Amino Acid Composition of Thirty Food Fishes of the Ganga Riverine Environment for Addressing Amino Acid Requirement through Fish Supplementation
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
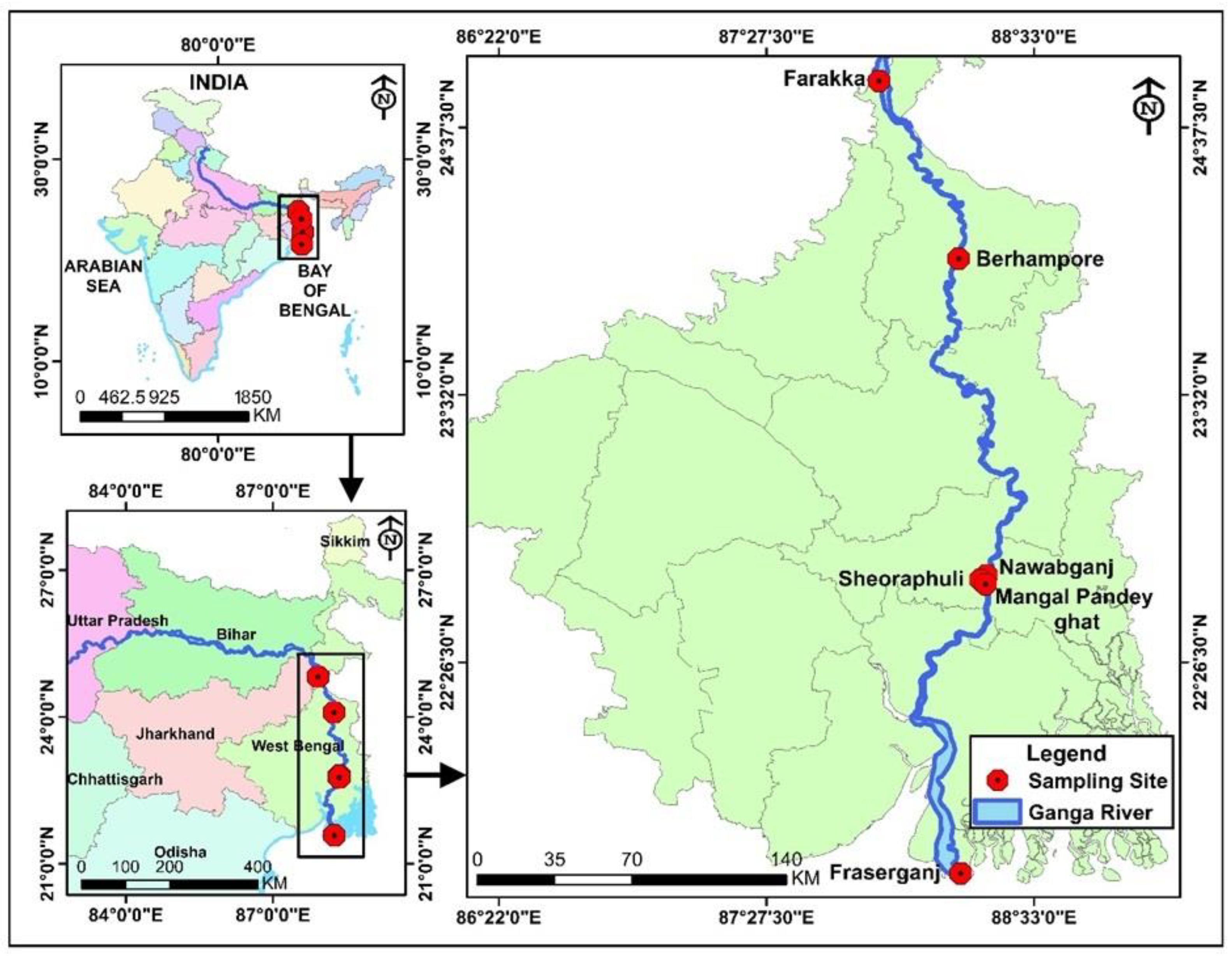
2.2. Study Area
2.3. Fish Sample Collection and Identification
2.4. Sample Processing
2.5. Analysis of Amino Acids
2.6. Statistical Analysis
2.7. Daily Value (DV%) and Potential Contribution to Recommended Dietary Allowance (RDA)
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhutia, D.T. Protein energy malnutrition in India: The plight of our under five children. J. Fam. Med. Prim. Care 2014, 3, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Rose, A.J. Amino acid nutrition and metabolism in health and disease. Nutrients 2019, 11, 2623. [Google Scholar] [CrossRef]
- Wu, G. Functional amino acids in growth, reproduction, and health. Adv. Nutr. 2010, 1, 31–37. [Google Scholar] [CrossRef]
- Cahill, G.F., Jr. Fuel metabolism in starvation. Annu. Rev. Nutr. 2006, 26, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Maida, A.; Zota, A.; Sjøberg, K.A.; Schumacher, J.; Sijmonsma, T.P.; Pfenninger, A.; Christensen, M.M.; Gantert, T.; Fuhrmeister, J.; Rothermel, U.; et al. A liver stress-endocrine nexus promotes metabolic integrity during dietary protein dilution. J. Clin. Investig. 2016, 126, 3263–3278. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Cummings, N.E.; Arriola Apelo, S.I.; Neuman, J.C.; Kasza, I.; Schmidt, B.A.; Cava, E.; Spelta, F.; Tosti, V.; Syed, F.A.; et al. Decreased consumption of branched-chain amino acids improves metabolic health. Cell Rep. 2016, 16, 520–530. [Google Scholar] [CrossRef]
