Development of a Time-Resolved Fluorescent Microsphere Test Strip for Rapid, On-Site, and Sensitive Detection of Picoxystrobin in Vegetables
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Instruments and Materials
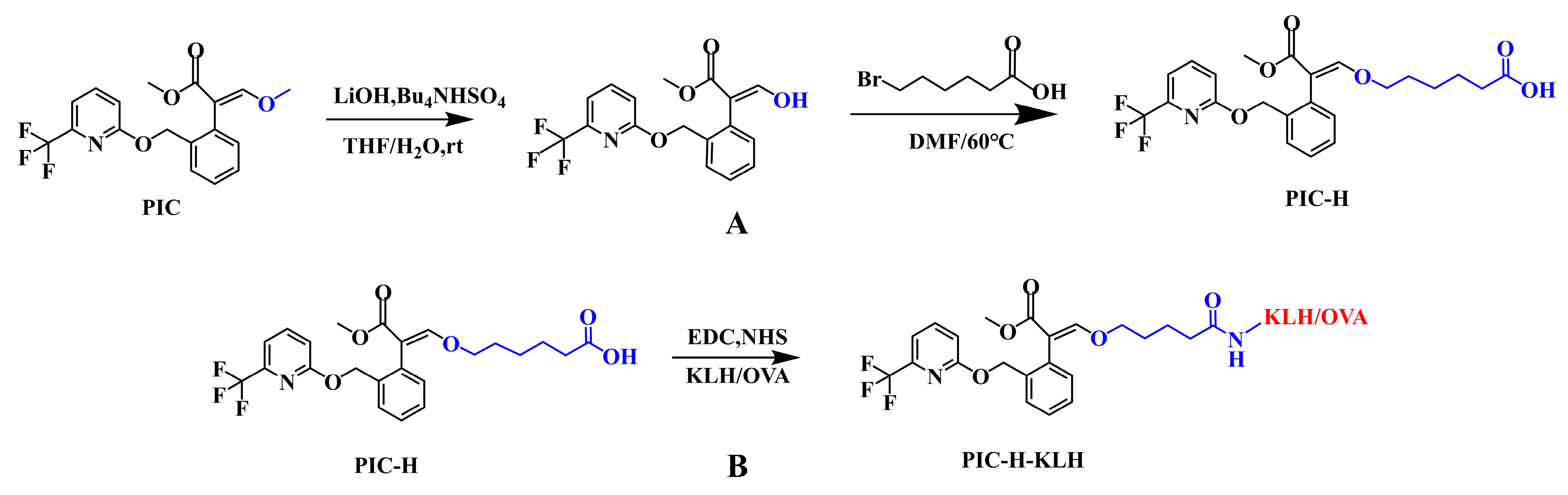
2.3. Preparation of Hapten and Artificial Antigen
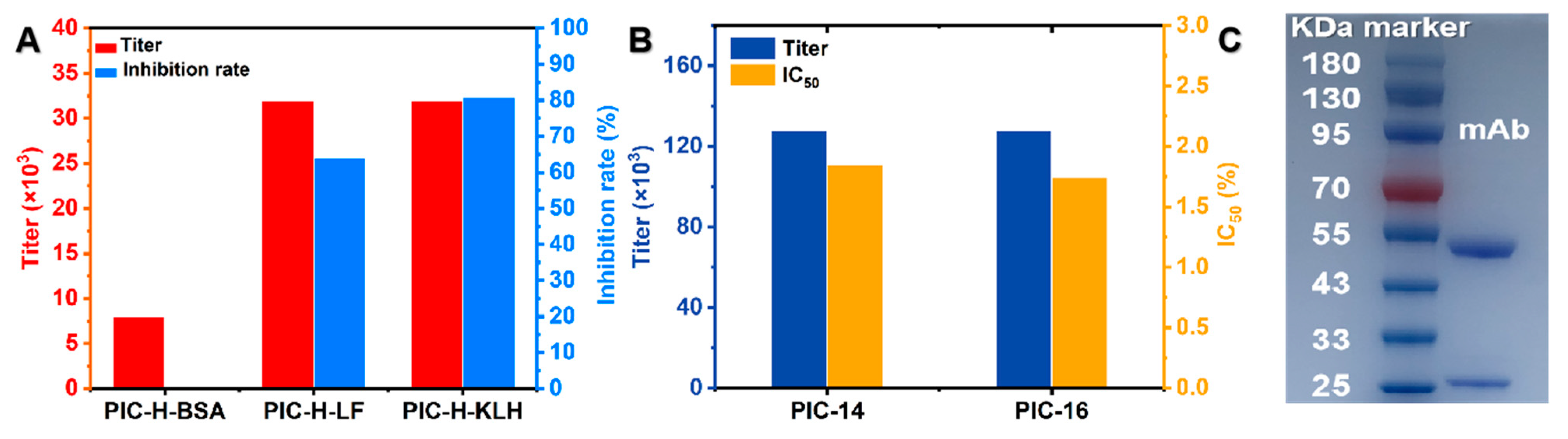
2.4. Preparation and Characterization of PIC Monoclonal Antibody
2.5. TRFM-Labeled PIC Antibody and Its Characterization
2.6. TRFICA Test Strip Assembly
2.7. TRFICA Detection Procedure and Principle
2.8. Performance of TRFICA
2.9. Recovery Test and Method Evaluation
3. Results and Discussion
3.1. Synthesis of Haptens and Antigens
3.2. Preparation and Identification of PIC-mAb
3.3. Characterization of the TRFM-mAb Probe
3.4. Optimization of TRFICA Working Conditions
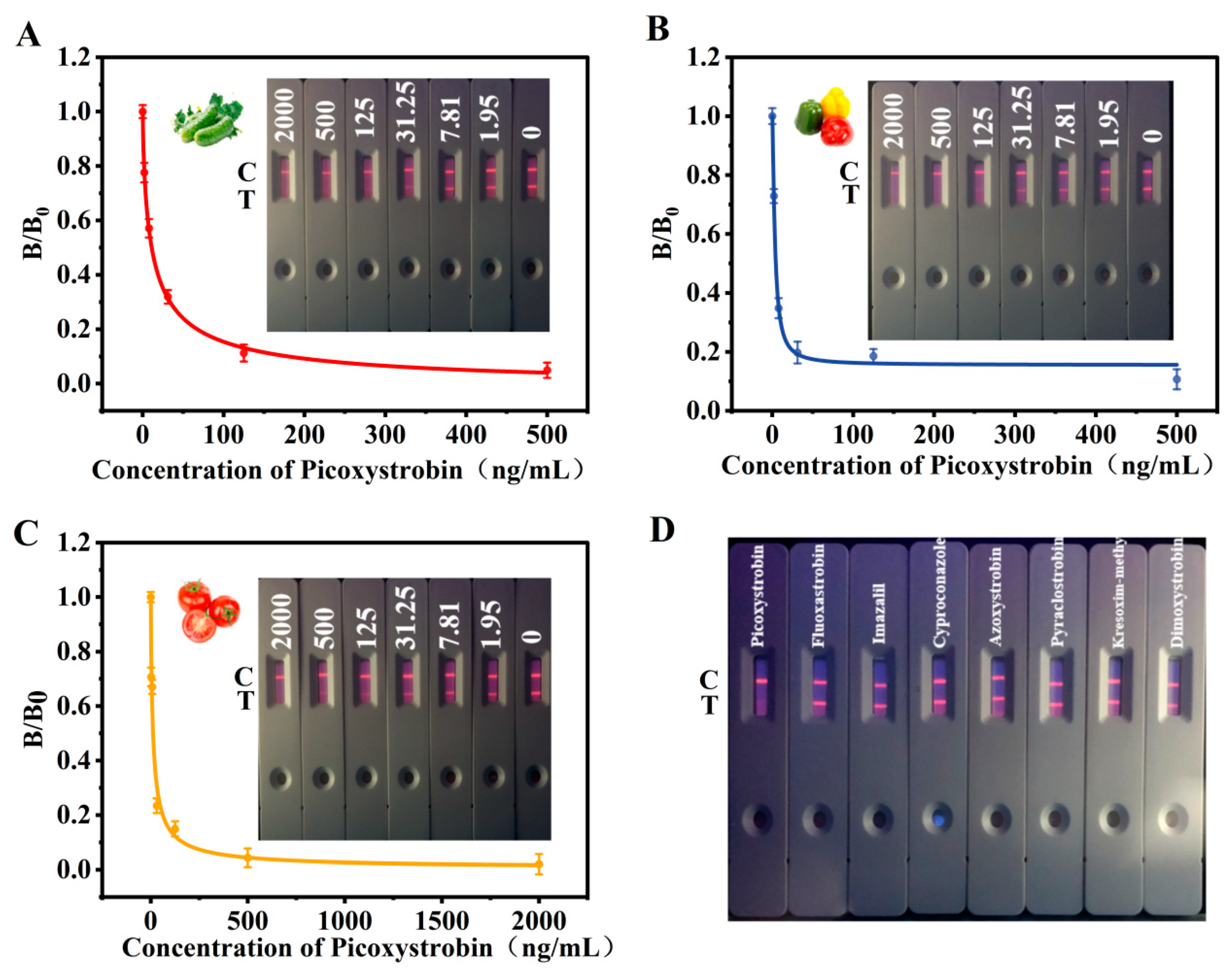
3.5. Sensitivity and Specificity
3.6. Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, Z.; Wang, Y.; Duan, Y.; He, Y.; Liu, S.; Chen, Y.; Deng, W.; Li, C.; Hu, W.; Gu, Y.; et al. Inhibitory Effect and Control Efficacy of Picoxystrobin against Neopestalotiopsis clavispora, Causing Vine Tea Leaf Blight. Agronomy 2023, 13, 1340. [Google Scholar] [CrossRef]
- GB 2763-2021; Limits of Mycotoxins in Food. National Standard for Food Safety. Official Document of SAC. Standardization Administration of China: Beijing, China, 2021; pp. 91–92.
- Malhat, F.; Saber, E.-S.; Amin, A.S.; Anagnostopoulos, C.; Shokr, S.A. Magnitude of picoxystrobin residues in strawberry under Egyptian conditions: Dissipation pattern and consumer risk assessment. Food Addit. Contam. Part A 2020, 37, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhao, Y.; Hu, J. Dissipation, residues and risk assessment of pyraclostrobin and picoxystrobin in cucumber under field conditions. J. Sci. Food Agric. 2020, 100, 5145–5151. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.H.; Rahman, M.M.; Abd El-Aty, A.M.; Chung, H.S.; Lee, H.S.; Park, S.-H.; Hacımüftüoğlu, F.; Chang, B.-J.; Shin, H.-C.; Shim, J.