Quantitative Analysis of 3-Monochloropropane-1,2-diol in Fried Oil Using Convolutional Neural Networks Optimizing with a Stepwise Hybrid Preprocessing Strategy Based on Fourier Transform Infrared Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Sample Preparation
2.2. Spectral Data Collection
2.3. Data Processing
2.3.1. Full Factorial Design
2.3.2. Max–Min Normalization
2.3.3. Savitzky–Golay Smoothing
2.3.4. Derivative
2.3.5. Other Methods and Data Analysis
2.4. Model Building
2.4.1. Data Splitting
2.4.2. Partial Least Squares Regression
2.4.3. Random Forest
2.4.4. Support Vector Regression
2.4.5. Convolutional Neural Networks
2.5. Performance Evaluation
2.6. 3-MCPD Quantification by GC-MS
3. Results and Discussion
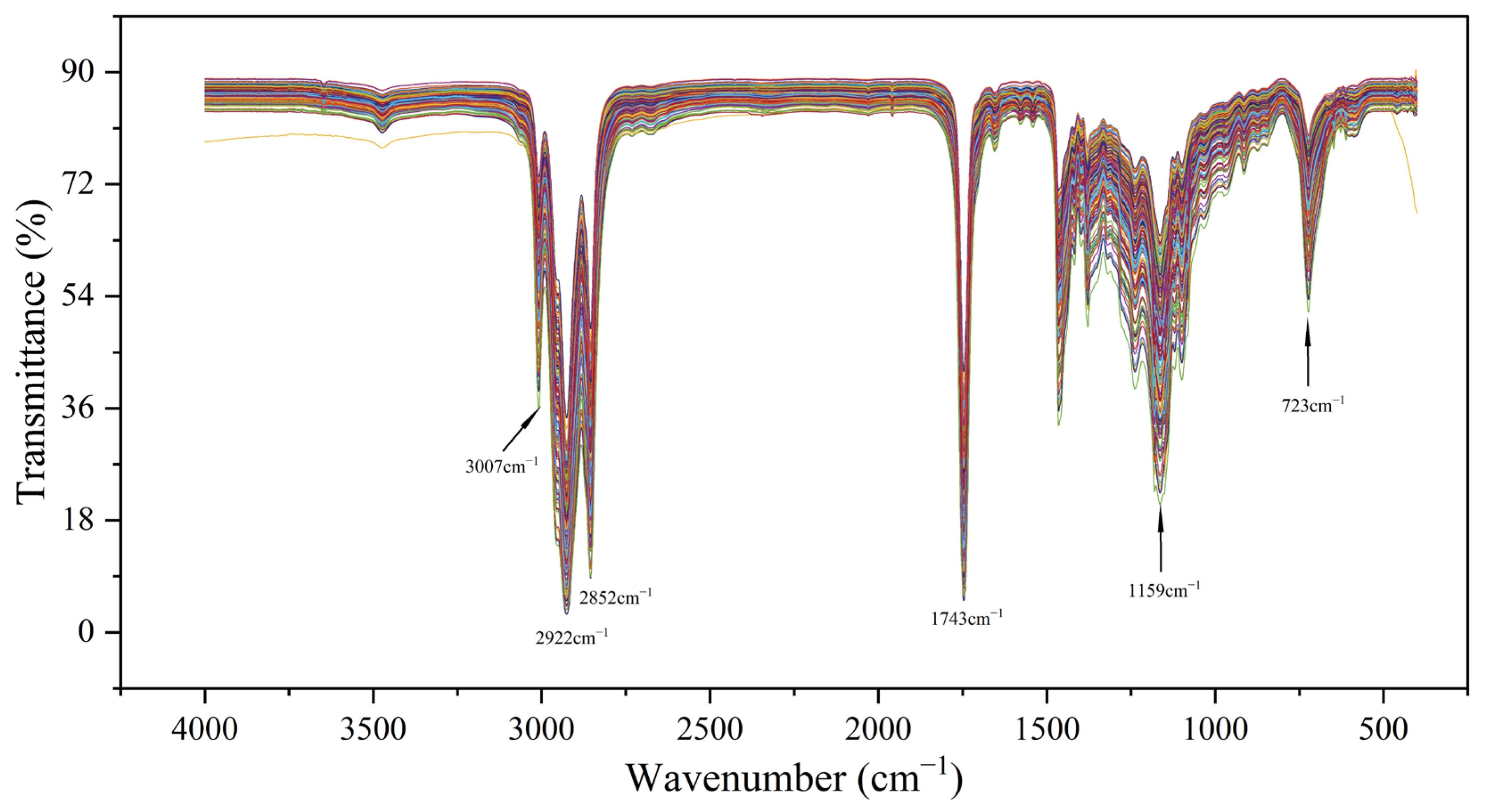
3.1. Spectral Analysis
3.2. Data Preprocessing and Model Development
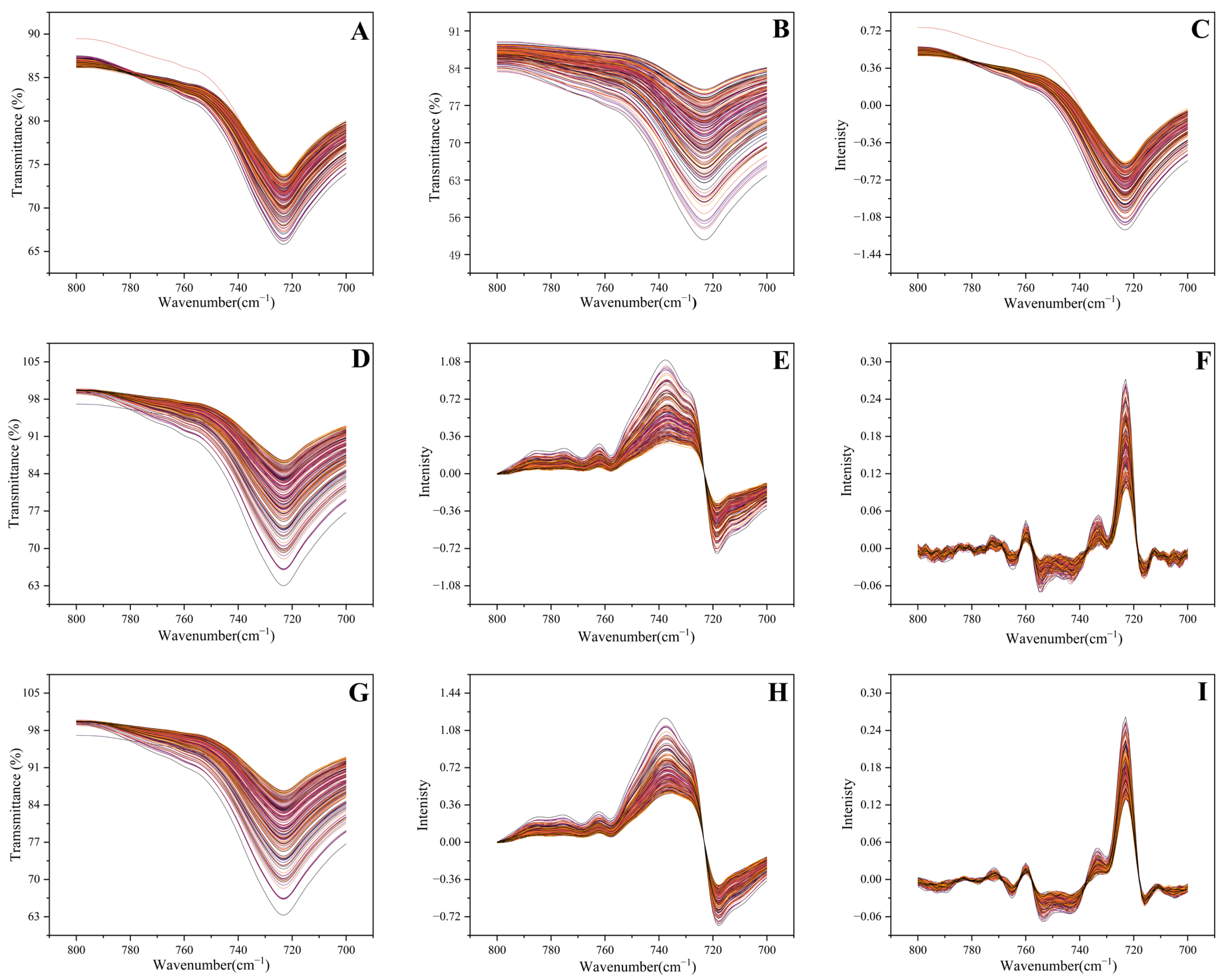
3.2.1. Impact of Data Preprocessing on Spectral Profile
3.2.2. Performance Comparison of Different Preprocessing Methods
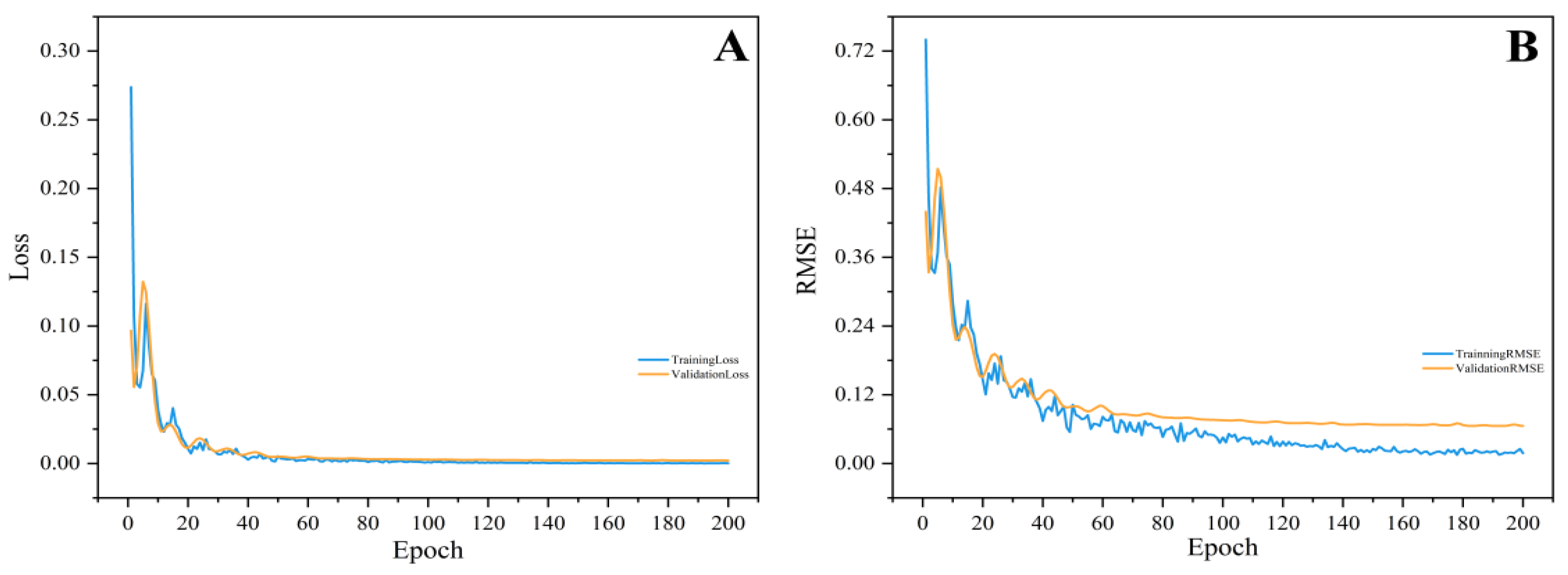
3.3. Optimizing the CNN Calibration Model
3.4. Visualization of the NL-SGS-D2-CNN
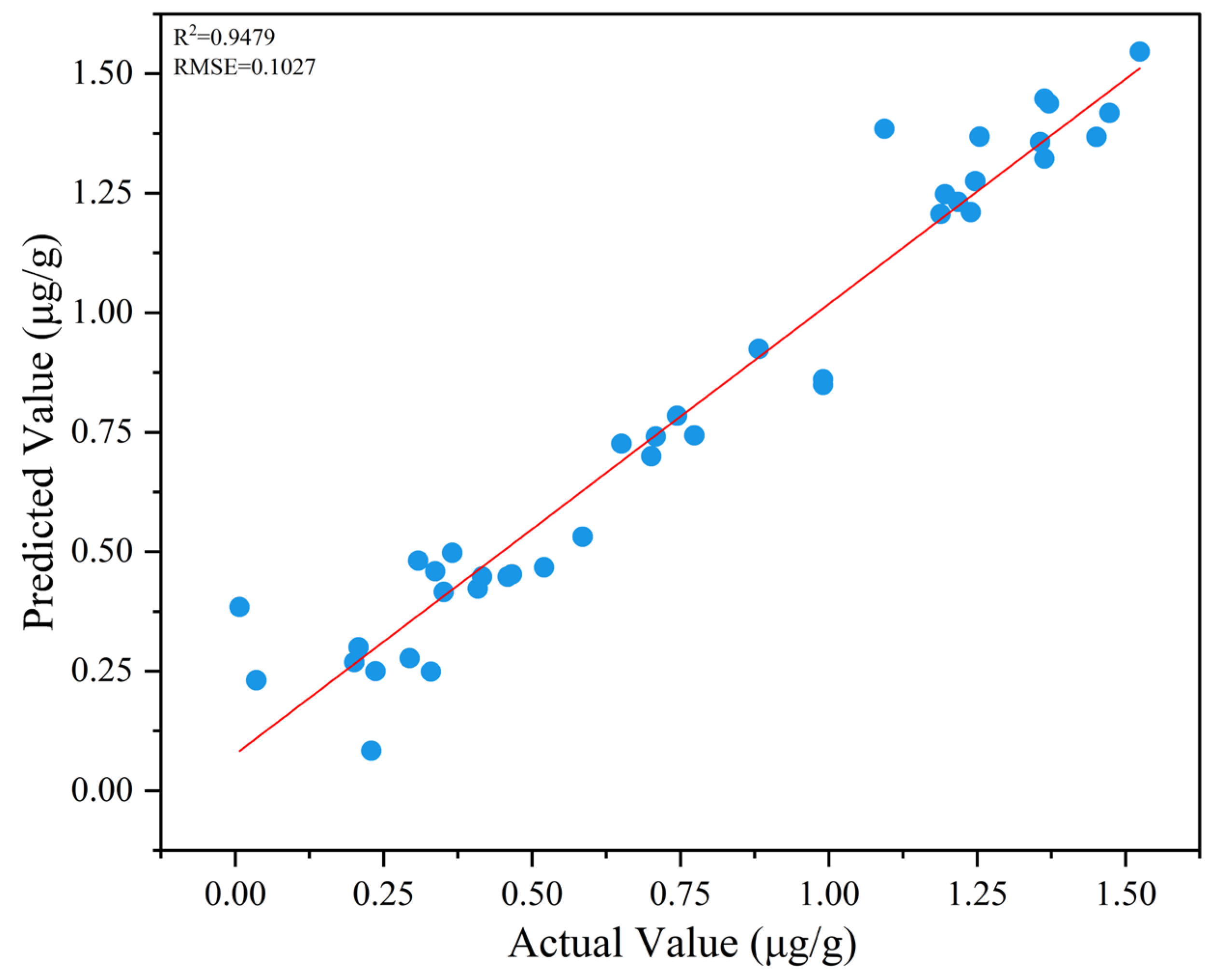
3.5. Quantitative Analysis of 3-MCPD by NL-SGS-D2-CNN
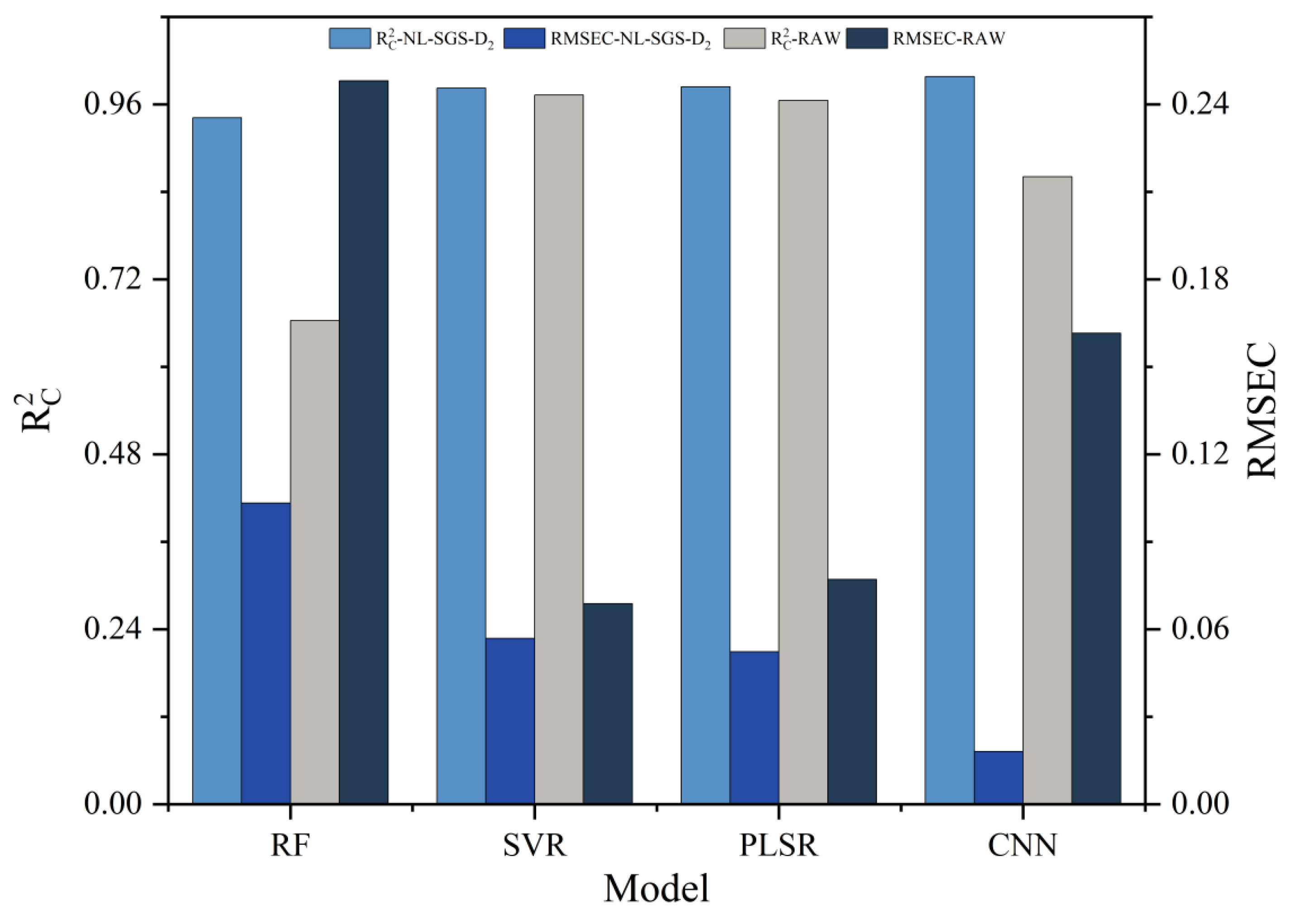
3.6. Comparison of Different Modeling Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisenreich, A.; Monien, B.H.; Götz, M.E.; Buhrke, T.; Oberemm, A.; Schultrich, K.; Abraham, K.; Braeuning, A.; Schäfer, B. 3-MCPD as contaminant in processed foods: State of knowledge and remaining challenges. Food Chem. 2023, 403, 134332. [Google Scholar] [CrossRef] [PubMed]
- Sevim, Ç.; Özkaraca, M.; Kara, M.; Ulaş, N.; Mendil, A.S.; Margina, D.; Tsatsakis, A. Apoptosis is induced by sub-acute exposure to 3-MCPD and glycidol on Wistar Albino rat brain cells. Environ. Toxicol. Pharmacol. 2021, 87, 103735. [Google Scholar] [CrossRef] [PubMed]
