, Z.; Zheng, W.; Zhang, H. Gut Microbiota Promotes Host Resistance to Low-Temperature Stress by Stimulating Its Arginine and Proline Metabolism Pathway in Adult Bactrocera Dorsalis. PLoS Pathog. 2020, 16, e1008441. [Google Scholar] [CrossRef]
- Hossain, A.; Habibullah-Al-Mamun, M.; Nagano, I.; Masunaga, S.; Kitazawa, D.; Matsuda, H. Antibiotics, Antibiotic-Resistant Bacteria, and Resistance Genes in Aquaculture: Risks, Current Concern, and Future Thinking. Environ. Sci. Pollut. Res. 2022, 29, 11054–11075. [Google Scholar] [CrossRef]
- Pham, D.K.; Chu, J.; Do, N.T.; Brose, F.; Degand, G.; Delahaut, P.; De Pauw, E.; Douny, C.; Van Nguyen, K.; Vu, T.D.; et al. Monitoring Antibiotic Use and Residue in Freshwater Aquaculture for Domestic Use in Vietnam. EcoHealth 2015, 12, 480–489. [Google Scholar] [CrossRef]
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global Trends in Antimicrobial Use in Aquaculture. Sci. Rep. 2020, 10, 21878. [Google Scholar] [CrossRef]
- Done, H.Y.; Venkatesan, A.K.; Halden, R.U. Does the Recent Growth of Aquaculture Create Antibiotic Resistance Threats Different from Those Associated with Land Animal Production in Agriculture? AAPS J. 2015, 17, 513–524. [Google Scholar] [CrossRef]
- Shao, Y.; Wang, Y.; Yuan, Y.; Xie, Y. A Systematic Review on Antibiotics Misuse in Livestock and Aquaculture and Regulation Implications in China. Sci. Total Environ. 2021, 798, 149205. [Google Scholar] [CrossRef]
- Thornber, K.; Bashar, A.; Ahmed, M.S.; Bell, A.; Trew, J.; Hasan, M.; Hasan, N.A.; Alam, M.M.; Chaput, D.L.; Haque, M.M.; et al. Antimicrobial Resistance in Aquaculture Environments: Unravelling the Complexity and Connectivity of the Underlying Societal Drivers. Environ. Sci. Technol. 2022, 56, 14891–14903. [Google Scholar] [CrossRef]
- Rimmer, M.A.; John Russell, D. Aspects of the Biology and Culture of Lates Calcarifer. In Tropical Mariculture; De Silva, S.S., Ed.; Elsevier: Amsterdam, The Netherlands, 1998; pp. 449–476. ISBN 978-0-12-210845-7. [Google Scholar]
- Asian Sea Bass Market Analysis—Size, Demand & Forecast 2024-2034. Available online: https://www.futuremarketinsights.com/reports/sea-bass-market (accessed on 3 February 2025).
- Jerry, D.R. Biology and Culture of Asian Seabass Lates Calcarifer; CRC Press: Boca Raton, FL, USA, 2013; ISBN 1482208075. [Google Scholar]
- Siddik, M.; Islam, M.; Hanif, M.; Chaklader, M.; Kleindienst, R. Barramundi, Lates Calcarifer (Bloch, 1790): A New Dimension to the Fish Farming in Coastal Bangladesh. J. Aquac. Res. Dev. 2016, 7, 1000461. [Google Scholar] [CrossRef]
- Dong, H.T.; Taengphu, S.; Sangsuriya, P.; Charoensapsri, W.; Phiwsaiya, K.; Sornwatana, T.; Khunrae, P.; Rattanarojpong, T.; Senapin, S. Recovery of Vibrio Harveyi from Scale Drop and Muscle Necrosis Disease in Farmed Barramundi, Lates Calcarifer in Vietnam. Aquaculture 2017, 473, 89–96. [Google Scholar] [CrossRef]
- Raharjo, H.M.; Budiyansah, H.; Mursalim, M.F.; Chokmangmeepisarn, P.; Sakulworakan, R.; Madyod, S.; Sewaka, M.; Sonthi, M.; Debnath, P.P.; Elayaraja, S.; et al. Distribution of Vibrionaceae in Farmed Asian Sea Bass, Lates Calcarifer in Thailand and Their High Prevalence of Antimicrobial Resistance. J. Fish Dis. 2022, 45, 1355–1371. [Google Scholar] [CrossRef] [PubMed]