-H. Residue analysis of picoxystrobin in oriental melon using gas chromatography coupled with electron capture detection and mass spectrometric confirmation: Application to dissipation kinetics and risk assessment. Food Sci. Biotechnol. 2017, 26, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Schnug, L.; Ergon, T.; Jakob, L.; Scott-Fordsmand, J.J.; Joner, E.J.; Leinaas, H.P. Responses of earthworms to repeated exposure to three biocides applied singly and as a mixture in an agricultural field. Sci. Total Environ. 2015, 505, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Mao, L.; Zhang, L.; Zhang, Y.; Jiang, H. Effects of two strobilurins (azoxystrobin and picoxystrobin) on embryonic development and enzyme activities in juveniles and adult fish livers of zebrafish (Danio rerio). Chemosphere 2018, 207, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cao, F.; Zhao, F.; Yang, Y.; Teng, M.; Wang, C.; Qiu, L. Developmental toxicity, oxidative stress and immunotoxicity induced by three strobilurins (pyraclostrobin, trifloxystrobin and picoxystrobin) in zebrafish embryos. Chemosphere 2018, 207, 781–790. [Google Scholar] [CrossRef]
- Domingues, C.E.; Abdalla, F.C.; Balsamo, P.J.; Pereira, B.V.; Hausen, M.D.A.; Costa, M.J.; Silva-Zacarin, E.C. Thiamethoxam and picoxystrobin reduce the survival and overload the hepato-nephrocitic system of the Africanized honeybee. Chemosphere 2017, 186, 994–1005. [Google Scholar] [CrossRef]
- Kwon, H.-Y.; Kim, C.-S.; Park, B.-J.; Kim, I.-H.; Hong, S.-M.; Son, K.-A.; Jin, Y.-D.; Lee, J.-B.; Im, G.-J.; Kim, D.-H. Development of Analytical Method for Picoxystrobin in Agricultural Commodities Using GC/ECD and GC/MS. Korean J. Environ. Agric. 2012, 31, 146–151. [Google Scholar] [CrossRef]
- Viñas, P.; Campillo, N.; Martínez-Castillo, N.; Hernández-Córdoba, M. Method development and validation for strobilurin fungicides in baby foods by solid-phase microextraction gas chromatography–mass spectrometry. J. Chromatogr. 2009, 1216, 140–146. [Google Scholar] [CrossRef]
- Xie, F.-F.; Hang, X.-D.; Jian, L.; Zhang, F. Determination of picoxystrobin in wheat by modified QuEchERS-HPLC. J. Anhui Polytech. Univ. 2015, 5, 8–12. [Google Scholar] [CrossRef]