- Ozluk, G.; González-Curbelo, M.Á.; Kabak, B. Chloropropanols and Their Esters in Food: An Updated Review. Foods 2024, 13, 2876. [Google Scholar] [CrossRef] [PubMed]
- Syed Putra, S.S.; Basirun, W.J.; Elgharbawy, A.A.M.; Hayyan, M.; Al Abdulmonem, W.; Aljohani, A.S.M.; Hayyan, A. 3-Monochloropropane-1,2-diol (3-MCPD): A review on properties, occurrence, mechanism of formation, toxicity, analytical approach and mitigation strategy. J. Food Meas. Charact. 2023, 17, 3592–3615. [Google Scholar] [CrossRef]
- Aka, M.A.; Fodjo, E.K.; Williams, I.B.I.; Narcisse, P.B.B.; Sylvestre, K.K.K.; Albert, T.; Yang, G.; Kong, C. Evaluation of 3-MCPD content of commonly consumed food in Côte d’Ivoire using bimodal UV-Vis/electrochemistry technique. Int. J. Biol. Chem. Sci. 2022, 16, 1795–1805. [Google Scholar] [CrossRef]
- He, B.; Wang, L.; Li, M. A biosensor for direct bioelectrocatalysis detection of 3-MCPD in soy sauce using Cyt-c incorporated in Au@AgNSs/FeMOF nanocomposite. J. Iran. Chem. Soc. 2021, 18, 151–158. [Google Scholar] [CrossRef]
- Xu, T.; Fu, Q.; Qingru, Z.; Wang, Z.; Liu, X.; Xiao, S.; Jiang, X.; Lu, Y.; Gong, Z.; Wu, Y.; et al. A simple fluorescence pyrocatechol–polyethyleneimine detection method for 3-MCPD. Anal. Methods 2024, 16, 276–283. [Google Scholar] [CrossRef]
- Goh, K.M.; Maulidiani, M.; Rudiyanto, R.; Wong, Y.H.; Ang, M.Y.; Yew, W.M.; Abas, F.; Lai, O.M.; Wang, Y.; Tan, C.P. Rapid assessment of total MCPD esters in palm-based cooking oil using ATR-FTIR application and chemometric analysis. Talanta 2019, 198, 215–223. [Google Scholar] [CrossRef]
- Karunathilaka, S.R.; Farris, S.; Mossoba, M.M. Rapid Prediction of Low (<1%) trans Fat Content in Edible Oils and Fast Food Lipid Extracts by Infrared Spectroscopy and Partial Least Squares Regression. J. Food Sci. 2018, 83, 2101–2108. [Google Scholar] [CrossRef]
