The Use of Explainable Machine Learning for the Prediction of the Quality of Bulk-Tank Milk in Sheep and Goat Farms
Abstract
1. Introduction
2. Materials and Methods
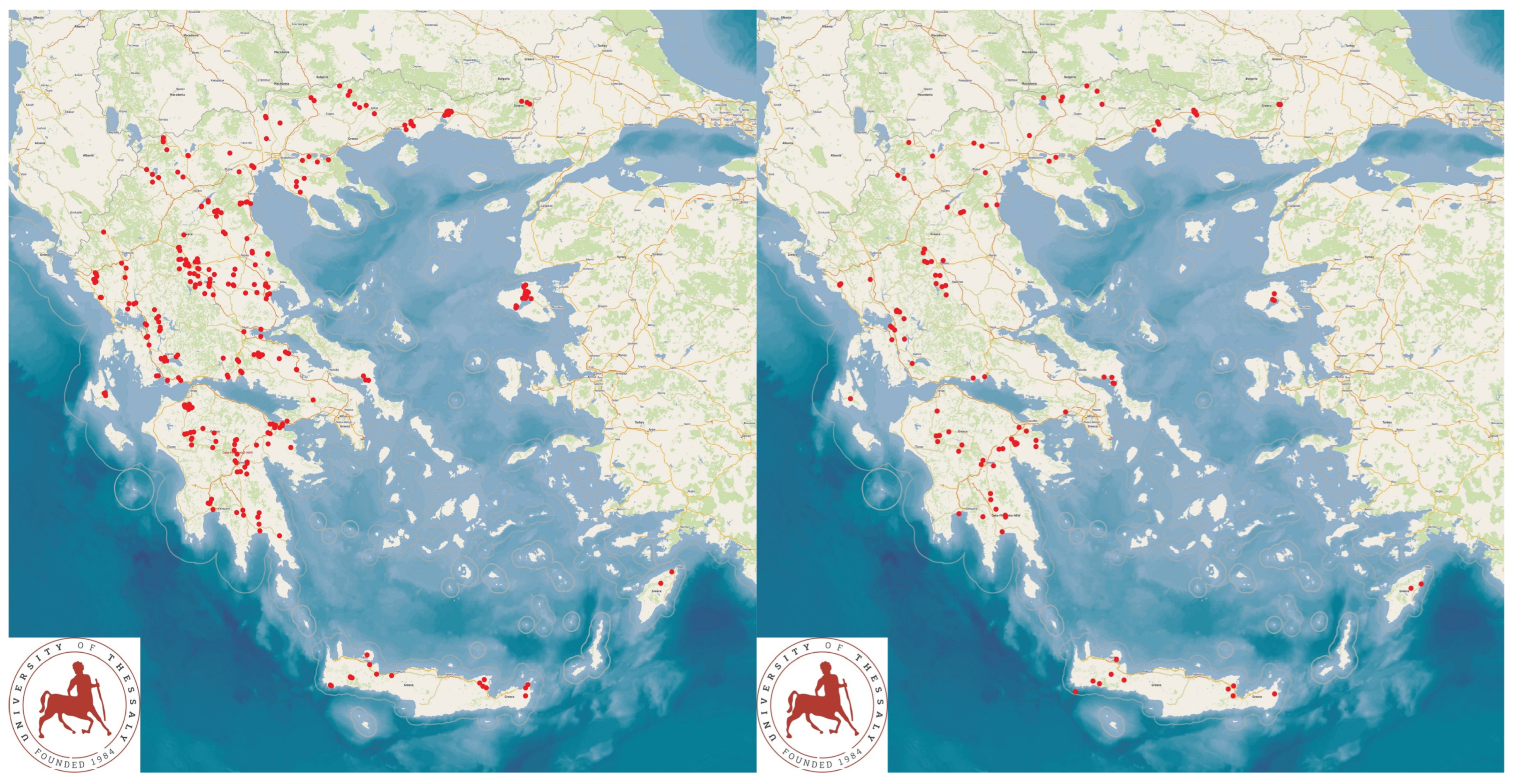
2.1. Collection of Field and Laboratory Data
2.2. Dataset Used for the Construction of the Computational Models
2.3. Implementation of Machine Learning Algorithms
2.4. Evaluations for Construction of Computational Model by Means of Supervised Learning
2.5. Combinations of Hyperparameters Employed in Each Machine Learning Tool
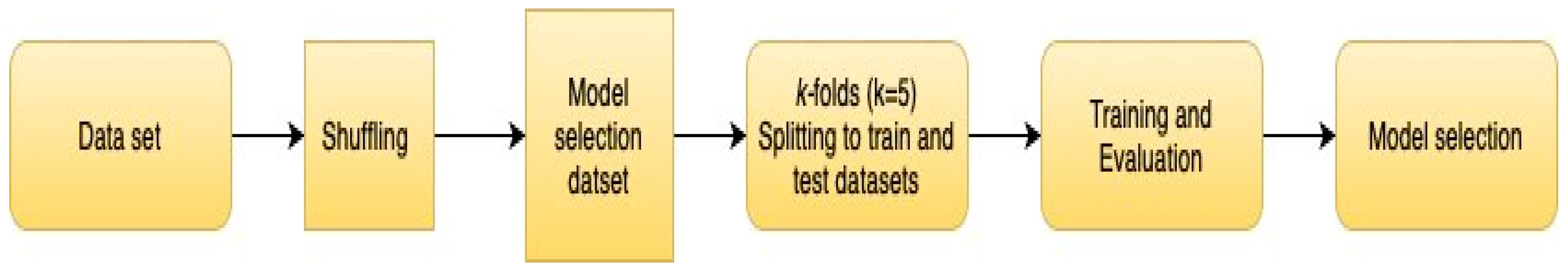
2.6. Data Management and Analysis
2.7. Analysis of the Importance of the Independent Variables in Predicting the Target Values in the Bulk-Tank Milk—Interpretation of Findings
3. Results
3.1. Selection of Best Computational Model
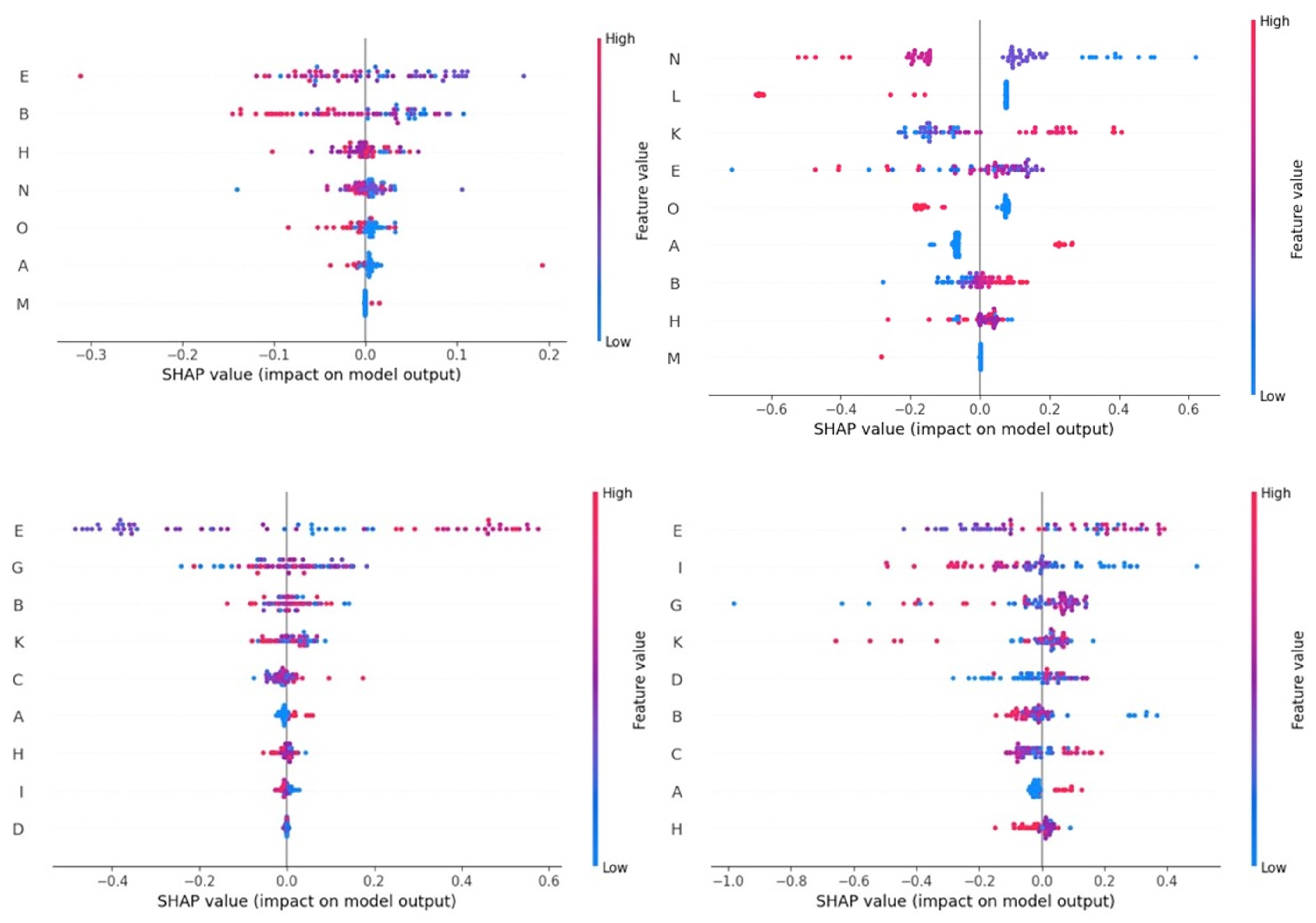
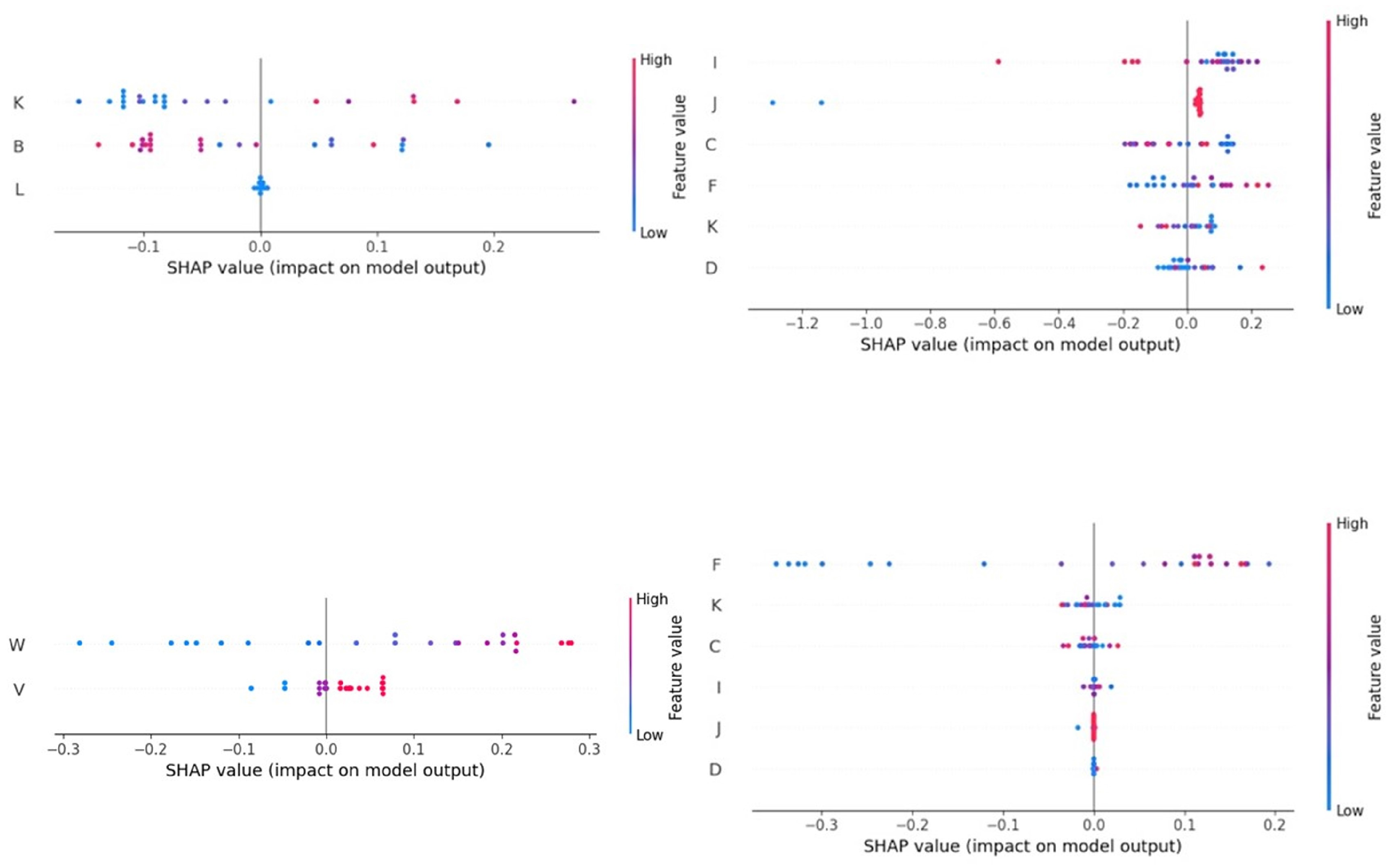
3.2. Results of Analysis of SHAP Values
4. Discussion
4.1. Preamble
4.2. Development of the Models Used
4.2.1. Independent Variables Used in the Development of the Models
4.2.2. Selection of Machine Learning Tools
- Decision trees provide interpretable models and are foundational for ensemble tools like random forests and XGBoost.
- Random forests add robustness in the data analysis by reducing overfitting through ensembling, making the tool versatile for both classification and regression supervised learning.
- XGBoost is a state-of-the-art gradient boosting algorithm that excels in accuracy and efficiency, particularly in structured data.
- k-nearest neighbours refer to a non-parametric and simple tool that leverages local information for effective predictions in spatially related data.
- Neural networks are tools capable of modelling highly complex and non-linear relationships, which extend problem-solving capability to multi-dimensional datasets.
4.2.3. Cross-Validation
4.2.4. Selection of Combinations of Hyperparameters
4.2.5. Selection of Mean Absolute Percentage Error as the Performance Metric
4.2.6. Predictions of Target Values
4.2.7. Possible Limitations
4.3. Importance of the Independent Variables in the Predictions
4.4. Application of the Findings in Small Ruminant Farms—Feasibility and Cost Effectiveness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barbano, D.M.; Lynch, J.M. Major advances in testing of dairy products: Milk component and dairy product attribute testing. J. Dairy Sci. 2006, 89, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.C.; Martin, N.H.; Barbano, D.M.; Wiedmann, M. Influence of raw milk quality on processed dairy products: How do raw milk quality test results relate to product quality and yield? J. Dairy Sci. 2016, 99, 10128–10149. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.T.; Gambrel-Lenarz, S.A.; Scher, F.M.; Graham, T.E.; Reddy, R. Microbiological Count Methods. In Standard Methods for the Examination of Dairy Products, 17th ed.; Wehr, H.M., Frank, J.F., Eds.; APHA Press: Washington, DC, USA, 2004; pp. 153–186. [Google Scholar]
- Park, Y.W.; Juárez, M.; Ramos, M.; Haenlein, G.F.W. Physico-chemical characteristics of goat and sheep milk. Small Rumin. Res. 2007, 68, 88–113. [Google Scholar] [CrossRef]