- Gao, Y.; Yang, S.; Li, X.; He, L.; Zhu, J.; Mu, W.; Liu, F. Residue determination of pyraclostrobin, picoxystrobin and its metabolite in pepper fruit via UPLC_MS/MS under open field conditions. Ecotoxicol. Environ. Saf. 2019, 182, 109445. [Google Scholar] [CrossRef] [PubMed]
- Preechakasedkit, P.; Pinwattana, K.; Dungchai, W. Development of a one-step immunochromatographic strip test using gold nanoparticles for the rapid detection of Salmonella typhi in human serum. Biosens. Bioelectron. 2012, 31, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Liang, J.; Zhang, Y.; Zhang, Y.; Teng, P.; Cao, D.; Zou, S.; Xu, T.; Zhao, J.; Tang, Y. CD8 cell counting in whole blood by a paper-based time-resolved fluorescence lateral flow immunoassay. Anal. Chim. Acta 2021, 1179, 338820. [Google Scholar] [CrossRef] [PubMed]
- Kolosova, A.; Maximova, K.; Eremin, S.A.; Zherdev, A.V.; Mercader, J.V.; Abad-Fuentes, A.; Dzantiev, B.B. Fluorescence polarisation immunoassays for strobilurin fungicides kresoxim-methyl, trifloxystrobin and picoxystrobin. Talanta 2017, 162, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Parra, J.; Abad-Fuentes, A.; Abad-Somovilla, A.; Mercader, J.V.; Esteve-Turrillas, F.A.; Agullo, C. Development of monoclonal antibody-based competitive immunoassays for the detection of picoxystrobin in cereal and oilseed flours. Food Control 2012, 26, 162–168. [Google Scholar] [CrossRef]
- Esteve-Turrillas, F.A.; Parra, J.; Abad-Fuentes, A.; Agulló, C.; Abad-Somovilla, A.; Mercader, J.V. Hapten synthesis, monoclonal antibody generation, and development of competitive immunoassays for the analysis of picoxystrobin in beer. Anal. Chim. Acta 2010, 682, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-Y.; Li, L.-H.; Wang, Y.; Wang, H.; Xu, Z.-L.; Tian, Y.-X.; Sun, Y.-M.; Yang, J.-Y.; Shen, Y.-D. Ultrasensitive and rapid colorimetric detection of paraquat via a high specific VHH nanobody. Biosens. Bioelectron. 2022, 205, 114089. [Google Scholar] [CrossRef]