- Harrou, F.; Saidi, A.; Sun, Y. Wind power prediction using bootstrap aggregating trees approach to enabling sustainable wind power integration in a smart grid. Energy Convers. Manag. 2019, 201, 112077. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Elizar, E.; Zulkifley, M.A.; Muharar, R.; Zaman, M.H.; Mustaza, S.M. A Review on Multiscale-Deep-Learning Applications. Sensors 2022, 22, 7384. [Google Scholar] [CrossRef]
- Weng, S.; Chu, Z.; Wang, M.; Han, K.; Zhu, G.; Liu, C.; Li, X.; Huang, L. Reflectance spectroscopy with operator difference for determination of behenic acid in edible vegetable oils by using convolutional neural network and polynomial correction. Food Chem. 2022, 367, 130668. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhang, X.; Li, M.; Jiang, H.; Chen, Q. Feasibility study on Raman spectra-based deep learning models for monitoring the contamination degree and level of aflatoxin B1 in edible oil. Microchem. J. 2022, 180, 107613. [Google Scholar] [CrossRef]
- Xu, Y.; Yang, W.; Wu, X.; Wang, Y.; Zhang, J. ResNet Model Automatically Extracts and Identifies FT-NIR Features for Geographical Traceability of Polygonatum kingianum. Foods 2022, 11, 3568. [Google Scholar] [CrossRef]
- Yi, L.; Dong, N.; Yun, Y.; Deng, B.; Ren, D.; Liu, S.; Liang, Y. Chemometric methods in data processing of mass spectrometry-based metabolomics: A review. Anal. Chim. Acta 2016, 914, 17–34. [Google Scholar] [CrossRef]
- Cozzolino, D.; Williams, P.J.; Hoffman, L.C. An overview of pre-processing methods available for hyperspectral imaging applications. Microchem. J. 2023, 193, 109129. [Google Scholar] [CrossRef]
- Spiegelman, C.H.; McShane, M.J.; Goetz, M.J.; Motamedi, M.; Yue, Q.L.; Coté, G.L. Theoretical Justification of Wavelength Selection in PLS Calibration: Development of a New Algorithm. Anal. Chem. 1998, 70, 35–44. [Google Scholar] [CrossRef]