- Maida, A.; Zota, A.; Vegiopoulos, A.; Appak-Baskoy, S.; Augustin, H.G.; Heikenwalder, M.; Herzig, S.; Rose, A.J. Dietary protein dilution limits dyslipidemia in obesity through fgf21-driven fatty acid clearance. J. Nutr. Biochem. 2018, 57, 189–196. [Google Scholar] [CrossRef]
- Kitada, M.; Ogura, Y.; Suzuki, T.; Monno, I.; Kanasaki, K.; Watanabe, A.; Koya, D. A low-protein diet exerts a beneficial effect on diabetic status and prevents diabetic nephropathy in wistar fatty rats, an animal model of type 2 diabetes and obesity. Nutr. Metab. 2018, 15, 20. [Google Scholar] [CrossRef]
- Mohanty, B.; Mahanty, A.; Ganguly, S.; Sankar, T.V.; Chakraborty, K.; Rangasamy, A.; Sharma, A.P. Amino acid compositions of 27 food fishes and their importance in clinical nutrition. J. Amino Acids 2014, 2014, 269797. [Google Scholar] [CrossRef]
- Arany, Z.; Neinast, M. Branched chain amino acids in metabolic disease. Curr. Diab. Rep. 2018, 18, 76. [Google Scholar] [CrossRef]
- Ribeiro, R.V.; Solon-Biet, S.M.; Pulpitel, T.; Senior, A.M.; Cogger, V.C.; Clark, X.; O’Sullivan, J.; Koay, Y.C.; Hirani, V.; Blyth, F.M.; et al. Of older mice and men: Branched-chain amino acids and body composition. Nutrients 2019, 11, 1882. [Google Scholar] [CrossRef]
- Joint WHO/FAO/UNU. Expert Consultation on Protein and Amino Acid Requirements in Human Nutrition; WHO Technical Report Series 935; World Health Organization: Geneva, Switzerland, 2007.