- Chen, B.; Shen, X.; Li, Z.; Wang, J.; Li, X.; Xu, Z.; Shen, Y.; Lei, Y.; Huang, X.; Wang, X.; et al. Antibody Generation and Rapid Immunochromatography Using Time-Resolved Fluorescence Microspheres for Propiconazole: Fungicide Abused as Growth Regulator in Vegetable. Foods 2022, 11, 324. [Google Scholar] [CrossRef]
- Liu, H.; Lin, T.; Mao, J.; Lu, H.; Yang, D.; Wang, J.; Li, Q. Paclobutrazol Residue Determination in Potato and Soil Using Low Temperature Partition Extraction and Ultrahigh Performance Liquid Chromatography Tandem Mass Spectrometry. J. Anal. Methods Chem. 2015, 2015, 404925. [Google Scholar] [CrossRef]
- Song, S.; Chen, Z.; Liu, J.; Xu, L.; Kuang, H.; Zhu, J.; Liu, L. Development of an immunocolloidal strip for rapid detection of picoxystrobin. Food Agric. Immunol. 2020, 31, 711–722. [Google Scholar] [CrossRef]
- Wang, X.; Lu, D.; Huang, Q.; Yang, J. Microfluidics-Based Time-Resolved Fluorescence Immunoassay for the On-Site Detection of Aflatoxins B1 Zearalenone and Deoxynivalenol in Cereals. Foods 2022, 11, 1319. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ma, B.; Chen, E.; Yu, X.; Ye, Z.; Sun, C.; Zhang, M. Dual fluorescent immunochromatographic assay for simultaneous quantitative detection of citrinin and zearalenone in corn samples. Food Chem. 2021, 336, 127713. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wen, K.; Wang, Z.; Jiang, H.; Beier, R.; Shen, J. An ultra-sensitive monoclonal antibody-based fluorescent microsphere immunochromatographic test strip assay for detecting aflatoxin M1 in milk. Food Control 2016, 60, 588–595. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Sun, J.; Ji, J.; Ye, Y.; Lu, X.; Zhang, Y.; Sun, X. Rapid, on-site, and sensitive detection of aflatoxin M1 in milk products by using time-resolved fluorescence microsphere test strip. Food Control 2021, 121, 107616. [Google Scholar] [CrossRef]
- Sun, J.; Wang, L.; Shao, J.; Yang, D.; Fu, X.; Sun, X. One-step time-resolved fluorescence microsphere immunochromatographic test strip for quantitative and simultaneous detection of DON and ZEN. Anal. Bioanal. Chem. 2021, 413, 6489–6502. [Google Scholar] [CrossRef]
