Determination of Bioactive Components in Chrysanthemum Tea (Gongju) Using Hyperspectral Imaging Technique and Chemometrics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Image Acquisition and Data Correction
2.3. Determination of Reference Values of Total Flavonoids and Chlorogenic Acid Content
2.3.1. Equipment and Chemicals
2.3.2. Determination of Total Flavonoid Content
2.3.3. Determination of Total Chlorogenic Acid Content
2.4. Spectra Extraction and Preprocessing
2.5. Feature Extraction Process
2.5.1. Optimal Wavelength Selection via VSPSO
2.5.2. Spatial Image Feature Extraction
2.6. Model Building and Evaluation
2.6.1. Conventional Models
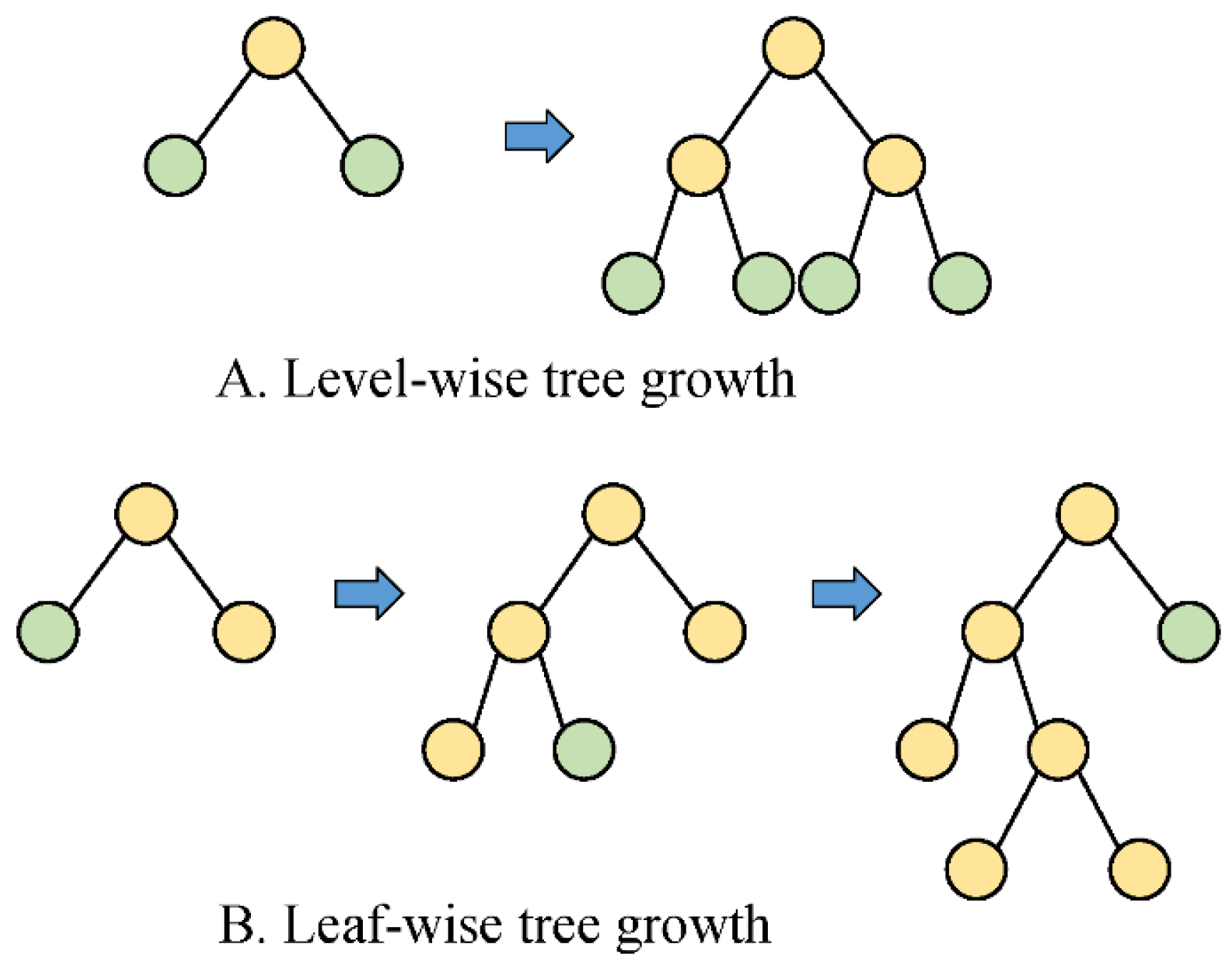
2.6.2. LightGBM
2.6.3. Evaluation Metrics for Model Performance
2.7. Software and Configurations
3. Results and Discussion
3.1. Statistical Information of Reference Values of TF and TCA Contents
3.2. Spectral Analysis
3.3. Analysis of the Preprocessing Methods
3.4. Regression Performance of Conventional Models
3.5. Regression Performance of LightGBM Model with Fused Features
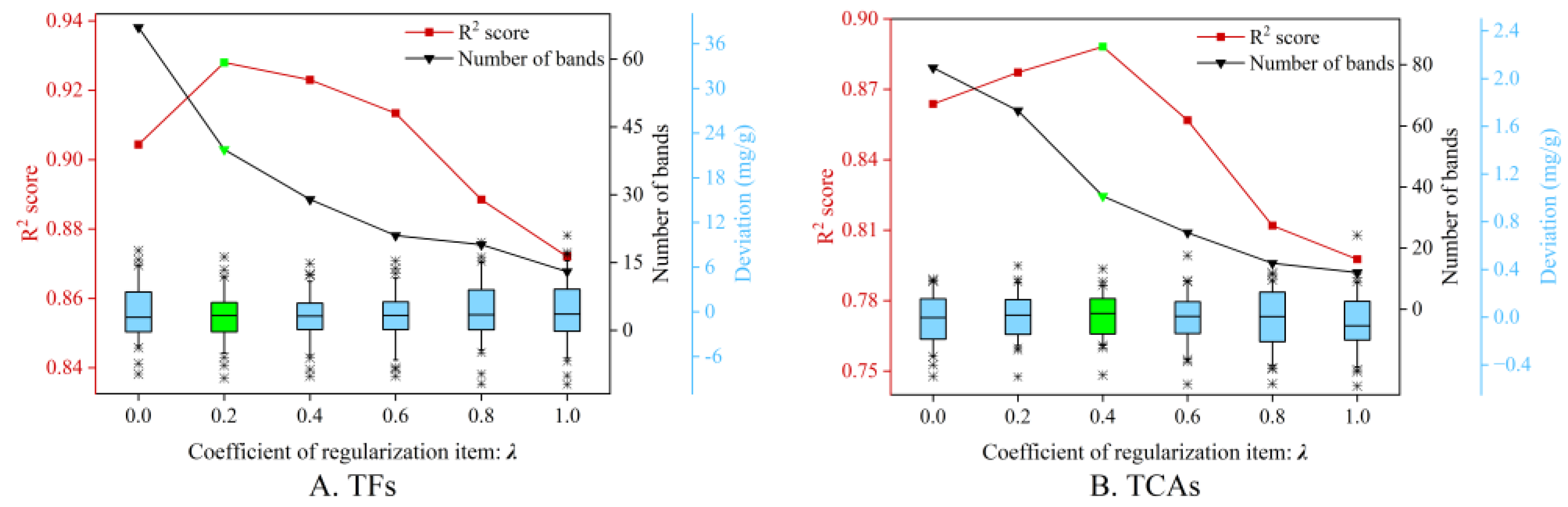
3.5.1. Analysis of the Coefficient of Regularization Item
3.5.2. Ablation Experiments
3.5.3. Model Results and Reliability Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kaczyński, P.; Iwaniuk, P.; Jankowska, M.; Orywal, K.; Socha, K.; Perkowski, M.; Farhan, J.; Łozowicka, B. Pesticide residues in common and herbal teas combined with risk assessment and transfer to the infusion. Chemosphere 2024, 367, 143550. [Google Scholar] [CrossRef]
- Ma, Y.L.; Sun, P.; Feng, J.; Yuan, J.; Wang, Y.; Shang, Y.F.; Niu, X.L.; Yang, S.H.; Wei, Z.J. Solvent effect on phenolics and antioxidant activity of Huangshan Gongju (Dendranthema morifolium (Ramat) Tzvel. cv. Gongju) extract. Food Chem. Toxicol. 2021, 147, 111875. [Google Scholar] [CrossRef] [PubMed]
- Hadizadeh, H.; Samiei, L.; Shakeri, A. Chrysanthemum, an ornamental genus with considerable medicinal value: A comprehensive review. S. Afr. J. Bot. 2022, 144, 23–43. [Google Scholar] [CrossRef]
- Peng, A.; Lin, L.; Zhao, M. Screening of key flavonoids and monoterpenoids for xanthine oxidase inhibitory activity-oriented quality control of Chrysanthemum morifolium Ramat. ‘Boju’ based on spectrum-effect relationship coupled with UPLC-TOF-MS and HS-SPME-GC/MS. Food Res. Int. 2020, 137, 109448. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.L.; Li, Z.; Wu, Z.F.; Wu, Q.L.; Guo, X.; Shang, Y.F.; Thakur, K.; Wei, Z.J. Amelioration activity of the high bioaccessible Chrysanthemum (Gongju) phenolics on alcohol-induced oxidative injury in AML-12 cells. Food Chem. 2024, 457, 140092. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Wang, E.; Gao, C.; Wen, Y.; Zhi, C.; Li, X.; Zhao, Y. Structural analysis, acetylcholinesterase inhibitory activity and immunoregulatory activity of two acidic polysaccharides from Chrysanthemum morifolium cv. Gongju. Int. J. Biol. Macromol. 2024, 279, 135073. [Google Scholar] [CrossRef]