- Shi, L.; Hao, G.; Chen, J.; Ma, S.; Weng, W. Nutritional evaluation of Japanese abalone (Haliotis discus hannai Ino) muscle: Mineral content, amino acid profile and protein digestibility. Food Res. Int. 2020, 129, 108876. [Google Scholar] [CrossRef]
- Ahmed, T.; Hossain, M.I.; Islam, M.; Ahmed, A.M.S.; Afroze, F.; Chisti, M.J. 143—Protein-energy malnutrition in children. In Hunter’s Tropical Medicine and Emerging Infectious Diseases, 10th ed.; Ryan, E.T., Hill, D.R., Solomon, T., Aronson, N.E., Endy, T.P., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 1034–1041. [Google Scholar]
- Muthayya, S.; Rah, J.H.; Sugimoto, J.D.; Roos, F.F.; Kraemer, K.; Robert, R.E. The global hidden hunger indices and maps: An advocacy tool for action. PLoS ONE 2013, 8, e67860. [Google Scholar] [CrossRef] [PubMed]
- Das, B.K.; Ray, A.; Johnson, C.; Verma, S.K.; Alam, A.; Baitha, R.; Manna, R.K.; Roy, S.; Sarkar, U.K. The present status of ichthyofaunal diversity of river Ganga India: Synthesis of present v/s past. Acta Ecol. Sin. 2023, 43, 307–332. [Google Scholar] [CrossRef]
- Talwar, P.K.; Jhingran, A.G. Inland Fishes of India and Adjacent Countries; Oxford and IBH Publishing Co. Pvt. Ltd.: New Delhi, India, 1991; Volume II. [Google Scholar]
- Nelson, J.S. Fishes of the World, 4th ed.; Wiley: New York, NY, USA, 2006. [Google Scholar]
- Jayaram, K.C. The freshwater fishes of India. In Handbook; Zoological Survey of India: Calcutta, India, 1981. [Google Scholar]
- AOAC. Official Methods of Analysis, 18th ed.; Association of Official Analytical Chemists: Gaithersburg, MD, USA, 2006. [Google Scholar]
- Ishida, Y.; Fujita, T.; Asai, K. New detection and separation method for amino acids by high-performance liquid chromatography. J. Chromatogr. 1981, 204, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.P.; Sankar, T.V.; Ganguly, S.; Mahanty, A.; Anandan, R.; Chakraborty, K.; Paul, B.N.; Sarma, D.; Dayal, J.S.; Mathew, S.; et al. Micronutrient composition of 35 food fishes from India and their significance in human nutrition. Biol. Trace Elem. Res. 2016, 174, 448–458. [Google Scholar] [CrossRef]
- Young, L.S.; Stoll, S. Proteins and amino acids. In Contemporary Nutrition Support Practice, 2nd ed.; Matarese, L.E., Gottschlich, M.M., Eds.; Saunders: New York, NY, USA, 2003; Volume 1, pp. 94–104. [Google Scholar]