- Huang, L.; Chen, H.; Cui, P.; Ding, Y.; Wang, M.; Hua, X. Development of immunoassay based on rational hapten design for sensitive detection of pendimethalin in environment. Sci. Total Environ. 2022, 830, 154690. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, B.; Liang, Y.; Liu, X.; Chen, H.; Li, J.; Lei, H.; Xiao, Z. A monoclonal antibody-based time-resolved fluorescence microsphere lateral flow immunoassay for paclobutrazol detection. Curr. Res. Food Sci. 2022, 5, 1395–1402. [Google Scholar] [CrossRef]
- He, F.; Zou, T.; Yang, J.; Wang, H.; Deng, L.; Tian, Y.; Xu, Z.; Sun, Y.; Lei, H.; Tan, X. Development of a skeleton-specific antibody and Au nanoparticle-based immunochromatographic sensor for simultaneous detection of various tadalafil adulterants in health food. Food Agric. Immunol. 2019, 30, 349–368. [Google Scholar] [CrossRef]
- Zhou, S.; Xu, X.; Wang, L.; Liu, L.; Kuang, H.; Xu, C. Rapid, on-site quantitative determination of higenamine in functional food using a time-resolved fluorescence microsphere test strip. Food Chem. 2022, 387, 132859. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hua, Q.; Wang, J.; Liang, Z.; Li, J.; Wu, J.; Shen, X.; Lei, H.; Li, X. A smartphone-based dual detection mode device integrated with two lateral flow immunoassays for multiplex mycotoxins in cereals. Biosens. Bioelectron. 2020, 158, 112178. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, H.; Zheng, K.; Meng, B.; Wang, F.; Cui, Y.; Zeng, S.; Zhang, K.; Hu, D. Simultaneous Determination and Method Validation of Difenoconazole, Propiconazole and Pyraclostrobin in Pepper and Soil by LC-MS/MS in Field Trial Samples from Three Provinces, China. Biomed. Chromatogr. 2018, 32, e4052. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-J.; Wu, H.-L.; Xiao, Z.-L.; Fu, H.-J.; Shen, Y.-D.; Luo, L.; Wang, H.; Lei, H.-T.; Hongsibsong, S.; Xu, Z.-L. Rational hapten design to produce high-quality antibodies against carbamate pesticides and development of immunochromatographic assays for simultaneous pesticide screening. J. Hazard. Mater. 2022, 421, 125241. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, R.; Wang, Y.; Tang, Y.; Deng, A. Hapten Design, Modification and Preparation of Artificial Antigens. Chin. J. Anal. Chem. 2010, 38, 1211–1218. [Google Scholar] [CrossRef]