- Nudda, A.; Atzori, A.S.; Correddu, F.; Battacone, G.; Lunesu, M.F.; Cannas, A.; Pulina, G. Effects of nutrition on main components of sheep milk. Small Rumin. Res. 2020, 184, 106015. [Google Scholar] [CrossRef]
- Ponnampalam, E.N.; Priyashantha, H.; Vidanarachchi, J.K.; Kiani, A.; Holman, B.W.B. Effects of nutritional factors on fat content, fatty acid composition, and sensorial properties of meat and milk from domesticated ruminants: An overview. Animals 2024, 14, 840. [Google Scholar] [CrossRef]
- Haenlein, G.F.W. About the evolution of goat and sheep milk production. Small Rumin. Res. 2007, 68, 3–6. [Google Scholar] [CrossRef]
- Nguyen, V.Q. Nutritional value and factors affecting milk production and milk composition from dairy sheep: A review. CTU J. Innov. Sustain. Dev. 2022, 14, 53–64. [Google Scholar] [CrossRef]
- Gonzalo, C. Milk hygiene in small ruminants: A review. Span. J. Agric. Res. 2017, 15, e05R02. [Google Scholar] [CrossRef]
- Kaskous, S.; Farschtschi, S.; Pfaffl, M.W. Physiological aspects of milk somatic cell count in small ruminants—A review. Dairy 2022, 4, 26–42. [Google Scholar] [CrossRef]
- Fthenakis, G.C. Correspondence of Somatic Cell Counts in Bulk-Tank Milk to Prevalence of Subclinical Mastitis in Sheep Flocks. Animals 2023, 13, 3541. [Google Scholar] [CrossRef]
- Albenzio, M.; Figliola, L.; Caroprese, M.; Marino, R.; Sevi, A.; Santillo, A. Somatic cell count in sheep milk. Small Rumin. Res. 2019, 176, 24–30. [Google Scholar] [CrossRef]
- Gonzalo, C.; Carriedo, J.A.; Beneitez, E.; Juárez, M.T.; De La Fuente, L.F.; Primitivo, F.S. Bulk tank total bacterial count in dairy sheep: Factors of variation and relationship with somatic cell count. J. Dairy Sci. 2006, 89, 549–552. [Google Scholar] [CrossRef] [PubMed]
- Quintana, Á.R.; Seseña, S.; Garzón, A.; Arias, R. Factors affecting levels of airborne bacteria in dairy farms: A review. Animals 2020, 10, 526. [Google Scholar] [CrossRef]
- Piras, C.; Hale, O.J.; Reynolds, C.K.; Jones, A.K.; Taylor, N.; Morris, M.; Cramer, R. Speciation and milk adulteration analysis by rapid ambient liquid MALDI mass spectrometry profiling using machine learning. Sci. Rep. 2021, 11, 3305. [Google Scholar] [CrossRef]
- Chu, C.; Wang, H.; Luo, X.; Wen, P.; Nan, L.; Du, C.; Fan, Y.; Gao, D.; Wang, D.; Yang, Z.; et al. Possible alternatives: Identifying and quantifying adulteration in buffalo, goat, and camel milk using mid-infrared spectroscopy combined with modern statistical machine learning methods. Foods 2023, 12, 3856. [Google Scholar] [CrossRef]
- Pérez-Calabuig, A.M.; Pradana-López, S.; Lopez-Ortega, S.; Sáenz, K.d.J.B.; Cancilla, J.C.; Torrecilla, J.S. Application of residual neural networks to detect and quantify milk adulterations. J. Food Comp. Anal. 2023, 122, 105427. [Google Scholar] [CrossRef]
- Xue, D.; Zhao, H. Machine learning and electrochemistry techniques for detecting adulteration of goat milk with cow milk. J. Food Meas. Charact. 2024, 18, 6012–6019. [Google Scholar] [CrossRef]