- Li, S.; Chen, L.; Zhou, Y. Essential oil obtained from Chrysanthemum indicum var. aromaticum leaf using solvent-less microwave irradiation-induced hydrodistillation and extraction in situ. Sustain. Chem. Pharm. 2023, 36, 101259. [Google Scholar] [CrossRef]
- Yuan, H.; Jiang, S.; Liu, Y.; Daniyal, M.; Jian, Y.; Peng, C.; Shen, J.; Liu, S.; Wang, W. The flower head of Chrysanthemum morifolium Ramat. (Juhua): A paradigm of flowers serving as Chinese dietary herbal medicine. J. Ethnopharmacol. 2020, 261, 113043. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, J.; Ma, D.; Li, X.; Gao, X.; Miao, J.; Gao, W. Improving the contents of the active components and bioactivities of Chrysanthemum morifolium Ramat.: The effects of drying methods. Food Biosci. 2019, 29, 9–16. [Google Scholar] [CrossRef]
- Wang, S.; Hao, L.; Zhu, J.; Zhang, Q.; Wang, Z.; Zhang, X.; Song, X. Study on the effects of sulfur fumigation on chemical constituents and antioxidant activity of Chrysanthemum morifolium cv. hang-ju. Phytomedicine 2014, 21, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Luo, J.; Lyu, M.; Jiang, S.; Qiu, Y.; Tian, X.; Liu, L.; Liu, S.; Ouyang, Y.; Wang, W. An integrated approach to Q-marker discovery and quality assessment of edible Chrysanthemum flowers based on chromatogram–effect relationship and bioinformatics analyses. Ind. Crops Prod. 2022, 188, 115745. [Google Scholar] [CrossRef]
- Vempatapu, B.P.; Kumar, J.; Upreti, B.; Kanaujia, P.K. Application of high-performance liquid chromatography in petroleum analysis: Challenges and opportunities. TrAC Trends Anal. Chem. 2024, 117, 117810. [Google Scholar] [CrossRef]
- Cortés, V.; Blasco, J.; Aleixos, N.; Cubero, S.; Talens, P. Monitoring strategies for quality control of agricultural products using visible and near-infrared spectroscopy: A review. Trends Food Sci. Technol. 2019, 85, 138–148. [Google Scholar] [CrossRef]
- De Beer, D.; Du Preez, B.; Joubert, E. Development of HPLC method for quantification of phenolic compounds in Cyclopia intermedia (honeybush) herbal tea infusions. J. Food Compos. Anal. 2021, 104, 104154. [Google Scholar] [CrossRef]
- Yu, P.; Wang, J.; Lao, F.; Shi, H.; Xu, X.; Wu, J. Investigation on sweaty off-flavors in small mill sesame oil and its formation mechanism via molecular sensory science, preparative gas chromatography, and microbiomics. Food Chem. 2024, 463, 141224. [Google Scholar] [CrossRef] [PubMed]