- Mari, G.M.; Li, H.; Dong, B.; Yang, H.; Talpur, A.; Mi, J.; Guo, L.; Yu, X.; Ke, Y.; Han, D.; et al. Hapten synthesis, monoclonal antibody production and immunoassay development for direct detection of 4-hydroxybenzehydrazide in chicken, the metabolite of nifuroxazide. Food Chem. 2021, 355, 129598. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhu, M.; Jiang, W.; Zhang, T.; Ma, M.; Shi, F. A simple synthesis method of microsphere immunochromatographic test strip for time-resolved luminescence detection of folic acid. Food Chem. 2023, 413, 135599. [Google Scholar] [CrossRef]
- Liu, Z.; Wei, X.; Ren, K.; Zhu, G.; Zhang, Z.; Wang, J.; Du, D. Highly efficient detection of paclobutrazol in environmental water and soil samples by time-resolved fluoroimmunoassay. Sci. Total Environ. 2016, 569, 1629–1634. [Google Scholar] [CrossRef]
- Wu, Q.; Yao, L.; Qin, P.; Xu, J.; Sun, X.; Yao, B.; Ren, F.; Chen, W. Time-resolved fluorescent lateral flow strip for easy and rapid quality control of edible oil. Food Chem. 2021, 357, 129739. [Google Scholar] [CrossRef]



| Method | Matrix | The Detection Limit (mg/kg) | References |
|---|---|---|---|
| GC-ECD/ GC-MS | Vegetables, fruits, and crops | 0.025 | [10] |
| SPME/GC-MS | Baby food | 0.005 | [11] |
| QuEchERS-HPLC | Wheat | 0.018 | [12] |
| UPLC-MS/MS | Pepper fruit | 0.036 | [13] |
| Ic ELISA | Flour | 0.0001 | [17] |
| Ic ELISA | Beer | 0.0002 | [18] |
| GICA | Cucumber | 0.1 | [22] |
| FPIAs | Red wine | 0.005 | [16] |
| TRFICA | Cucumber | 0.00026 | This Work |
| Green pepper | 0.00061 | ||
| Tomato | 0.0034 |
| Sample | Spiked Level (ng/mL) | Detection Level (Mean ± SD) (ng/mL) | Recovery (%) | CV (%) |
|---|---|---|---|---|
| Cucumber | 2 | 1.60 ± 0.23 | 79.8 | 14.2 |
| 12 | 11.77 ± 1.23 | 98.1 | 10.5 | |
| 72 | 71.70 ± 3.05 | 99.6 | 4.3 | |
| Green pepper | 2 | 1.64 ± 0.11 | 81.8 | 6.6 |
| 8 | 7.34 ± 0.63 | 91.8 | 8.5 | |
| 32 | 33.59 ± 1.66 | 105.0 | 4.9 | |