- Ottesen, A.; Kocurek, B.; Ramachandran, P.; Reed, E.; Commichaux, S.; Engelbach, G.; Mammel, M.; Fleurant, S.S.; Zhao, S.; Kabera, C.; et al. Advancing Antimicrobial Resistance Monitoring in Surface Waters with Metagenomic and Quasimetagenomic Methods. PLoS Water 2022, 1, e0000067. [Google Scholar] [CrossRef]
- Takeda-Nishikawa, K.; Palanichamy, R.; Miyazato, N.; Suzuki, T. What Samples Are Suitable for Monitoring Antimicrobial-Resistant Genes? Using NGS Technology, a Comparison between eDNA and mrDNA Analysis from Environmental Water. Front. Microbiol. 2023, 14, 954783. [Google Scholar] [CrossRef] [PubMed]
- Kwon, T.; Gebhardt, J.T.; Lyoo, E.L.; Gaudreault, N.N.; Trujillo, J.D.; Woodworth, J.C.; Paulk, C.B.; Jones, C.K.; Richt, J.A. Development and Optimization of Sampling Techniques for Environmental Samples from African Swine Fever Virus-Contaminated Surfaces with No Organic Contaminants. Front. Vet. Sci. 2024, 11, 1425928. [Google Scholar] [CrossRef]
- Galvin, S.; Dolan, A.; Cahill, O.; Daniels, S.; Humphreys, H. Microbial Monitoring of the Hospital Environment: Why and How? J. Hosp. Infect. 2012, 82, 143–151. [Google Scholar] [CrossRef]
- Rheman, S.; Hossain, S.; Sarker, M.S.; Akter, F.; Khor, L.; Gan, H.M.; Powell, A.; Card, R.M.; Hounmanou, Y.M.G.; Dalsgaard, A.; et al. Nanopore Sequencing for Identification and Characterization of Antimicrobial-Resistant Escherichia Coli and Salmonella Spp. from Tilapia and Shrimp Sold at Wet Markets in Dhaka, Bangladesh. Front. Microbiol. 2024, 15, 1329620. [Google Scholar] [CrossRef]
- Bourdonnais, E.; Le Bris, C.; Brauge, T.; Midelet, G. Tracking Antimicrobial Resistance Indicator Genes in Wild Flatfish from the English Channel and the North Sea Area: A One Health Concern. Environ. Pollut. 2024, 343, 123274. [Google Scholar] [CrossRef]
- Ong, H.M.G.; Zhong, Y.; Hu, C.C.; Ong, K.H.; Khor, W.C.; Schlundt, J.; Aung, K.T. Quantitative Risk Evaluation of Antimicrobial-Resistant Vibrio Parahaemolyticus Isolated from Farmed Grey Mullets in Singapore. Pathogens 2023, 12, 93. [Google Scholar] [CrossRef]
- Tskhay, F.; Köbsch, C.; Elena, A.X.; Bengtsson-Palme, J.; Berendonk, T.U.; Klümper, U. Fish Are Poor Sentinels for Surveillance of Riverine Antimicrobial Resistance. One Health 2025, 20, 101026. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. Ultrafast One-Pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Abueg, L.A.L.; Afgan, E.; Allart, O.; Awan, A.H.; Bacon, W.A.; Baker, D.; Bassetti, M.; Batut, B.; Bernt, M.; Blankenberg, D.; et al. The Galaxy Platform for Accessible, Reproducible, and Collaborative Data Analyses: 2024 Update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020.
- Oksanen, J.; Simpson, G.; Blanchet, F.; Kindt, R.; Legendre, P.; Minchin, P.; O’Hara, R.; Solymos, P.; Stevens, M.; Szoecs, E.; et al. Vegan: Community Ecology Package, R Package Version 2.6-6.1. 2024. Available online: https://github.com/vegandevs/vegan (accessed on 5 February 2025).