- Young, V.R.; Pellett, P.L. Background paper 5: Amino acid composition in relation to protein nutritional quality of meat and poultry products. Am. J. Clin. Nutr. 1984, 40, 737–742. [Google Scholar] [CrossRef]
- Wu, G. Functional amino acids in nutrition and health. Amino Acids 2013, 45, 407–411. [Google Scholar] [CrossRef]
- Ganguly, S.; Mahanty, A.; Mitra, T.; Mohanty, S.; Das, B.K.; Mohanty, B.P. Nutrigenomic studies on hilsa to evaluate flesh quality attributes and genes associated with fatty acid metabolism from the rivers Hooghly and Padma. Food Res. Int. 2018, 103, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Uhe, A.M.; Collier, G.R.; O’Dea, K. A comparison of the effects of beef, chicken and fish protein on satiety and amino acid profiles in lean male subjects. J. Nutr. 1992, 122, 467–472. [Google Scholar] [CrossRef]
- Shahidi, F.; Han, X.-Q.; Synowiecki, J. Production and characteristics of protein hydrolysates from capelin (Mallotus villosus). Food Chem. 1995, 53, 285–293. [Google Scholar] [CrossRef]
- Etzel, M.R. Manufacture and use of dairy protein fractions. J. Nutr. 2004, 134, 996S–1002S. [Google Scholar] [CrossRef]
- De Bandt, J.P.; Cynober, L. Therapeutic use of branched-chain amino acids in burn, trauma, and sepsis. J. Nutr. 2006, 185, 308S–313S. [Google Scholar] [CrossRef]
- Juneta-Nor, A.S.; Noordin, N.M.; Azra, M.N.; Ma, H.Y.; Husin, N.M.; Ikhwanuddin, M. Amino acid compounds released by the giant freshwater prawn Macrobrachium rosenbergii during ecdysis: A factor attracting cannibalistic behaviour. J. Zhejiang Univ. Sci. B 2020, 21, 823–834. [Google Scholar] [CrossRef]
- Kaushik, S.J. Whole body amino acid composition of European seabass (Dicentrarchus labrax), gilthead seabream (Sparus aurata) and turbot (Psetta maxima) with an estimation of their IAA requirement profiles. Aquat. Living Resour. 1998, 11, 355–358. [Google Scholar] [CrossRef]
- Brosnan, J.T.; Brosnan, M.E. The sulfur-containing amino acids: An overview. J. Nutr. 2006, 136, 1636–1640. [Google Scholar] [CrossRef]
- Stipanuk, M.H.; Ueki, I. Dealing with methionine/homocysteine sulfur: Cysteine metabolism to taurine and inorganic sulfur. J. Inherit. Metab. Dis. 2011, 34, 17–32. [Google Scholar] [CrossRef]
- Srivastava, A. Micro–level estimation of methionine using inhibitory kinetic spectrophotometric method. Biointerface Res. Appl. Chem. 2021, 11, 10654–10663. [Google Scholar]