- Kiouvrekis, Y.; Vasileiou, N.G.C.; Katsarou, E.I.; Lianou, D.T.; Michael, C.K.; Zikas, S.; Katsafadou, A.I.; Bourganou, M.V.; Liagka, D.V.; Chatzopoulos, D.C.; et al. The use of machine learning to predict prevalence of subclinical mastitis in dairy sheep farms. Animals 2024, 14, 2295. [Google Scholar] [CrossRef]
- Leitner, G.; Rovai, M.; Merin, U. Clinical and subclinical intramammary infection caused by coagulase negative staphylococci negatively affect milk yield and its quality in dairy sheep. Small Rumin. Res. 2019, 180, 74–78. [Google Scholar] [CrossRef]
- Novac, C.S.; Andrei, S. The impact of mastitis on the biochemical parameters, oxidative and nitrosative stress markers in goat’s milk: A review. Pathogens 2020, 9, 882. [Google Scholar] [CrossRef]
- Gonzalo, C.; Juárez, M.T.; García-Jimeno, M.C.; De La Fuente, L.F. Bulk tank somatic cell count and total bacterial count are affected by target practices and milking machine features in dairy sheep flocks in Castilla y Leon region, Spain. Small Rumin. Res. 2019, 178, 22–29. [Google Scholar] [CrossRef]
- van den Brom, R.; de Jong, A.; van Engelen, E.; Heuvelink, A.; Vellema, P. Zoonotic risks of pathogens from sheep and their milk borne transmission. Small Rumin. Res. 2020, 189, 106123. [Google Scholar] [CrossRef] [PubMed]
- Lianou, D.T.; Chatziprodromidou, I.P.; Vasileiou, N.G.C.; Michael, C.K.; Mavrogianni, V.S.; Politis, A.P.; Kordalis, N.G.; Billinis, C.; Giannakopoulos, A.; Papadopoulos, E.; et al. A detailed questionnaire for the evaluation of health management in dairy sheep and goats. Animals 2020, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Lianou, D.T.; Michael, C.K.; Vasileiou, N.G.C.; Petinaki, E.; Cripps, P.J.; Tsilipounidaki, K.; Katsafadou, A.I.; Politis, A.P.; Kordalis, N.G.; Ioannidi, K.S.; et al. Extensive countrywide field investigation of somatic cell counts and total bacterial counts in bulk-tank raw milk in sheep flocks in Greece. Foods 2021, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Lianou, D.T.; Michael, C.K.; Vasileiou, N.G.C.; Petinaki, E.; Cripps, P.J.; Tsilipounidaki, K.; Katsafadou, A.I.; Politis, A.P.; Kordalis, N.G.; Ioannidi, K.S.; et al. Extensive countrywide field investigation of somatic cell counts and total bacterial counts in bulk-tank raw milk in goat herds in Greece. J. Dairy Res. 2021, 88, 307–313. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Wiggans, G.R.; Shook, G.E. A lactation measure of somatic cell count. J. Dairy Sci. 1987, 70 (Suppl. S13), 2666–2672. [Google Scholar] [CrossRef]
- Franzoi, M.; Manuelian, C.L.; Penasa, M.; De Marchi, M. Effects of somatic cell score on milk yield and mid-infrared predicted composition and technological traits of Brown Swiss, Holstein Friesian, and Simmental cattle breeds. J. Dairy Sci. 2020, 103, 791–804. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31th International Conference on Neural Information Processing Systems (NIPS’17), Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Berthelot, X.; Lagriffoul, G.; Concordet, D.; Barillet, F.; Bergonier, D. Physiological and pathological thresholds of somatic cell counts in ewe milk. Small Rumin. Res. 2006, 62, 27–31. [Google Scholar] [CrossRef]