- Melicherová, N.; Vaculovič, T.; Kočí, R.; Trtílek, M.; Lavická, J.; Foret, F. Determination of nutrient concentration in liquid culture of cyanobacteria Nostoc sp. by capillary electrophoresis and inductively coupled plasma mass spectrometry. Anal. Biochem. 2024, 694, 115630. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Li, X.; Cheng, W.; Zhao, G.; Tang, L.; Yang, Y.; Wu, Y.; Zhang, P.; Wang, Q. Data fusion strategy based on ultraviolet–visible spectra and near-infrared spectra for simultaneous and accurate determination of key parameters in surface water. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 302, 123007. [Google Scholar] [CrossRef] [PubMed]
- Lolli, V.; Caligiani, A. How nuclear magnetic resonance contributes to food authentication: Current trends and perspectives. Curr. Opin. Food Sci. 2024, 58, 101200. [Google Scholar] [CrossRef]
- Hao, N.; Gao, X.; Zhao, Q.; Miao, P.; Cheng, J.; Li, Z.; Liu, C.; Li, W. Rapid origin identification of Chrysanthemum morifolium using laser-induced breakdown spectroscopy and chemometrics. Postharvest Biol. Technol. 2023, 197, 112226. [Google Scholar] [CrossRef]
- Hu, H.; Mei, Y.; Wei, Y.; Xu, Z.; Zhao, Y.; Xu, H.; Mao, X.; Huang, L. Chemical composition prediction in goji (Lycium barbarum) using hyperspectral imaging and multi-task 1DCNN with attention mechanism. LWT Food Sci. Technol. 2024, 204, 116436. [Google Scholar] [CrossRef]
- Sharma, S.; Goyal, P.; Devi, J.; Atri, C.; Kumar, R.; Banga, S. Using near-infrared reflectance spectroscopy (NIRS) to predict the nitrogen levels in the stem and root tissues of Brassica juncea (Indian mustard). Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 322, 124755. [Google Scholar] [CrossRef]
- Singh, N.; Kaur, S.; Jain, A.; Kumar, A.; Bhardwaj, R.; Pandey, R.; Riar, A. Comparative analysis of deep learning and machine learning-based models for simultaneous prediction of minerals in perilla (Perilla frutescens L.) seeds using near-infrared reflectance spectroscopy. J. Food Compos. Anal. 2024, 136, 106824. [Google Scholar] [CrossRef]
- Nanda, M.A.; Amaru, K.; Rosalinda, S.; Novianty, I.; Sholihah, W.; Mindara, G.P.; Faricha, A.; Park, T. Higuchi fractal dimension and deep learning on near-infrared spectroscopy for determination of free fatty acid (FFA) content in oil palm fruit. J. Agric. Food Res. 2024, 18, 101437. [Google Scholar] [CrossRef]
- Hong, F.W.; Chia, K.S. A review on recent near infrared spectroscopic measurement setups and their challenges. Measurement 2021, 171, 108732. [Google Scholar] [CrossRef]