- Guo, T.; Chang, L.; Xiao, Y.; Liu, Q. S-adenosyl-L-methionine for the treatment of chronic liver disease: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0122124. [Google Scholar] [CrossRef]
- Strain, J.J.; Lynch, S.M. Excess dietary methionine decreases indices of copper status in the rat. Ann. Nutr. Metab. 1990, 34, 93–97. [Google Scholar] [CrossRef]
- Finkelstein, J.D. Methionine metabolism in mammals. J. Nutr. Biochem. 1990, 1, 228–237. [Google Scholar] [CrossRef]
- Löest, C.A.; Ferreira, A.V.; van der Merwe, H.J.; Fair, M.D. Chemical and essential amino acid composition of South African Mutton Merino lamb carcasses. S. Afr. J. Anim. Sci. 1997, 27, 7–12. [Google Scholar]
- Brosnan, J.T.; Brosnan, M.E. Glutamate: A truly functional amino acid. Amino Acids 2013, 45, 413–418. [Google Scholar] [CrossRef]
- Hou, H.; Li, B.; Zhao, X. Enzymatic hydrolysis of defatted mackerel protein with low bitter taste. J. Ocean. Univ. China 2011, 10, 85–92. [Google Scholar] [CrossRef]
- Sathivel, S.; Smiley, S.; Prinyawiwatkul, W.; Bechtel, P.J. Functional and nutritional properties of red salmon (Oncorhynchus nerka) enzymatic hydrolysates. J. Food Sci. 2005, 70, C401–C406. [Google Scholar] [CrossRef]
- Wang, W.; Wu, Z.; Dai, Z.; Yang, Y.; Wang, J.; Wu, G. Glycine metabolism in animals and humans: Implications for nutrition and health. Amino Acids 2013, 45, 463–477. [Google Scholar] [CrossRef] [PubMed]
- Mischoulon, D.; Fava, M. Role of S-adenosyl-L-methionine in the treatment of depression: A review of the evidence. Am. J. Clin. Nutr. 2002, 76, 1158S–1161S. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.M.; Du, Q.S.; Meng, J.Z.; Pang, Z.W.; Huang, R.B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef]
- Heimann, W. Fundamental of Food Chemistry; AVI Publishing Company: Westport, CT, USA, 1982. [Google Scholar]
- Mahanty, A.; Ganguly, S.; Verma, A.; Sahoo, S.; Mitra, P.; Paria, P.; Sharma, A.P.; Singh, B.K.; Mohanty, B.P. Nutrient profile of small indigenous fish Puntius sophore: Proximate composition, amino acid, fatty acid and micronutrient profiles. Natl. Acad. Sci. Lett. 2014, 37, 39–44. [Google Scholar] [CrossRef]
- Chen, C.; Sander, J.E.; Dale, N.M. The effect of dietary lysine deficiency on the immune response to Newcastle disease vaccination in chickens. Avian Dis. 2003, 47, 1346–1351. [Google Scholar] [CrossRef]