- Leitner, G.; Silanikove, N.; Merin, U. Estimate of milk and curd yield loss of sheep and goats with intrammamary infection and its relation to somatic cell count. Small Rumin. Res. 2008, 74, 221–225. [Google Scholar] [CrossRef]
- Skeie, S. Characteristics in milk influencing the cheese yield and cheese quality. J. Anim. Feed Sci. 2007, 16, 130–142. [Google Scholar] [CrossRef]
- Fenelon, M.A.; Guinee, T.P. The effect of milk fat on Cheddar cheese yield and its prediction, using modifications of the Van Slyke cheese yield formula. J. Dairy Sci. 1999, 82, 2287–2299. [Google Scholar] [CrossRef]
- Pazzola, M. Coagulation traits of sheep and goat milk. Animals 2019, 9, 540. [Google Scholar] [CrossRef] [PubMed]
- Chilliard, Y.; Ferlay, A.; Rouel, J.; Lamberet, G. A review of nutritional and physiological factors affecting goat milk lipid synthesis and lipolysis. J. Dairy Sci. 2003, 86, 1751–1770. [Google Scholar] [CrossRef]
- Roche, J.R.; Friggens, N.C.; Kay, J.K.; Fisher, M.W.; Stafford, K.J.; Berry, D.P. Body condition score and its association with dairy cow productivity, health, and welfare. J. Dairy Sci. 2009, 92, 5769–5801. [Google Scholar] [CrossRef]
- Kenyon, P.R.; Maloney, S.K.; Blache, D. Review of sheep body condition score in relation to production characteristics. N. Z. J. Agric. Res. 2014, 57, 38–64. [Google Scholar] [CrossRef]
- Matar, A.M.; Aljummah, R.S. Postpartum body condition score (BCS) and lactation stage (30 and 60 days) affecting essential fatty acids (efa) and milk quality of Najdi sheep. Vet. Sci. 2023, 10, 552. [Google Scholar] [CrossRef]
- Signorelli, F.; Contarini, G.; Annicchiarico, G.; Napolitano, F.; Orru, L.; Catillo, G.; Haenlein, G.F.W.; Moioli, B. Breed differences in sheep milk fatty acid profiles: Opportunities for sustainable use of animal genetic resources. Small Rumin. Res. 2008, 78, 24–31. [Google Scholar] [CrossRef]
- Talpur, F.N.; Bhanger, M.I.; Memon, N.N. Milk fatty acid composition of indigenous goat and ewe breeds from Sindh, Pakistan. J. Food Compos. Anal. 2009, 22, 59–64. [Google Scholar] [CrossRef]
- Sinanoglou, V.J.; Koutsouli, P.; Fotakis, C.; Sotiropoulou, G.; Cavouras, D.; Bizelis, I. Assessment of lactation stage and breed effect on sheep milk fatty acid profile and lipid quality indices. Dairy Sci. Technol. 2015, 95, 509–531. [Google Scholar] [CrossRef]
- Lianou, D.T.; Michael, C.K.; Vasileiou, N.G.C.; Liagka, D.V.; Mavrogianni, V.S.; Caroprese, M.; Fthenakis, G.C. Association of breed of sheep or goats with somatic cell counts and total bacterial counts of bulk-tank milk. Appl. Sci. 2021, 11, 7356. [Google Scholar] [CrossRef]
- Puggioni, G.M.G.; Tedde, V.; Uzzau, S.; Dore, S.; Liciardi, M.; Cannas, E.A.; Pollera, C.; Moroni, P.; Bronzo, V.; Addis, M.F. Relationship of late lactation milk somatic cell count and cathelicidin with intramammary infection in small ruminants. Pathogens 2020, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Sakul, H.; Boylan, W.J. Evaluation of U.S. sheep breeds for milk production and milk composition. Small Rumin. Res. 1992, 7, 195–201. [Google Scholar] [CrossRef]