- Lu, X.; Wei, S.; Chen, B.; Wei, Z.; Kang, T. Rapid and nondestructive detection of multiple adulterants in kudzu starch by near infrared (NIR) spectroscopy and chemometrics. LWT Food Sci. Technol. 2015, 61, 290–595. [Google Scholar] [CrossRef]
- Diao, Z.; Guo, P.; Zhang, B.; Yan, J.; He, Z.; Zhao, S.; Zhao, C.; Zhang, J. Spatial-spectral attention-enhanced Res-3D-OctConv for corn and weed identification utilizing hyperspectral imaging and deep learning. Comput. Electron. Agric. 2023, 212, 108092. [Google Scholar] [CrossRef]
- León-Ecay, S.; Insausti, K.; Arazuri, S.; Goenaga, I.; López-Maestresalas, A. Combination of spectral and textural features of hyperspectral imaging for the authentication of the diet supplied to fattening cattle. Food Control 2024, 159, 110284. [Google Scholar] [CrossRef]
- Varela, J.I.; Miller, N.D.; Infante, V.; Kaeppler, S.M.; de Leon, N.; Spalding, E.P. A novel high-throughput hyperspectral scanner and analytical methods for predicting maize kernel composition and physical traits. Food Chem. 2022, 391, 133264. [Google Scholar] [CrossRef] [PubMed]
- Malegori, C.; Oliveri, P.; Mustorgi, E.; Boggiani, M.A.; Pastorini, G.; Casale, M. An in-depth study of cheese ripening by means of NIR hyperspectral imaging: Spatial mapping of dehydration, proteolysis and lipolysis. Food Chem. 2021, 343, 128547. [Google Scholar] [CrossRef]
- Pullanagari, R.R.; Li, M. Uncertainty assessment for firmness and total soluble solids of sweet cherries using hyperspectral imaging and multivariate statistics. J. Food Eng. 2021, 289, 110177. [Google Scholar] [CrossRef]
- Long, W.; Zhang, Q.; Wang, S.R.; Suo, Y.; Chen, H.; Bai, X.; Yang, X.; Zhou, Y.P.; Yang, J.; Fu, H. Fast and non-destructive discriminating the geographical origin of Hangbaiju by hyperspectral imaging combined with chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 284, 121786. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhang, C.; Zhou, L.; He, Y. Simultaneous determination of five micro-components in Chrysanthemum morifolium (hangbaiju) using near-infrared hyperspectral imaging coupled with deep learning with wavelength selection. Infrared Phys. Technol. 2021, 116, 103802. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Ahmed, M.T.; Monjur, O.; Kamruzzaman, M. Deep learning-based hyperspectral image reconstruction for quality assessment of agro-product. J. Food Eng. 2024, 382, 8128–8134. [Google Scholar] [CrossRef]
- Cruz-Tirado, J.; dos Santos Vieira, M.S.; Correa, O.O.V.; Delgado, D.R.; Angulo-Tisoc, J.M.; Barbin, D.F.; Siche, R. Detection of adulteration of Alpaca (Vicugna pacos) meat using a portable NIR spectrometer and NIR-hyperspectral imaging. J. Food Compos. Anal. 2024, 126, 105901. [Google Scholar] [CrossRef]