- Zuraini, A.; Somchit, M.N.; Solihah, M.H.; Goh, Y.M.; Arifah, A.K.; Zakaria, M.S.; Somchit, N.; Rajion, M.A.; Zakaria, Z.A.; MatJais, A.M. Fatty acid and amino acid composition of three local Malaysian Channa spp. fish. Food Chem. 2006, 97, 674–678. [Google Scholar] [CrossRef]
- Hyland, K. Inherited disorders affecting dopamine and serotonin: Critical neurotransmitters derived from aromatic amino acids. J. Nutr. 2007, 137, 1568S–1572S. [Google Scholar] [CrossRef] [PubMed]
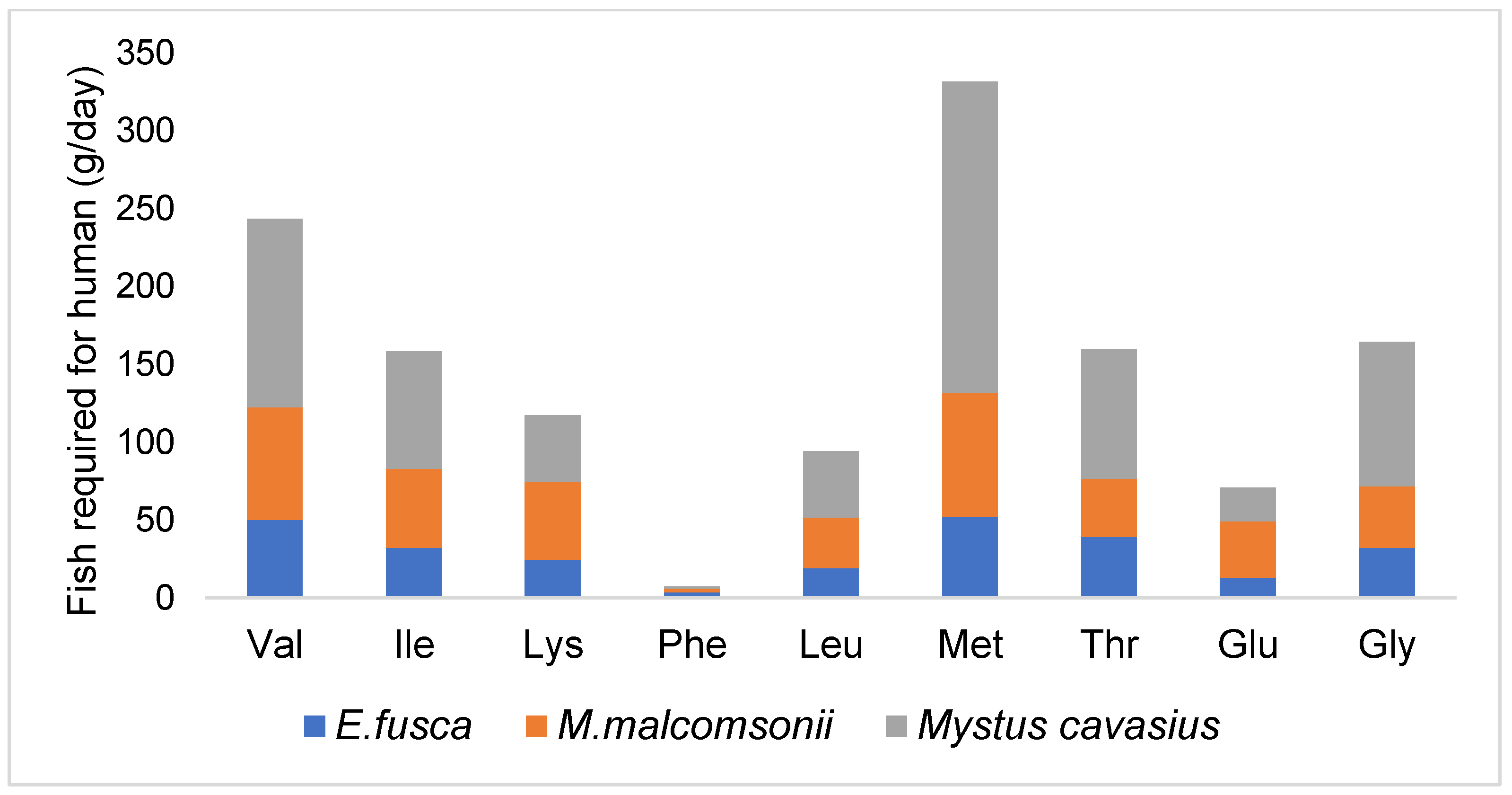

| Amino Acid (g/100 g) | L. guntea | S. ruconius | G. cenia | C. morar | M. malcomsonii † | R. corsola | Requirement for Adult Human (g/kg body wt./day) (WHO, 2007) [12] | |
|---|---|---|---|---|---|---|---|---|
| Crude protein (%) | 15.8 ± 0.01 | 13.7 ± 0.5 | 13.5 ± 0.11 | 15.6 ± 0.04 | 15.5 ± 0.09 | 17.3 ± 0.24 | ||
| Essential amino acids (EAAs) | ||||||||
| ARG | 1.10 ± 0.14 | 0.35 ± 0.01 | 0.16 ± 0.01 | 1.62 ± 0.14 | 0.35 ± 0.01 | 0.09 ± 0.0 | - | |
| VAL | 0.46 ± 0.02 | 0.59 ± 0.02 | 0.51 ± 0.04 | 0.84 ± 0.09 | 2.15 ± 0.19 | 0.51 ± 0.4 | 0.026 | |
| HIS | 0.19 ± 0.01 | 0.89 ± 0.06 | 0.23 ± 0.01 | 1.21 ± 0.11 | 3.30 ± 0.32 | 0.28 ± 0.02 | 0.01 | |
| ILE | 0.43 ± 0.01 | 0.51 ± 0.03 | 0.49 ± 0.02 | 0.41 ± 0.02 | 1.97 ± 0.21 | 0.46 ± 0.06 | 0.02 | |
| LEU | 0.71 ± 0.04 | 0.92 ± 0.06 | 0.80 ± 0.06 | 0.78 ± 0.12 | 3.08 ± 0.32 | 0.85 ± 0.12 | 0.039 | |
| LYS | 0.95 ± 0.03 | 0.97 ± 0.05 | 0.91 ± 0.08 | 0.93 ± 0.06 | 2.01 ± 0.19 | 0.92 ± 0.11 | 0.030 | |
| MET | 0.02 ± 0.00 | 0.11 ± 0.01 | 0.04 ± 0.0 | 0.08 ± 0.0 | 1.26 ± 0.12 | 0.16 ± 0.01 | 0.010 | |
| PHE | 0.40 ± 0.01 | 0.49 ± 0.02 | 0.46 ± 0.02 | 0.40 ± 0.01 | 2.51 ± 0.19 | 0.45 ± 0.06 | Tryo + phyla = 0.025 | |
| THR | 0.47 ± 0.05 | 0.71 ± 0.03 | 0.45 ± 0.04 | 0.06 ± 0.0 | 2.67 ± 0.24 | 0.58 ± 0.09 | 0.015 | |
| Nonessential amino acids (NEAAs) | ||||||||
| CYS | - | - | 1.90 ± 0.11 | - | 0.07 ± 0.0 | - | 0.004 | |
| GLU | 1.91 ± 0.21 | 0.82 ± 0.05 | 1.83 ± 0.14 | 1.16 ± 0.12 | 2.85 ± 0.22 | 1.62 ± 0.14 | - | |
| GLY | 1.01 ± 0.11 | 0.85 ± 0.06 | 0.48 ± 0.02 | 0.11 ± 0.01 | 2.53 ± 0.14 | 0.34 ± 0.02 | - | |
| PRO | 0.55 ± 0.02 | 0.47 ± 0.01 | 0.36 ± 0.02 | 0.39 ± 0.01 | 0.08 ± 0.0 | 0.38 ± 0.06 | - | |
| TYR | 0.29 ± 0.01 | 0.35 ± 0.01 | nd | nd | 8.06 ± 1.14 | 0.38 ± 0.01 | Tryo + phyla = 0.025 | |
| ALA | 0.74 ± 0.06 | 0.70 ± 0.04 | 0.59 ± 0.03 | 0.59 ± 0.03 | 2.02 ± 0.18 | 0.65 ± 0.11 | - | |
| ASP | 1.16 ± 0.12 | 0.09 ± 0.01 | 1.14 ± 0.20 | 0.53 ± 0.02 | 0.10 ± 0.01 | 0.88 ± 0.09 | - | |