- Bencini, R.; Pulina, B. The quality of sheep milk: A review. Aus. J. Exp. Agric. 1997, 37, 485–504. [Google Scholar] [CrossRef]
- Amills, M.; Jordana, J.; Zidi, A.; Serradilla, J.M. Genetic factors that regulate milk protein and lipid composition in goats. In Milk Production. Advanced Genetic Traits, Cellular Mechanism, Animal Management and Health; Chaiyabutr, N., Ed.; Intech: Zagreb, Croatia, 2012; pp. 3–30. [Google Scholar]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; p. 598. [Google Scholar]
- Ranganathan, S.; Gribskov, M.; Nakai, K.; Schönbach, C. Encyclopedia of Bioinformatics and Computational Biology; Elsevier: Amsterdam, The Netherlands, 2019; Volume 3, p. 3280. [Google Scholar]
- Frizzarin, M.; Gormley, C.; Berry, D.P.; Murphy, T.B.; Casa, A.; Lynch, A.; McParland, S. Predicting cow milk quality traits from routinely available milk spectra using statistical machine learning methods. J. Dairy Sci. 2021, 104, 7438–7447. [Google Scholar] [CrossRef]
- Calin, O. Deep Learning Architectures: A Mathematical Approach; Springer: Cham, Switzeraland, 2020; 760p. [Google Scholar]
- Xu, Y.; Goodacre, R. On splitting training and validation set: A comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J. Anal. Test. 2018, 2, 249–262. [Google Scholar] [CrossRef]
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014; 449p. [Google Scholar]
- Schröder, T.; Schulz, M. Monitoring machine learning models: A categorization of challenges and methods. Data Sci. Manag. 2022, 5, 105–116. [Google Scholar] [CrossRef]
- Bramer, M. Principles of Data Mining, 4th ed.; Springer: London, UK, 2020; 571p. [Google Scholar]
- Bowerman, B.L.; O’Connell, R.T.; Koehler, A.B. Forecasting, Time Series and Regression: An Applied Approach, 4th ed.; Thomson Brooks/Cole: Belmont, CA, USA, 2004; 704p. [Google Scholar]
- Murphy, K.P. Probabilistic Machine Learning: An Introduction; MIT Press: Cambridge, MA, USA, 2022; 830p. [Google Scholar]
- Jenkins, T.C.; McGuire, M.A. Major advances in nutrition: Impact on milk composition. J. Dairy Sci. 2006, 89, 1302–1310. [Google Scholar] [CrossRef]
- Rajini, A.; Sravani, T. Machine learning approaches for dairy(milk) quality assurance. In Proceedings of the 5th International Conference on Data Science, Machine Learning and Applications, Hyderabad, India, 15–16 December 2023; Volume 1, pp. 896–902. [Google Scholar]
- Samad, A.; Taze, S.; Uçar, M.K. Enhancing milk quality detection with machine learning: A comparative analysis of knn and distance-weighted knn algorithms. Int. J. Innov. Sci. Res. Technol. 2024, 9, 2021–2029. [Google Scholar] [CrossRef]
- DePeters, E.J.; Cant, J.P. Nutritional factors influencing the nitrogen composition of bovine milk: A review. J. Dairy Sci. 1992, 75, 2043–2070. [Google Scholar] [CrossRef]
- Beardshall, M. Understanding Explainable AI (XAI): Enhancing Transparency with SHAP and LIME. Available online: https://www.linkedin.com/pulse/understanding-explainable-ai-xai-enhancing-shap-lime-mike-beardshall-tmjyf/ (accessed on 3 December 2024).