- Edris, M.; Ghasemi-Varnamkhasti, M.; Kiani, S.; Yazdanpanah, H.; Izadi, Z. Identifying the authenticity and geographical origin of rice by analyzing hyperspectral images using unsupervised clustering algorithms. J. Food Compos. Anal. 2024, 125, 105737. [Google Scholar] [CrossRef]
- Tan, K.; Wang, S.; Song, Y.; Liu, Y.; Gong, Z. Estimating nitrogen status of rice canopy using hyperspectral reflectance combined with BPSO-SVR in cold region. Chemom. Intell. Lab. Syst. 2018, 172, 68–79. [Google Scholar] [CrossRef]
- Fu, J.; Feng, L.; Wei, S.; Ma, X.; Huang, R.; Feng, S.; Dong, Q.; Yan, Z. Distinctive morphological characteristics contribute to the identification of Artemisia annua L. germplasms with high yield and high artemisinin content. J. Appl. Res. Med. Aromat. Plants 2016, 3, 43–47. [Google Scholar] [CrossRef]
- Gao, S.; Xu, J. Hyperspectral image information fusion-based detection of soluble solids content in red globe grapes. Comput. Electron. Agric. 2022, 196, 106822. [Google Scholar] [CrossRef]
- Zhang, Y.; Zareef, M.; Rong, Y.; Lin, H.; Chen, Q.; Ouyang, Q. Application of colorimetric sensor array coupled with chemometric methods for monitoring the freshness of snakehead fillets. Food Chem. 2024, 439, 138172. [Google Scholar] [CrossRef] [PubMed]
- Nyasulu, C.; Diattara, A.; Traore, A.; Ba, C.; Diedhiou, P.M.; Sy, Y.; Raki, H.; Peluffo-Ordóñez, D.H. A comparative study of Machine Learning-based classification of Tomato fungal diseases: Application of GLCM texture features. Heliyon 2023, 9, e21697. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.; Rao, P.S. Predictive modeling of allowable storage time of finger millet grains using artificial neural network and support vector regression approaches. J. Food Eng. 2024, 383, 112224. [Google Scholar] [CrossRef]
- Rayyad, A.; Elderderi, S.; Massot, V.; Chourpa, I. Comparison of SVMR and PLSR for ATR-IR data treatment: Application to AQC of mAbs in clinical solutions. Vib. Spectrosc. 2023, 129, 103594. [Google Scholar] [CrossRef]
- Wen, Y.; Li, Z.; Ning, Y.; Yan, Y.; Li, Z.; Wang, N.; Wang, H. Portable Raman spectroscopy coupled with PLSR analysis for monitoring and predicting of the quality of fresh-cut Chinese yam at different storage temperatures. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 310, 123956. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Wang, T.; Zheng, Y.; Zhang, W.; Yun, Y.H. Deep learning in food authenticity: Recent advances and future trends. Trends Food Sci. Technol. 2024, 144, 104344. [Google Scholar] [CrossRef]