- Bian, X.; Wang, K.; Tan, E.; Diwu, P.; Zhang, F.; Guo, Y. A selective ensemble preprocessing strategy for near-infrared spectral quantitative analysis of complex samples. Chemom. Intell. Lab. Syst. 2020, 197, 103916. [Google Scholar] [CrossRef]
- Tiu, E.S.K.; Huang, Y.F.; Ng, J.L.; AlDahoul, N.; Ahmed, A.N.; Elshafie, A. An evaluation of various data pre-processing techniques with machine learning models for water level prediction. Nat. Hazards 2022, 110, 121–153. [Google Scholar] [CrossRef]
- Wang, Z.; Deng, J.; Ding, Z.; Jiang, H. Quantification of heavy metal Cd in peanut oil using near-infrared spectroscopy combined with chemometrics: Analysis and comparison of variable selection methods. Infrared Phys. Technol. 2024, 141, 105447. [Google Scholar] [CrossRef]
- Taylan, O.; Cebi, N.; Tahsin Yilmaz, M.; Sagdic, O.; Bakhsh, A.A. Detection of lard in butter using Raman spectroscopy combined with chemometrics. Food Chem. 2020, 332, 127344. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhu, C.; Jiang, H. Comparison of the performance of different one-dimensional convolutional neural network models-based near-infrared spectra for determination of chlorpyrifos residues in corn oil. Infrared Phys. Technol. 2023, 132, 104734. [Google Scholar] [CrossRef]
- Kuhlmann, J. Collaborative study for the quantification of total contents of 2- and 3-monochloropropanediol and glycidol in food emulsifiers by GC–MS. J. Am. Oil Chem. Soc. 2021, 98, 1131–1142. [Google Scholar] [CrossRef]
- Wong, Y.H.; Goh, K.M.; Abas, F.; Maulidiani, M.; Nyam, K.L.; Nehdi, I.A.; Sbihi, H.M.; Gewik, M.M.; Tan, C.P. Rapid quantification of 3-monochloropropane-1,2-diol in deep-fat frying using palm olein: Using ATR-FTIR and chemometrics. LWT 2019, 100, 404–408. [Google Scholar] [CrossRef]