- Abiri, R.; Rizan, N.; Balasundram, S.K.; Shahbazi, A.B.; Abdul-Hamid, H. Application of digital technologies for ensuring agricultural productivity. Heliyon 2023, 9, e22601. [Google Scholar] [CrossRef] [PubMed]
- Khanna, A.; Kaur, S. Evolution of Internet of Things (IoT) and its significant impact in the field of Precision Agriculture. Comp. Electron. Agric. 2019, 157, 218–231. [Google Scholar] [CrossRef]
- Leveau, L.; Bénel, A.; Cahier, J.P.; Pinet, F.; Salembier, P.; Soulignac, V.; Bergez, J.E. Information and communication technology (ICT) and the agroecological transition. In Agroecological Transitions: From Theory to Practice in Local Participatory Design; Bergez, J.E., Audouin, E., Therond, O., Eds.; Springer: Cham, Switzerland, 2019; pp. 263–287. [Google Scholar]
- Schneider, I. Democratic governance of digital platforms and artificial intelligence? Exploring governance models of China, the US, the EU and Mexico. JeDEM-Ej. Edemocr. Open Gov. 2020, 12, 1–24. [Google Scholar] [CrossRef]
| Target Values in Bulk-Tank Milk in Sheep Farms | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Independent Variables | Fat Content (%) | Protein Content (%) | Fat and Protein Content (%) | Somatic Cell Counts (cells mL−1) | Total Bacterial Counts (c.f.u. mL−1) | |||||
| Type of Farm | ||||||||||
| Sheep | Goats | Sheep | Goats | Sheep | Goats | Sheep | Goats | Sheep | Goats | |
| Month into lactation period at sampling (numerical) | • 1 | • | • | • | • | • | • | |||
| Month of start of milking period (numerical) | • | • | ||||||||
| Management system applied in farm (categorical) | • | • | ||||||||
| Presence of milking parlour (categorical) | • | • | • | • | • | |||||
| Type of milking parlour (categorical) | • | |||||||||
| Availability of ventilators in main barn (categorical) | • | |||||||||
| No. of animals in farm (numerical) | • | |||||||||
| Animal breed (categorical) | • | • | • | • | • | • | ||||
| Grazing of animals (categorical) | • | • | • | • | ||||||
| Number of daily milkingsessions (categorical) | • | |||||||||
| Cleaning of parlour with water after milking (categorical) | • | |||||||||
| Provision of concentrates to animals (categorical) | • | • | • | • | • | |||||
| Age of newborns taken away from dam (numerical) | • | • | • | • | • | • | ||||
| Administration of anthelmintics at last stage of pregnancy (categorical) | • | • | ||||||||
| Annual milk production per animal (numerical) | • | |||||||||
| Annual incidence rate of clinical mastitis (numerical) | • | • | ||||||||
| Average number of newborns per dam (numerical) | • | |||||||||
| Body condition score of female animals (numerical) | • | • | ||||||||
| Age of farmer (numerical) | • | |||||||||
| Length of experience of farmer (numerical) | • | |||||||||