- Guo, X.; Gui, X.; Xiong, H.; Hu, X.; Li, Y.; Cui, H.; Qiu, Y.; Ma, C. Critical role of climate factors for groundwater potential mapping in arid regions: Insights from random forest, XGBoost, and LightGBM algorithms. J. Hydrol. 2023, 621, 129599. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Zhu, S.; Li, Y.; He, Y.; Liu, F. Shape induced reflectance correction for non-destructive determination and visualization of soluble solids content in winter jujubes using hyperspectral imaging in two different spectral ranges. Postharvest Biol. Technol. 2020, 161, 111080. [Google Scholar] [CrossRef]
- Berardo, N.; Pisacane, V.; Battilani, P.; Scandolara, A.; Pietri, A.; Marocco, A. Rapid detection of kernel rots and mycotoxins in maize by near-infrared reflectance spectroscopy. J. Agric. Food Chem. 2005, 53, 8128–8134. [Google Scholar] [CrossRef]
- Hu, H.; Mei, Y.; Zhou, Y.; Zhao, Y.; Fu, L.; Xu, H.; Mao, X.; Huang, L. Optimizing starch content prediction in kudzu: Integrating hyperspectral imaging and deep learning with WGAN-GP. Food Control 2024, 166, 110762. [Google Scholar] [CrossRef]
- Yin, Y.; Li, J.; Ling, C.; Zhang, S.; Liu, C.; Sun, X.; Wu, J. Fusing spectral and image information for characterization of black tea grade based on hyperspectral technology. LWT Food Sci. Technol. 2023, 185, 115150. [Google Scholar] [CrossRef]
- Wang, W.; Lawrence, K.C.; Ni, X.; Yoon, S.C.; Heitschmidt, G.W.; Feldner, P. Near-infrared hyperspectral imaging for detecting Aflatoxin B1 of maize kernels. Food Control 2015, 51, 347–355. [Google Scholar] [CrossRef]
- Li, Y.; Ma, B.; Li, C.; Yu, G. Accurate prediction of soluble solid content in dried Hami jujube using SWIR hyperspectral imaging with comparative analysis of models. Comput. Electron. Agric. 2022, 193, 106655. [Google Scholar] [CrossRef]
- Luo, W.; Zhang, J.; Liu, S.; Huang, H.; Zhan, B.; Fan, G.; Zhang, H. Prediction of soluble solid content in Nanfeng mandarin by combining hyperspectral imaging and effective wavelength selection. J. Food Compos. Anal. 2024, 126, 105939. [Google Scholar] [CrossRef]
- Vallese, F.; Paoloni, S.; Springer, V.; de Sousa Fernandes, D.; Diniz, P.; Pistonesi, M. Exploiting the successive projections algorithm to improve the quantification of chemical constituents and discrimination of botanical origin of Argentinean bee-pollen. J. Food Compos. Anal. 2024, 126, 105925. [Google Scholar] [CrossRef]
- Hassan, M.; Jiao, T.; Ahmad, W.; Yi, X.; Zareef, M.; Ali, S.; Li, H.; Chen, Q. Cellulose paper-based SERS sensor for sensitive detection of 2,4-D residue levels in tea coupled uninformative variable elimination-partial least squares. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119198. [Google Scholar] [CrossRef]