- Gerretzen, J.; Szymańska, E.; Jansen, J.J.; Bart, J.; van Manen, H.-J.; van den Heuvel, E.R.; Buydens, L.M.C. Simple and Effective Way for Data Preprocessing Selection Based on Design of Experiments. Anal. Chem. 2015, 87, 12096–12103. [Google Scholar] [CrossRef]
- Ali, P.J.M. Investigating the Impact of min-max data normalization on the regression performance of K-nearest neighbor with different similarity measurements. ARO-Sci. J. Koya Univ. 2022, 10, 85–91. [Google Scholar] [CrossRef]
- Steinier, J.; Termonia, Y.; Deltour, J. Smoothing and differentiation of data by simplified least square procedure. Anal. Chem. 1972, 44, 1906–1909. [Google Scholar] [CrossRef]
- Hong, Y.; Liu, Y.; Chen, Y.; Liu, Y.; Yu, L.; Liu, Y.; Cheng, H. Application of fractional-order derivative in the quantitative estimation of soil organic matter content through visible and near-infrared spectroscopy. Geoderma 2019, 337, 758–769. [Google Scholar] [CrossRef]
- Maleki, M.R.; Mouazen, A.M.; Ramon, H.; De Baerdemaeker, J. Multiplicative Scatter Correction during On-line Measurement with Near Infrared Spectroscopy. Biosyst. Eng. 2007, 96, 427–433. [Google Scholar] [CrossRef]
- Jahani, T.; Kashaninejad, M.; Ziaiifar, A.M.; Golzarian, M.; Akbari, N.; Soleimanipour, A. Effect of selected pre-processing methods by PLSR to predict low-fat mozzarella texture measured by hyperspectral imaging. J. Food Meas. Charact. 2024, 18, 5060–5072. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Prasad, A.M.; Iverson, L.R.; Liaw, A. Newer Classification and Regression Tree Techniques: Bagging and Random Forests for Ecological Prediction. Ecosystems 2006, 9, 181–199. [Google Scholar] [CrossRef]