| Education of farmer (categorical) | • | • | • | • | ||||||
| Supervised Learning Tool | Hyperparameters Employed | No. of Different Models Evaluated |
|---|---|---|
| Decision trees | (i) minimum number of split samples, (ii) maximum depth of the tree | 18 |
| Random forests | (i) number of trees in the forest, (ii) criteria for measuring split quality | 16 |
| XGBoost | (i) L2 regularization term (lambda), (ii) number of estimators, (iii) learning rate, (iv) maximum depth of the trees | 240 |
| k-nearest neighbours | (i) p, (ii) number of neighbours (k), (iii) distance metric | 72 |
| Neural networks | (i) activation function, (ii) hidden layers, (iii) learning rate, (iv) solver | 576 |
| Target Value | Supervised Learning Tool | Details of Model Employed | MAPE |
|---|---|---|---|
| Fat content | Random forests | (i) number of trees in the forest = 100, (ii) criteria for measuring split quality = ‘absolute_error’ | 11.37% (11.13%–11.61%) |
| k-nearest neighbours | (i) p = 1, (ii) number of neighbours (k) = 9, (iii) metric = ‘distance’ | 11.39% (11.15%–11.62%) | |
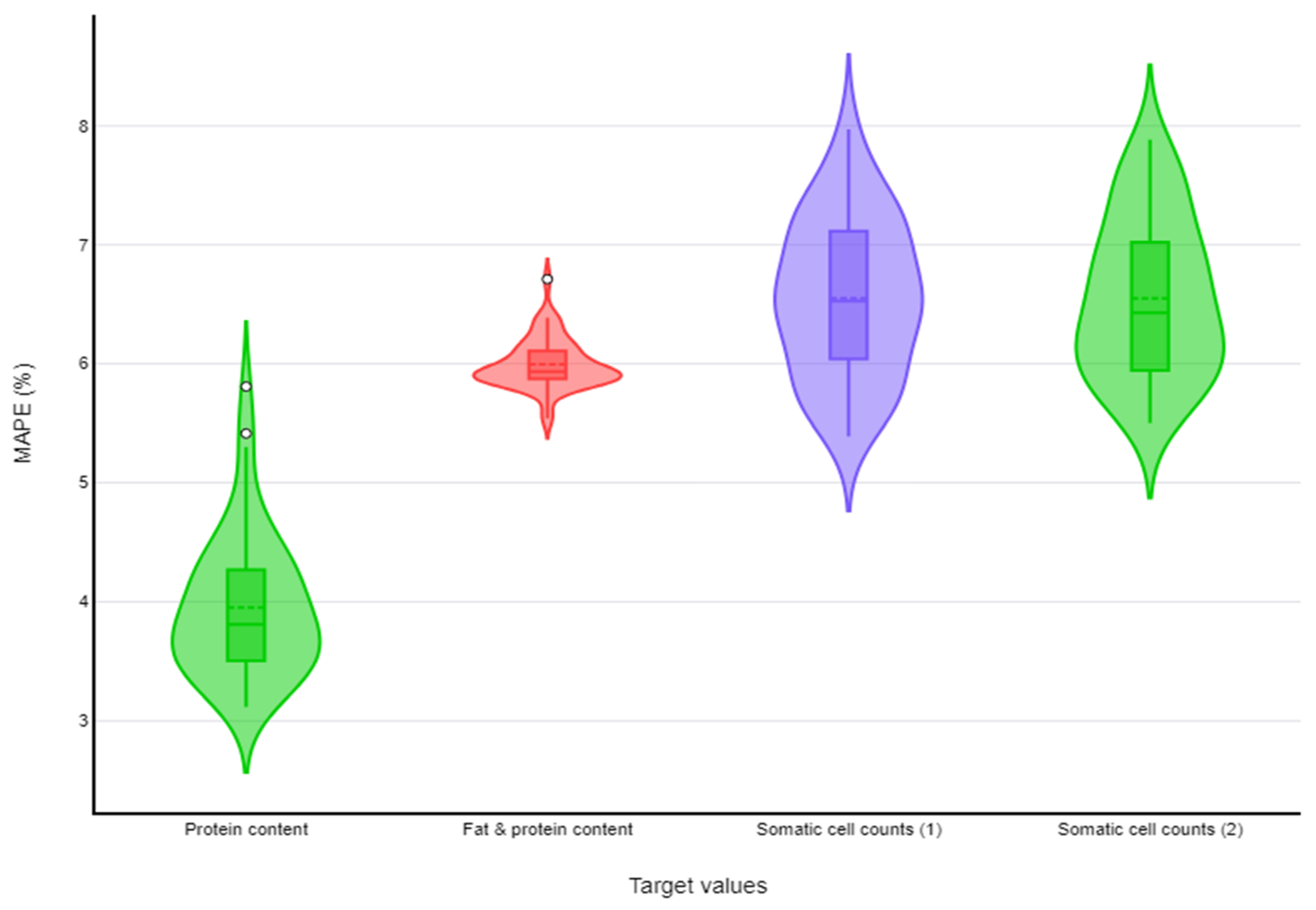
| Protein content | k-nearest neighbours | (i) p = 1, (ii) number of neighbours (k) = 20, (iii) metric = ‘uniform’ | 3.95% (3.87%–4.03%) |
| Fat and protein content | Neural networks | (i) activation function = ‘logistics’, (ii) hidden layers = 100, (iii) learning rate = 1, (iv) solver = ‘adam’ | 6.00% (5.96%–6.02%) |
| Somatic cell counts | Random forests | (i) number of trees in the forest = 100, (ii) criteria for measuring split quality = ‘absolute_error’ | 6.55% (6.5%–6.6%) |
| k-nearest neighbours | (i) p = 1, (ii) number of neighbours (k) = 20, (iii) metric = ‘uniform’ | 6.55% (6.46%–6.64%) | |
| Total bacterial counts | Neural networks | (i) activation function = ‘logistic’, (ii) hidden layers = 500, (iii) learning rate = 0.20, (iv) solver = ‘adam’ | 10.25% (10.13%–10.37%) |
| Target Value | Supervised Learning Tool | Details of Model Employed | MAPE |
|---|---|---|---|
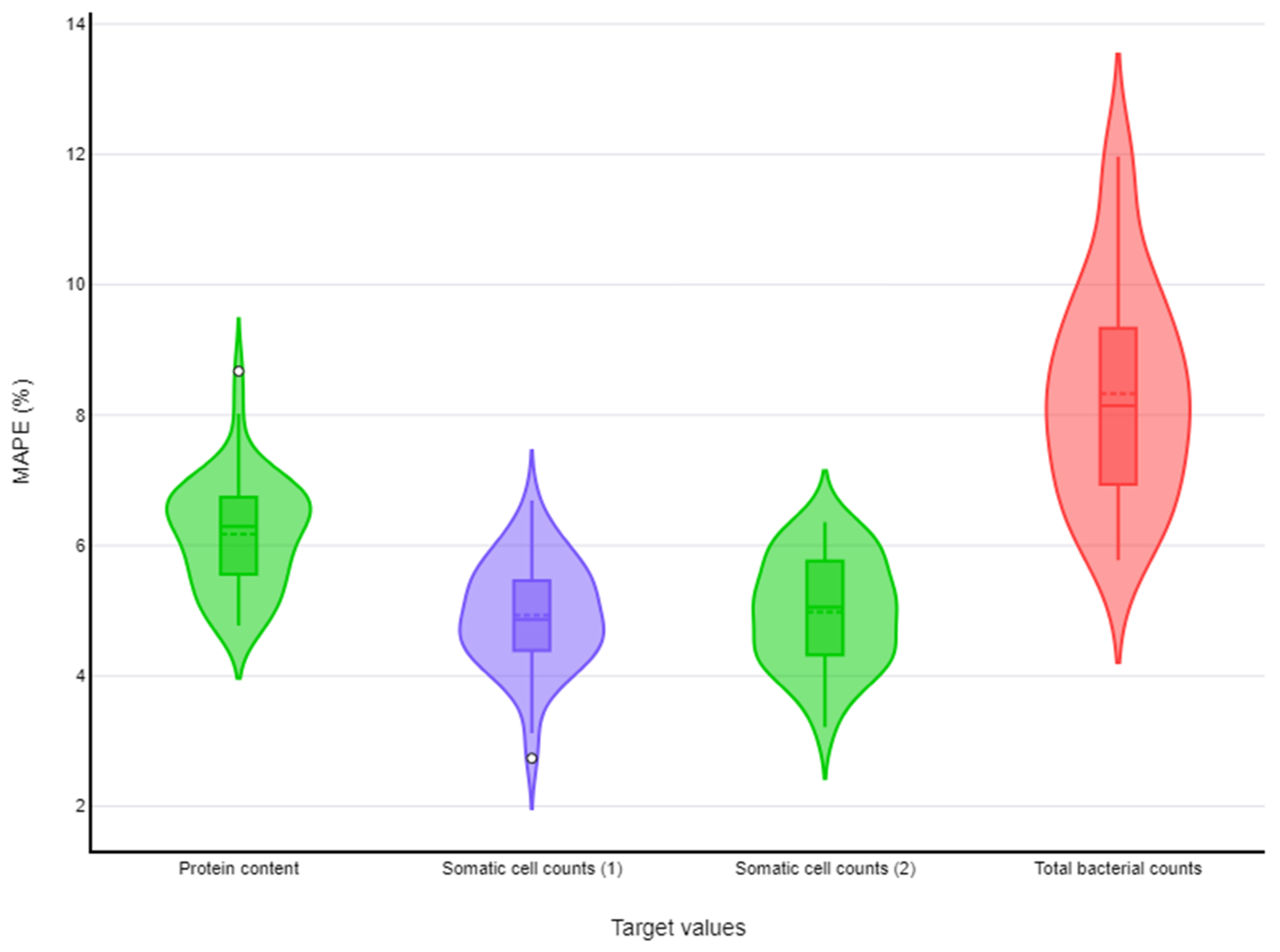
| Protein content | k-nearest neighbours | (i) p = 1, (ii) number of neighbours (k) = 8, (iii) metric = ‘distance’ | 6.17% (6.06%–6.29%) |
| Fat and protein content | Neural networks | (i) activation function = ‘tanh’, (ii) hidden layers = 50, (iii) learning rate = 1, (iv) solver = ‘adam’ | 10.62% (10.26%–10.98%) |
| Somatic cell counts | Random forests | (i) number of trees in the forest = 200, (ii) criteria for measuring split quality = ‘absolute_error’ | 4.93% (4.82%–5.04%) |
| k-nearest neighbours | (i) p = 1, (ii) number of neighbours (k) = 15, (iii) distance metric = ‘uniform’ | 4.98% (4.87%–5.09%) | |
| Total bacterial counts | Neural networks | (i) activation function = ‘logistic’, (ii) hidden layers = 20, (iii) learning rate = 0.001, (iv) solver = ‘adam’ | 8.33% (8.11%–8.55%) |