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2021–2022. EFSA J. 2024, 22, e8583. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2022 Zoonoses Report. EFSA J. 2023, 21, e8442. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of Long, Error-Prone Reads Using Repeat Graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Research 2023, 51, D690–D699. [Google Scholar] [CrossRef]
- Said, M.M.; Abo-Al-Ela, H.G.; El-Barbary, Y.A.; Ahmed, O.M.; Dighiesh, H.S. Influence of Stocking Density on the Growth, Immune and Physiological Responses, and Cultivation Environment of White-Leg Shrimp (Litopenaeus Vannamei) in Biofloc Systems. Sci. Rep. 2024, 14, 11147. [Google Scholar] [CrossRef] [PubMed]
- Corral-Jara, K.F.; Skírnisdóttir, S.; Knobloch, S.; Briem, H.; Cobo-Díaz, J.F.; Carlino, N.; Bergsten, P.; Armanini, F.; Asnicar, F.; Pinto, F.; et al. Inter-Facility Characterization of Bacteria in Seafood Processing Plants: Exploring Potential Reservoirs of Spoilage Organisms and the Resistome. Heliyon 2024, 10, e33866. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, H.; Abdelrahman, H.; Kelly, A.; Roy, L.; Wang, L. Profiling and Source Tracking of the Microbial Populations and Resistome Present in Fish Products. Int. J. Food Microbiol. 2024, 413, 110591. [Google Scholar] [CrossRef]
- Park, J.; Bae, D.; Kim, S.A. Microbial Trace Investigation throughout the Entire Chicken Supply Chain Based on Metagenomic High-Throughput Sequencing. Food Res. Int. 2023, 169, 112775. [Google Scholar] [CrossRef] [PubMed]
- Bi, B.; Yuan, Y.; Jia, D.; Jiang, W.; Yan, H.; Yuan, G.; Gao, Y. Identification and Pathogenicity of Emerging Fish Pathogen Acinetobacter Johnsonii from a Disease Outbreak in Rainbow Trout (Oncorhynchus Mykiss). Aquac. Res. 2023, 2023, 1995494. [Google Scholar] [CrossRef]
- Hasiri, Z.; Rahimi, E.; Momtaz, H.; Shakerian, A. Prevalence and Phenotypic and Genotypic Patterns of Antibiotic Resistance of Acinetobacter Baumannii Strains Isolated from Fish, Shrimp, and Lobster Samples. J. Food Process. Preserv. 2023, 2023, 6245225. [Google Scholar] [CrossRef]
- Paździor, E. Shewanella Putrefaciens—A New Opportunistic Pathogen of Freshwater Fish. J. Vet. Res. 2016, 60, 429–434. [Google Scholar] [CrossRef]
- Ringø, E.; Jutfelt, F.; Kanapathippillai, P.; Bakken, Y.; Sundell, K.; Glette, J.; Mayhew, T.M.; Myklebust, R.; Olsen, R.E. Damaging Effect of the Fish Pathogen Aeromonas Salmonicida Ssp. Salmonicida on Intestinal Enterocytes of Atlantic Salmon (Salmo Salar L.). Cell Tissue Res. 2004, 318, 305–311. [Google Scholar] [CrossRef]
- Novoslavskij, A.; Terentjeva, M.; Eizenberga, I.; Valciņa, O.; Bartkevičs, V.; Bērziņš, A. Major Foodborne Pathogens in Fish and Fish Products: A Review. Ann. Microbiol. 2016, 66, 1–15. [Google Scholar] [CrossRef]
- Dewi, R.R.; Hassan, L.; Daud, H.M.; Matori, M.F.; Nordin, F.; Ahmad, N.I.; Zakaria, Z. Prevalence and Antimicrobial Resistance of Escherichia Coli, Salmonella and Vibrio Derived from Farm-Raised Red Hybrid Tilapia (Oreochromis Spp.) and Asian Sea Bass (Lates Calcarifer Bloch 1970) on the West Coast of Peni. Antibiotics 2022, 11, 136. [Google Scholar] [CrossRef]