- Qin, Y.; Song, K.; Zhang, N.; Wang, M.; Zhang, M.; Peng, B. Robust NIR quantitative model using MIC-SPA variable selection and GA-ELM. Infrared Phys. Technol. 2023, 128, 104534. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.; Jiang, Q.; Cui, M.; Li, N.; Ou, Y.; Diao, Z.; Zhang, B. Early identification of strawberry leaves disease utilizing hyperspectral imaging combing with spectral features, multiple vegetation indices and textural features. Comput. Electron. Agric. 2023, 204, 107553. [Google Scholar] [CrossRef]





| Parameters | Search Space | Optimal Parameters | |
|---|---|---|---|
| TFs | TCAs | ||
| num_boost_round | [1, 3000] | 412 | 121 |
| max_depth | [−1, 10] | 3 | 3 |
| learning_rate | [0.01, 0.3] | 0.08 | 0.19 |
| lambda_l1 | [0, 1] | 0.39 | 0.47 |
| lambda_l2 | [0, 1] | 0.07 | 0 |
| metric | mse | ||
| boosting_type | gbdt | ||
| early_stopping_rounds | 100 | ||
| Components | Data Sets | Samples | Content Indicators (mg/g) | |||
|---|---|---|---|---|---|---|
| Max | Min | Mean | Std | |||
| TFs | Calibration | 140 | 146.20 | 98.24 | 125.71 | 10.39 |
| Prediction | 60 | 145.45 | 98.98 | 127.92 | 11.37 | |
| Total | 200 | 146.20 | 98.24 | 126.77 | 9.97 | |
| TCAs | Calibration | 140 | 21.10 | 18.45 | 19.56 | 0.52 |
| Prediction | 60 | 20.75 | 18.58 | 19.62 | 0.53 | |
| Total | 200 | 21.10 | 18.45 | 19.57 | 0.52 | |
| Bioactive Components | Preprocessing | Evaluation Metrics | |||
|---|---|---|---|---|---|
| R2 | RMSE | MAE | RPD | ||
| TFs | Raw | 0.8454 | 4.4339 | 3.6182 | 2.5651 |
| MSC | 0.8721 | 4.0340 | 3.2663 | 2.8194 | |
| SGCS | 0.8560 | 4.2797 | 3.3018 | 2.6576 | |
| MSC + SGCS | 0.8885 | 3.7658 | 3.0546 | 3.0202 | |
| TCAs | Raw | 0.7977 | 0.2377 | 0.1952 | 2.2419 |
| MSC | 0.8569 | 0.1999 | 0.1610 | 2.6655 | |
| SGCS | 0.8239 | 0.2218 | 0.1830 | 2.4031 | |
| MSC + SGCS | 0.8674 | 0.1924 | 0.1576 | 2.7695 | |
| Bioactive Components | Models | Evaluation Metrics | |||
|---|---|---|---|---|---|
| R2 | RMSE | MAE | RPD | ||
| TFs | SVR | 0.8496 | 4.3734 | 3.3320 | 2.6006 |
| SPA(29)-SVR | 0.8137 | 4.8679 | 3.7432 | 2.3364 | |
| UVE(96)-SVR | 0.8398 | 4.5136 | 3.5128 | 2.5198 | |
| PLSR | 0.8885 | 3.7658 | 3.0546 | 3.0202 | |
| SPA(29)-PLSR | 0.8492 | 4.3793 | 3.5278 | 2.5971 | |
| UVE(96)-PLSR | 0.8567 | 4.2694 | 3.4015 | 2.6640 | |
| 1DCNN | 0.9233 | 3.1235 | 2.7053 | 3.6413 | |
| TCAs | SVR | 0.8329 | 0.2160 | 0.1783 | 2.4670 |
| SPA(38)-SVR | 0.8071 | 0.2321 | 0.1929 | 2.2959 | |
| UVE(89)-SVR | 0.8152 | 0.2272 | 0.1843 | 2.3456 | |
| PLSR | 0.8674 | 0.1924 | 0.1576 | 2.7695 | |
| SPA(35)-PLSR | 0.8382 | 0.2126 | 0.1790 | 2.5068 | |
| UVE(89)-PLSR | 0.8581 | 0.1991 | 0.1696 | 2.6771 | |