| Target Value/Supervised Learning Tool | |||
|---|---|---|---|
| Protein Content/ k-Nearest Neighbours | Fat and Protein Content/ Neural Networks | Somatic Cell Counts/ Random Forests | Somatic Cell Counts/ k-Nearest Neighbours |
| Age of newborns taken away from dam | Management system applied in farm | Age of newborns taken away from dam | Age of newborns taken away from dam |
| Month into lactation period at sampling | Grazing of animals | Body condition score of female animals | Age of farmer |
| Education of farmer | Animal breed | Age of farmer | Month into lactation period at sampling |
| Target Value/Supervised Learning Tool | |||
|---|---|---|---|
| Protein Content/ k-Nearest Neighbours | Somatic Cell Counts/ Random Forests | Somatic Cell Counts/ k-Nearest Neighbours | Total Bacterial Counts/ Neural Networks |
| Animal breed | Body condition score of female animals | Annual milk production per animal | No. of animals in farm |
| Month into lactation period at sampling | Number of daily milking sessions | Animal breed | Type of milking parlour |
| Grazing of animals | Month of start of milking period | Month of start of milking period | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lianou, D.T.; Kiouvrekis, Y.; Michael, C.K.; Vasileiou, N.G.C.; Psomadakis, I.; Politis, A.P.; Katsafadou, A.I.; Katsarou, E.I.; Bourganou, M.V.; Liagka, D.V.; et al. The Use of Explainable Machine Learning for the Prediction of the Quality of Bulk-Tank Milk in Sheep and Goat Farms. Foods 2024, 13, 4015. https://doi.org/10.3390/foods13244015
Lianou DT, Kiouvrekis Y, Michael CK, Vasileiou NGC, Psomadakis I, Politis AP, Katsafadou AI, Katsarou EI, Bourganou MV, Liagka DV, et al. The Use of Explainable Machine Learning for the Prediction of the Quality of Bulk-Tank Milk in Sheep and Goat Farms. Foods. 2024; 13(24):4015. https://doi.org/10.3390/foods13244015
Chicago/Turabian StyleLianou, Daphne T., Yiannis Kiouvrekis, Charalambia K. Michael, Natalia G. C. Vasileiou, Ioannis Psomadakis, Antonis P. Politis, Angeliki I. Katsafadou, Eleni I. Katsarou, Maria V. Bourganou, Dimitra V. Liagka, and et al. 2024. "The Use of Explainable Machine Learning for the Prediction of the Quality of Bulk-Tank Milk in Sheep and Goat Farms" Foods 13, no. 24: 4015. https://doi.org/10.3390/foods13244015
APA StyleLianou, D. T., Kiouvrekis, Y., Michael, C. K., Vasileiou, N. G. C., Psomadakis, I., Politis, A. P., Katsafadou, A. I., Katsarou, E. I., Bourganou, M. V., Liagka, D. V., Chatzopoulos, D. C., Solomakos, N. M., & Fthenakis, G. C. (2024). The Use of Explainable Machine Learning for the Prediction of the Quality of Bulk-Tank Milk in Sheep and Goat Farms. Foods, 13(24), 4015. https://doi.org/10.3390/foods13244015