- Otchere, D.A.; Arbi Ganat, T.O.; Gholami, R.; Ridha, S. Application of supervised machine learning paradigms in the prediction of petroleum reservoir properties: Comparative analysis of ANN and SVM models. J. Pet. Sci. Eng. 2021, 200, 108182. [Google Scholar] [CrossRef]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A survey of the recent architectures of deep convolutional neural networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, C.; Sun, Z.; Hu, X.; Shen, Q.; Wu, J. Authentication of edible vegetable oils adulterated with used frying oil by Fourier Transform Infrared Spectroscopy. Food Chem. 2012, 132, 1607–1613. [Google Scholar] [CrossRef]
- Rohman, A.; Man, Y.B.C. Fourier transform infrared (FTIR) spectroscopy for analysis of extra virgin olive oil adulterated with palm oil. Food Res. Int. 2010, 43, 886–892. [Google Scholar] [CrossRef]
- Li, S.; Yang, Y.; Gao, S.; Lin, D.; Li, G.; Hu, Y.; Yang, W. Research on LIBS online monitoring criteria for aircraft skin laser paint removal based on OPLS-DA. Opt. Express 2024, 32, 4122–4136. [Google Scholar] [CrossRef]
- Zeng, G.; Ma, Y.; Du, M.; Chen, T.; Lin, L.; Dai, M.; Luo, H.; Hu, L.; Zhou, Q.; Pan, X. Deep convolutional neural networks for aged microplastics identification by Fourier transform infrared spectra classification. Sci. Total Environ. 2024, 913, 169623. [Google Scholar] [CrossRef]
- Dogo, E.M.; Afolabi, O.J.; Nwulu, N.I.; Twala, B.; Aigbavboa, C.O. A Comparative Analysis of Gradient Descent-Based Optimization Algorithms on Convolutional Neural Networks. In Proceedings of the 2018 International Conference on Computational Techniques, Electronics and Mechanical Systems (CTEMS), Belgaum, India, 21–22 December 2018; pp. 92–99. [Google Scholar]
- Martínez Vidal, J.L.; Arrebola, F.J.; Mateu-Sánchez, M. Application of gas chromatography–tandem mass spectrometry to the analysis of pesticides in fruits and vegetables. J. Chromatogr. A 2002, 959, 203–213. [Google Scholar] [CrossRef]