- Falcão, J.P.; Falcão, D.P.; Gomes, T.A.T. Ice as a Vehicle for Diarrheagenic Escherichia Coli. Int. J. Food Microbiol. 2004, 91, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.G.; Thornber, K.; Chaput, D.L.; Hasan, N.A.; Mehedi Alam, M.; Haque, M.M.; Cable, J.; Temperton, B.; Tyler, C.R. Metagenomic Assessment of the Diversity and Ubiquity of Antimicrobial Resistance Genes in Bangladeshi Aquaculture Ponds. Aquac. Rep. 2023, 29, 101462. [Google Scholar] [CrossRef]
- World Health Organization. Stop TB Initiative (World Health Organization) Treatment of Tuberculosis: Guidelines, 4th ed.; World Health Organization: Geneva, Switzerland, 2010; ISBN 9241547839. [Google Scholar]
- Smittipat, N.; Juthayothin, T.; Billamas, P.; Jaitrong, S.; Rukseree, K.; Dokladda, K.; Chaiyasirinroje, B.; Disratthakit, A.; Chaiprasert, A.; Mahasirimongkol, S.; et al. Mutations in Rrs, rpsL and gidB in Streptomycin-Resistant Mycobacterium Tuberculosis Isolates from Thailand. J. Glob. Antimicrob. Resist. 2016, 4, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Rui, Y.; Qiu, G. Analysis of Antibiotic Resistance Genes in Water Reservoirs and Related Wastewater from Animal Farms in Central China. Microorganisms 2024, 12, 396. [Google Scholar] [CrossRef]
- Spänig, S.; Eick, L.; Nuy, J.K.; Beisser, D.; Ip, M.; Heider, D.; Boenigk, J. A Multi-Omics Study on Quantifying Antimicrobial Resistance in European Freshwater Lakes. Environ. Int. 2021, 157, 106821. [Google Scholar] [CrossRef]
- Zhou, S.; Zhu, Y.; Yan, Y.; Wang, W.; Wang, Y. Deciphering Extracellular Antibiotic Resistance Genes (eARGs) in Activated Sludge by Metagenome. Water Res. 2019, 161, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Koutsoumanis, K.; Allende, A.; Álvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Role Played by the Environment in the Emergence and Spread of Antimicrobial Resistance (AMR) through the Food Chain. EFSA J. 2021, 19, e06651. [Google Scholar] [CrossRef]
- Brunton, L.A.; Desbois, A.P.; Garza, M.; Wieland, B.; Mohan, C.V.; Häsler, B.; Tam, C.C.; Le, P.N.T.; Phuong, N.T.; Van, P.T.; et al. Identifying Hotspots for Antibiotic Resistance Emergence and Selection, and Elucidating Pathways to Human Exposure: Application of a Systems-Thinking Approach to Aquaculture Systems. Sci. Total Environ. 2019, 687, 1344–1356. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Adamiak, J.W.; Leus, I.V. Making Sense of Drug-Efflux Transporters in the Physiological Environment. Curr. Opin. Microbiol. 2022, 69, 102179. [Google Scholar] [CrossRef]
- Ramírez-Castillo, F.Y.; Guerrero-Barrera, A.L.; Avelar-González, F.J. An Overview of Carbapenem-Resistant Organisms from Food-Producing Animals, Seafood, Aquaculture, Companion Animals, and Wildlife. Front. Vet. Sci. 2023, 10, 1158588. [Google Scholar] [CrossRef]
- Li, L.; Xiao, Y.; Wang, C.; Olsen, R.H.; Meng, H.; Shi, L. Exploring the Resistome, Virulome, Mobilome and Microbiome along Pork Production Chain Using Metagenomics. Int. J. Food Microbiol. 2022, 371, 109674. [Google Scholar] [CrossRef]
- Guan, Y.; Xue, X.; Jia, J.; Li, X.; Xing, H.; Wang, Z. Metagenomic Assembly and Binning Analyses the Prevalence and Spread of Antibiotic Resistome in Water and Fish Gut Microbiomes along an Environmental Gradient. J. Environ. Manag. 2022, 318, 115521. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Wang, L.; Li, Z.-H.; Zhang, M.-Q.; Shao, W.; Sheng, G.-P. Antibiotic Resistance and Microbiota in the Gut of Chinese Four Major Freshwater Carp from Retail Markets. Environ. Pollut. 2019, 255, 113327. [Google Scholar] [CrossRef] [PubMed]