| SER | 0.51 ± 0.05 | 0.71 ± 0.04 | 0.47 ± 0.02 | 1.08 ± 0.09 | 2.17 ± 0.22 | 0.56 ± 0.08 | - | |
| Amino Acid (g/100 g) | C. marulius | A. bato | M. panculus | C. cynoglossus | S. phasa | P. ranga | Requirement for Adult Human (g/kg body wt./day) [12] | |
| Crude protein (%) | 17 ± 0.33 | 16.95 ± 0.08 | 18.1 ± 0.03 | 17.1 ± 0.12 | 22.2 ± 0.04 | 11.7 ± 0.21 | ||
| Essential amino acids (EAAs) | ||||||||
| ARG | 0.99 ± 0.22 | 0.07 ± 0.00 | 1.94 ± 0.25 | 1.80 ± 0.12 | 0.69 ± 0.06 | 0.91 ± 0.07 | - | |
| VAL | 0.47 ± 0.03 | 0.40 ± 0.03 | 0.88 ± 0.09 | 1.04 ± 0.08 | 0.35 ± 0.02 | 0.38 ± 0.02 | 0.026 | |
| HIS | 0.24 ± 0.01 | 0.70 ± 0.08 | 0.45 ± 0.04 | 0.51 ± 0.04 | 0.17 ± 0.01 | 0.18 ± 0.01 | 0.01 | |
| ILE | 0.42 ± 0.03 | 0.02 ± 0 | 0.84 ± 0.10 | 0.99 ± 0.08 | 0.29 ± 0.01 | 0.34 ± 0.01 | 0.02 | |
| LEU | 0.75 ± 0.08 | 0.73 ± 0.08 | 1.45 ± 0.14 | 1.65 ± 0.14 | 0.58 ± 0.03 | 0.66 ± 0.03 | 0.039 | |
| LYS | 0.74 ± 0.09 | 0.93 ± 0.09 | 1.46 ± 0.11 | 1.92 ± 0.21 | 0.66 ± 0.04 | 0.32 ± 0.01 | 0.030 | |
| MET | 0.04 ± 0.00 | 0.12 ± 0.01 | 0.25 ± 0.02 | 0.33 ± 0.01 | 0.14 ± 0.01 | 0.14 ± 0.11 | 0.010 | |
| PHE | 0.42 ± 0.03 | 0.38 ± 0.04 | 0.79 ± 0.08 | 0.90 ± 0.09 | 0.31 ± 0.01 | 0.34 ± 0.01 | Tryo + phyla = 0.025 | |
| THR | 0.42 ± 0.01 | 0.87 ± 0.10 | 0.83 ± 0.10 | 0.86 ± 0.10 | 0.40 ± 0.01 | 0.39 ± 0.01 | 0.015 | |
| Nonessential amino acids (NEAAs) | ||||||||
| CYS | nd | 0.21 ± 0.01 | nd | nd | nd | nd | 0.004 | |
| GLU | 1.52 ± 0.22 | 0.75 ± 0.09 | 3.22 ± 0.33 | 3.36 ± 0.31 | 1.45 ± 0.11 | 1.56 ± 0.11 | - | |
| GLY | 0.54 ± 0.06 | 0.76 ± 0.10 | 1.65 ± 0.14 | 1.04 ± 0.09 | 0.47 ± 0.02 | 0.63 ± 0.02 | - | |
| PRO | 0.44 ± 0.02 | 0.37 ± 0.02 | 0.97 ± 0.12 | 3.24 ± 0.24 | 0.41 ± 0.02 | 0.55 ± 0.04 | - | |
| TYR | 0.28 ± 0.01 | 0.34 ± 0.02 | 0.63 ± 0.08 | 0.64 ± 0.04 | 0.27 ± 0.01 | 0.31 ± 0.02 | Tryo + phyla = 0.025 | |
| ALA | 0.53 ± 0.95 | 0.63 ± 0.06 | 1.32 ± 0.15 | 1.18 ± 0.11 | 0.49 ± 0.02 | 0.58 ± 0.03 | - | |
| ASP | 0.91 ± 0.1 | 0.08 ± 0.01 | 1.88 ± 0.14 | 2.09 ± 0.21 | 0.85 ± 0.08 | 0.93 ± 0.03 | - | |
| SER | 0.41 ± 0.04 | 0.66 ± 0.08 | 0.85 ± 0.09 | 0.89 ± 0.09 | 0.35 ± 0.01 | 0.41 ± 0.02 | - | |
| Amino Acid (g/100 g) | S. silondia * | S. bacaila | C. dussumeri | A. chacunda | G. telchitta | M. cavasius | Requirement for Adult Human (g/kg body wt./day) [12] | |
| Crude protein (%) | 18.35 ± 0.13 | 15.05 ± 0.09 | 18.22 ± 0.08 | 17.2 ± 0.04 | 14.9 ± 0.1 | 16.7 ± 0.01 | ||
| Essential amino acids (EAAs) | ||||||||