- Nicolaï, B.M.; Beullens, K.; Bobelyn, E.; Peirs, A.; Saeys, W.; Theron, K.I.; Lammertyn, J. Nondestructive measurement of fruit and vegetable quality by means of NIR spectroscopy: A review. Postharvest Biol. Technol. 2007, 46, 99–118. [Google Scholar] [CrossRef]
- Godoy, J.L.; Vega, J.R.; Marchetti, J.L. Relationships between PCA and PLS-regression. Chemom. Intell. Lab. Syst. 2014, 130, 182–191. [Google Scholar] [CrossRef]
- Tin Kam, H. The random subspace method for constructing decision forests. IEEE Trans. Pattern Anal. Mach. Intell. 1998, 20, 832–844. [Google Scholar] [CrossRef]
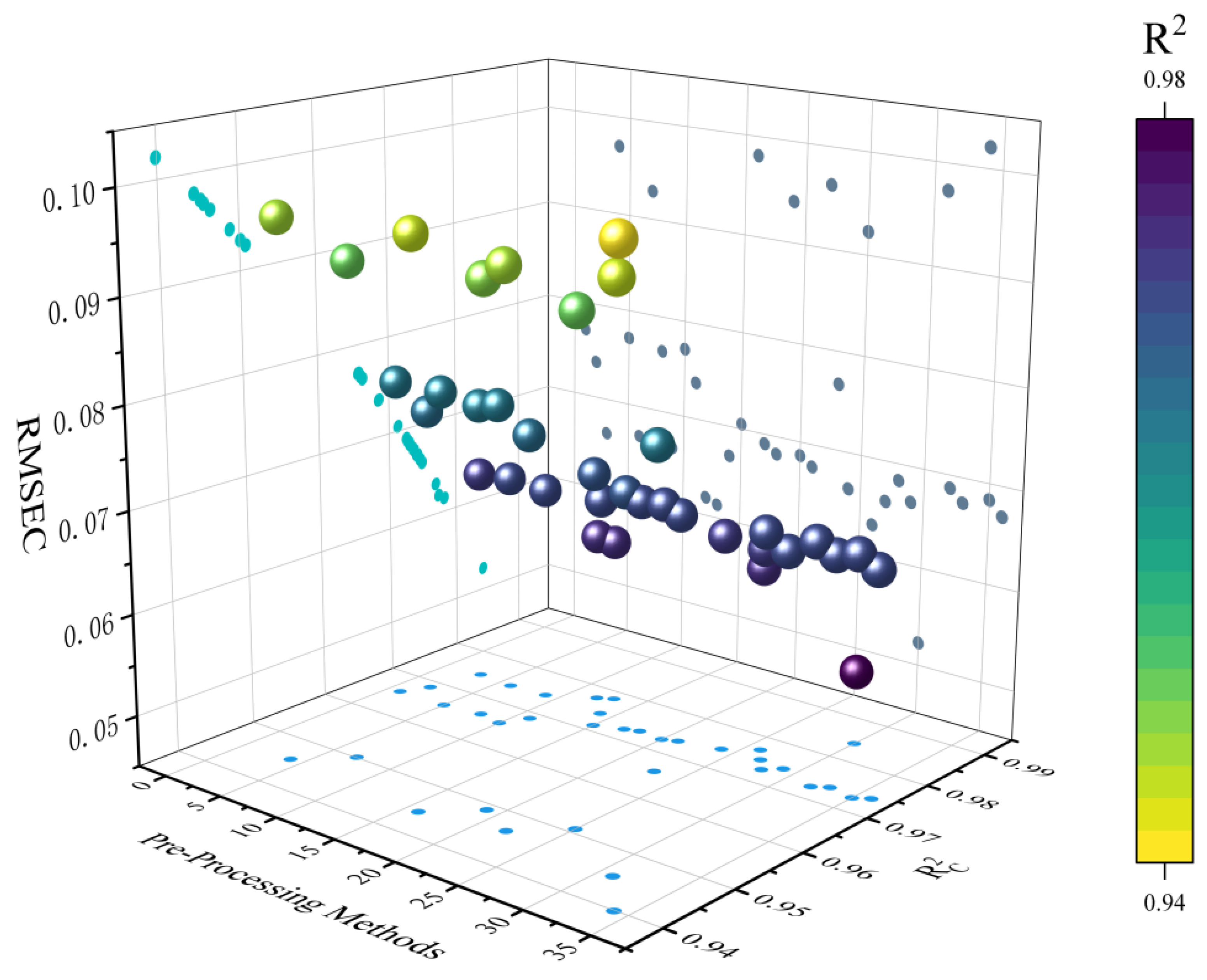

| NO. | Method | NO. | Method | NO. | Method |
|---|---|---|---|---|---|
| 1 | RAW | 13 | SGS | 25 | NL-SNV |
| 2 | RAW-D1 | 14 | SGS-D1 | 26 | NL-SNV-D1 |
| 3 | RAW-D2 | 15 | SGS-D2 | 27 | NL-SNV-D2 |
| 4 | MSC | 16 | SGS-MSC | 28 | NL-SGS |
| 5 | MSC-D1 | 17 | SGS-MSC-D1 | 29 | NL-SGS-D1 |
| 6 | MSC-D2 | 18 | SGS-MSC-D2 | 30 | NL-SGS-D2 |
| 7 | SNV | 19 | SGS-SNV | 31 | NL-SGS-MSC |
| 8 | SNV-D1 | 20 | SGS-SNV-D1 | 32 | NL-SGS-MSC-D1 |
| 9 | SNV-D2 | 21 | SGS-SNV-D2 | 33 | NL-SGS-MSC-D2 |
| 10 | NL | 22 | NL-MSC | 34 | NL-SGS-SNV |
| 11 | NL-D1 | 23 | NL-MSC-D1 | 35 | NL-SGS-SNV-D1 |
| 12 | NL-D2 | 24 | NL-MSC-D2 | 36 | NL-SGS-SNV-D2 |
| Convolutional Layer | Dropout | Max Number of Epochs | R2C | RMSEC | R2V | RMSEV |
|---|---|---|---|---|---|---|
| 1 | No | 60 | 0.9675 | 0.0781 | 0.9257 | 0.1160 |
| 1 | No | 80 | 0.9790 | 0.0628 | 0.9335 | 0.1097 |
| 1 | No | 100 | 0.9868 | 0.0497 | 0.9373 | 0.1065 |
| 1 | No | 120 | 0.9967 | 0.0241 | 0.9179 | 0.1149 |
| 1 | YES | 100 | 0.9859 | 0.0515 | 0.9384 | 0.1056 |
| 2 | YES | 100 | 0.9959 | 0.0279 | 0.9302 | 0.1124 |
| 3 | YES | 100 | 0.9982 | 0.0181 | 0.9464 | 0.0985 |
| 4 | YES | 100 | 0.9993 | 0.0117 | 0.9436 | 0.1010 |
| 5 | YES | 100 | 0.9823 | 0.0575 | 0.9304 | 0.1122 |
| Model | Methods | Calibration Set | Validation Set | ||
|---|---|---|---|---|---|
| R2C | RMSEC | R2V | RMSEV | ||