- Tavares, J.; Martins, A.; Fidalgo, L.G.; Lima, V.; Amaral, R.A.; Pinto, C.A.; Silva, A.M.; Saraiva, J.A. Fresh Fish Degradation and Advances in Preservation Using Physical Emerging Technologies. Foods 2021, 10, 780. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Parente, E.; Ianniello, R.G.; De Filippis, F.; Ricciardi, A. Dynamics of Bacterial Communities and Interaction Networks in Thawed Fish Fillets during Chilled Storage in Air. Int. J. Food Microbiol. 2019, 293, 102–113. [Google Scholar] [CrossRef]
- Lee, A.W.T.; Ng, I.C.F.; Wong, E.Y.K.; Wong, I.T.F.; Sze, R.P.P.; Chan, K.Y.; So, T.Y.; Zhang, Z.; Ka-Yee Fung, S.; Choi-Ying Wong, S.; et al. Comprehensive Identification of Pathogenic Microbes and Antimicrobial Resistance Genes in Food Products Using Nanopore Sequencing-Based Metagenomics. Food Microbiol. 2024, 121, 104493. [Google Scholar] [CrossRef]


| Sample | Location | Coordinates | Date | Note |
|---|---|---|---|---|
| Farm | ||||
| Wi-A, Wo-A | Farm A | 7.151493 N, 100.550689 E | 18 June 2024 | Cage, fish farm |
| Wi-B, Wo-B | Farm B | 7.167891 N, 100.583121 E | 21 June 2024 | Cage, fish farm |
| Wi-C, Wo-C | Farm C | 7.262975 N, 100.423502 E | 25 June 2024 | Recirculating aquaculture |
| Container | ||||
| Co-A | Farm A container | 7.151493 N, 100.550689 E | 18 June 2024 | |
| Co-B | Farm B container | 7.167891 N, 100.583121 E | 21 June 2024 | |
| Co-C | Farm C container | 7.262975 N, 100.423502 E | 25 June 2024 | |
| Market | ||||
| Fi-1 | Plaza market | 7.012232 N, 100.466658 E | 9 July 2024 | |
| Fi-2 | Plaza market | 7.012232 N, 100.466658 E | 9 July 2024 | |
| Fi-3 | Koh Mhee market | 7.052951 N, 100.506005 E | 10 July 2024 | |
| Fi-4 | Koh Mhee market | 7.052951 N, 100.506005 E | 10 July 2024 | |
| Fi-5 | Songkhla market | 7.200978 N, 100.593816 E | 11 July 2024 | |
| Fi-6 | Khokrai market | 7.150168 N, 100.570592 E | 11 July 2024 | |
| Fi-7 | Supermarket A | 6.993322 N, 100.486395 E | 11 July 2024 | |
| Fi-8 | Klongrian market | 7.000037 N, 100.489579 E | 17 July 2024 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Detcharoen, M.; Khrueakaew, P.; Benjakul, S.; Romyasamit, C.; Suyapoh, W.; Saetang, J. Surveillance of Antimicrobial Resistance in the Asian Seabass (Lates calcarifer) Supply Chain Using Nanopore Sequencing. Foods 2025, 14, 1691. https://doi.org/10.3390/foods14101691
Detcharoen M, Khrueakaew P, Benjakul S, Romyasamit C, Suyapoh W, Saetang J. Surveillance of Antimicrobial Resistance in the Asian Seabass (Lates calcarifer) Supply Chain Using Nanopore Sequencing. Foods. 2025; 14(10):1691. https://doi.org/10.3390/foods14101691
Chicago/Turabian StyleDetcharoen, Matsapume, Panatda Khrueakaew, Soottawat Benjakul, Chonticha Romyasamit, Watcharapol Suyapoh, and Jirakrit Saetang. 2025. "Surveillance of Antimicrobial Resistance in the Asian Seabass (Lates calcarifer) Supply Chain Using Nanopore Sequencing" Foods 14, no. 10: 1691. https://doi.org/10.3390/foods14101691
APA StyleDetcharoen, M., Khrueakaew, P., Benjakul, S., Romyasamit, C., Suyapoh, W., & Saetang, J. (2025). Surveillance of Antimicrobial Resistance in the Asian Seabass (Lates calcarifer) Supply Chain Using Nanopore Sequencing. Foods, 14(10), 1691. https://doi.org/10.3390/foods14101691