| 1DCNN | 0.8956 | 0.1708 | 0.1464 | 3.1209 | |
| Case | Modules 1 | Total Flavonoids | Total Chlorogenic Acids | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pre | PSO | VS | Tex | Col | R2 | RMSE | MAE | RPD | R2 | RMSE | MAE | RPD | |
| 1 | × | × | × | × | × | 0.8721 | 4.0340 | 3.2663 | 2.8194 | 0.8260 | 0.2204 | 0.1866 | 2.4177 |
| 2 | √ | × | × | × | × | 0.9376 | 2.8173 | 2.3618 | 4.0370 | 0.9090 | 0.1594 | 0.1339 | 3.3435 |
| 3 | √ | √ | × | × | × | 0.9044 | 3.4879 | 2.8294 | 3.2608 | 0.8638 | 0.1950 | 0.1637 | 2.7328 |
| 4 | √ | √ | √ | × | × | 0.9280 | 3.0263 | 2.6422 | 3.7582 | 0.8882 | 0.1767 | 0.1516 | 3.0161 |
| 5 | × | × | × | √ | √ | 0.7276 | 5.8860 | 4.7830 | 1.9323 | 0.6938 | 0.2925 | 0.2317 | 1.8223 |
| 6 | √ | √ | √ | √ | × | 0.9433 | 2.6891 | 2.2955 | 4.2369 | 0.9105 | 0.1581 | 0.1329 | 3.3710 |
| 7 | √ | √ | √ | × | √ | 0.9315 | 2.9526 | 2.5104 | 3.8521 | 0.8948 | 0.1714 | 0.1418 | 3.1093 |
| 8 | √ | √ | √ | √ | √ | 0.9541 | 2.4150 | 2.0353 | 4.7095 | 0.9137 | 0.1553 | 0.1319 | 3.4326 |
| Bioactive Components | Models | Evaluation Metrics | |||
|---|---|---|---|---|---|
| R2 | RMSE | MAE | RPD | ||
| TFs | SVR | 0.8774 | 3.9490 | 3.1268 | 2.8801 |
| PLSR | 0.9026 | 3.5198 | 2.8071 | 3.2312 | |
| 1DCNN | 0.9359 | 2.8855 | 2.4218 | 3.9831 | |
| LightGBM | 0.9541 | 2.4150 | 2.0353 | 4.7095 | |
| TCAs | SVR | 0.8555 | 0.2009 | 0.1685 | 2.6529 |
| PLSR | 0.8740 | 0.1876 | 0.1560 | 2.8409 | |
| 1DCNN | 0.9043 | 0.1634 | 0.1353 | 3.2621 | |
| LightGBM | 0.9137 | 0.1553 | 0.1319 | 3.4326 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, Y.; Hu, H.; Yuan, M.; Xu, H.; Mao, X.; Zhao, Y.; Huang, L. Determination of Bioactive Components in Chrysanthemum Tea (Gongju) Using Hyperspectral Imaging Technique and Chemometrics. Foods 2024, 13, 4145. https://doi.org/10.3390/foods13244145
Wei Y, Hu H, Yuan M, Xu H, Mao X, Zhao Y, Huang L. Determination of Bioactive Components in Chrysanthemum Tea (Gongju) Using Hyperspectral Imaging Technique and Chemometrics. Foods. 2024; 13(24):4145. https://doi.org/10.3390/foods13244145
Chicago/Turabian StyleWei, Yunpeng, Huiqiang Hu, Minghua Yuan, Huaxing Xu, Xiaobo Mao, Yuping Zhao, and Luqi Huang. 2024. "Determination of Bioactive Components in Chrysanthemum Tea (Gongju) Using Hyperspectral Imaging Technique and Chemometrics" Foods 13, no. 24: 4145. https://doi.org/10.3390/foods13244145
APA StyleWei, Y., Hu, H., Yuan, M., Xu, H., Mao, X., Zhao, Y., & Huang, L. (2024). Determination of Bioactive Components in Chrysanthemum Tea (Gongju) Using Hyperspectral Imaging Technique and Chemometrics. Foods, 13(24), 4145. https://doi.org/10.3390/foods13244145