| RF | NL-SGS-D2 | 0.9416 cd | 0.1033 bc | 0.9295 a | 0.1176 cd |
| RAW | 0.6636 e | 0.2481 a | 0.5424 d | 0.2995 a | |
| SVR | NL-SGS-D2 | 0.9823 ab | 0.0569 de | 0.8096 b | 0.1932 bc |
| RAW | 0.9727 bc | 0.0688 cd | 0.6705 c | 0.2440 ab | |
| PLSR | NL-SGS-D2 | 0.9842 ab | 0.0523 e | 0.9485 a | 0.1101 de |
| RAW | 0.9656 bc | 0.0771 cd | 0.9435 a | 0.0997 ef | |
| CNN | NL-SGS-D2 | 0.9982 a | 0.0181 f | 0.9464 a | 0.0985 f |
| RAW | 0.8607 d | 0.1615 ab | 0.5607 d | 0.2819 a | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wang, S.; Zhang, S.; Yin, J.; Zhao, Q. Quantitative Analysis of 3-Monochloropropane-1,2-diol in Fried Oil Using Convolutional Neural Networks Optimizing with a Stepwise Hybrid Preprocessing Strategy Based on Fourier Transform Infrared Spectroscopy. Foods 2025, 14, 1670. https://doi.org/10.3390/foods14101670
Wang X, Wang S, Zhang S, Yin J, Zhao Q. Quantitative Analysis of 3-Monochloropropane-1,2-diol in Fried Oil Using Convolutional Neural Networks Optimizing with a Stepwise Hybrid Preprocessing Strategy Based on Fourier Transform Infrared Spectroscopy. Foods. 2025; 14(10):1670. https://doi.org/10.3390/foods14101670
Chicago/Turabian StyleWang, Xi, Siyi Wang, Shibing Zhang, Jiping Yin, and Qi Zhao. 2025. "Quantitative Analysis of 3-Monochloropropane-1,2-diol in Fried Oil Using Convolutional Neural Networks Optimizing with a Stepwise Hybrid Preprocessing Strategy Based on Fourier Transform Infrared Spectroscopy" Foods 14, no. 10: 1670. https://doi.org/10.3390/foods14101670
APA StyleWang, X., Wang, S., Zhang, S., Yin, J., & Zhao, Q. (2025). Quantitative Analysis of 3-Monochloropropane-1,2-diol in Fried Oil Using Convolutional Neural Networks Optimizing with a Stepwise Hybrid Preprocessing Strategy Based on Fourier Transform Infrared Spectroscopy. Foods, 14(10), 1670. https://doi.org/10.3390/foods14101670