| Tomato | 12 | 10.52 ± 0.79 | 87.7 | 7.5 |
| 42 | 34.89 ± 2.41 | 83.1 | 6.9 | |
| 150 | 136.51 ± 9.13 | 91.0 | 6.7 |
| Assay | TRFICA | UPLC-MS/MS | |||
|---|---|---|---|---|---|
| Samples | Number | Test Value (Mean ± SD, ng/g) | CV (%) | Test Value (Mean ± SD, ng/g) | CV (%) |
| Cucumber | Sample 1 | ND | - | ND | - |
| Sample 2 | 40.34 ± 5.04 | 12.4 | 53.99 ± 6.95 | 12.8 | |
| Sample 3 | ND | - | ND | - | |
| Sample 4 | ND | - | ND | - | |
| Sample 5 | ND | - | ND | - | |
| Green pepper | Sample 1 | ND | - | ND | - |
| Sample 2 | ND | - | ND | - | |
| Sample 3 | ND | - | ND | - | |
| Sample 4 | ND | - | ND | - | |
| Sample 5 | 47.90 ± 8.06 | 16.8 | 49.77 ± 3.26 | 6.5 | |
| Tomato | Sample 1 | ND | - | ND | - |
| Sample 2 | ND | - | ND | - | |
| Sample 3 | ND | - | ND | - | |
| Sample 4 | ND | - | ND | - | |
| Sample 5 | ND | - | ND | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Chen, L.; Zhang, Y.; Xiang, S.; Zhang, R.; Shen, Y.; Liao, J.; Xie, H.; Yang, J. Development of a Time-Resolved Fluorescent Microsphere Test Strip for Rapid, On-Site, and Sensitive Detection of Picoxystrobin in Vegetables. Foods 2024, 13, 423. https://doi.org/10.3390/foods13030423
Chen J, Chen L, Zhang Y, Xiang S, Zhang R, Shen Y, Liao J, Xie H, Yang J. Development of a Time-Resolved Fluorescent Microsphere Test Strip for Rapid, On-Site, and Sensitive Detection of Picoxystrobin in Vegetables. Foods. 2024; 13(3):423. https://doi.org/10.3390/foods13030423
Chicago/Turabian StyleChen, Junjie, Lidan Chen, Yongyi Zhang, Siyi Xiang, Ruizhou Zhang, Yudong Shen, Jiaming Liao, Huahui Xie, and Jinyi Yang. 2024. "Development of a Time-Resolved Fluorescent Microsphere Test Strip for Rapid, On-Site, and Sensitive Detection of Picoxystrobin in Vegetables" Foods 13, no. 3: 423. https://doi.org/10.3390/foods13030423
APA StyleChen, J., Chen, L., Zhang, Y., Xiang, S., Zhang, R., Shen, Y., Liao, J., Xie, H., & Yang, J. (2024). Development of a Time-Resolved Fluorescent Microsphere Test Strip for Rapid, On-Site, and Sensitive Detection of Picoxystrobin in Vegetables. Foods, 13(3), 423. https://doi.org/10.3390/foods13030423