| ARG | 2.00 ± 0.21 | 0.93 ± 0.04 | 0.88 ± 0.05 | 1.00 ± 0.08 | 0.96 ± 0.08 | 2.28 ± 018 | - | |
| VAL | 0.92 ± 0.10 | 0.48 ± 0.03 | 0.47 ± 0.02 | 0.59 ± 0.03 | 0.53 ± 0.04 | 1.29 ± 0.09 | 0.026 | |
| HIS | 0.46 ± 0.02 | 0.25 ± 0.01 | 0.18 ± 0.01 | 0.34 ± 0.01 | 0.21 ± 0.02 | 0.59 ± 0.04 | 0.01 | |
| ILE | 0.93 ± 0.10 | 0.45 ± 0.02 | 0.43 ± 0.02 | 0.53 ± 0.04 | 0.53 ± 0.03 | 1.33 ± 0.14 | 0.02 | |
| LEU | 1.67 ± 0.11 | 0.85 ± 0.07 | 0.80 ± 0.05 | 0.97 ± 0.08 | 0.95 ± 0.05 | 2.34 ± 0.23 | 0.039 | |
| LYS | 1.66 ± 0.12 | 0.86 ± 0.07 | 0.82 ± 0.08 | 1.04 ± 0.10 | 0.93 ± 0.05 | 2.34 ± 0.23 | 0.030 | |
| MET | 0.31 ± 0.01 | 0.16 ± 0.01 | 0.14 ± 0.01 | 0.19 ± 0.01 | 0.13 ± 0.01 | 0.50 ± 0.04 | 0.010 | |
| PHE | 0.83 ± 0.07 | 0.44 ± 0.02 | 0.42 ± 0.01 | 0.52 ± 0.04 | 0.52 ± 0.02 | 1.28 ± 0.11 | Tryo + phyla = 0.025 | |
| THR | 0.89 ± 0.07 | 0.45 ± 0.01 | 0.40 ± 0.03 | 0.53 ± 0.04 | 0.48 ± 0.04 | 1.20 ± 0.12 | 0.015 | |
| Nonessential amino acids (NEAAs) | ||||||||
| CYS | nd | 0.11 ± 0.01 | nd | nd | nd | nd | 0.004 | |
| GLU | 0.95 ± 0.09 | 1.77 ± 0.14 | 1.66 ± 0.12 | 1.92 ± 0.18 | 1.79 ± 0.14 | 4.62 ± 0.31 | - | |
| GLY | 0.74 ± 0.04 | 0.46 ± 0.02 | 0.49 ± 0.02 | 0.56 ± 0.02 | 0.48 ± 0.02 | 1.08 ± 0.09 | - | |
| PRO | 0.66 ± 0.04 | 0.39 ± 0.01 | 0.39 ± 0.01 | 0.46 ± 0.02 | 0.55 ± 0.02 | 0.98 ± 0.07 | - | |
| TYR | 0.73 ± 0.06 | 0.40 ± 0.02 | 0.36 ± 0.02 | 0.44 ± 0.02 | 0.41 ± 0.02 | 1.00 ± 0.08 | Tryo + phyla = 0.025 | |
| ALA | 1.12 ± 0.11 | 0.63 ± 0.02 | 0.57 ± 0.03 | 0.69 ± 0.05 | 0.65 ± 0.04 | 1.55 ± 0.11 | - | |
| ASP | 0.59 ± 0.03 | 1.07 ± 0.11 | 0.96 ± 0.05 | 1.14 ± 0.11 | 1.09 ± 0.08 | 2.77 ± 0.18 | - | |
| SER | 0.91 ± 0.09 | 0.44 ± 0.02 | 0.40 ± 0.02 | 0.51 ± 0.04 | 0.47 ± 0.02 | 1.20 ± 0.02 | - | |
| Amino Acid (g/100 g) | A. botia | E. fusca | C. garua | C. soborna | G. manmina | Requirement for Adult Human (g/kg body wt./day) [12] | ||
| Crude protein (%) | 15.6 ± 0.08 | 14.5 ± 0.13 | 18.75 ± 0.10 | 16 ± 0.09 | 17.3 ± 0.02 | |||
| Essential amino acids (EAAs) | ||||||||
| ARG | 2.22 ± 0.11 | 4.90 ± 0.36 | 0.58 ± 0.04 | 0.99 ± 0.11 | 1.02 ± 0.09 | - | ||
| VAL | 0.97 ± 0.08 | 3.12 ± 0.34 | 0.33 ± 0.02 | 0.47 ± 0.04 | 0.54 ± 0.02 | 0.026 | ||
| HIS | 0.43 ± 0.02 | 1.77 ± 0.021 | 0.89 ± 0.06 | 0.24 ± 0.01 | 0.25 ± 0.03 | 0.01 | ||
| ILE | 0.87 ± 0.09 | 3.10 ± 0.24 | 0.30 ± 0.01 | 0.42 ± 0.06 | 0.50 ± 0.06 | 0.02 | ||
| LEU | 1.53 ± 0.16 | 5.24 ± 0.41 | 0.64 ± 0.09 | 0.75 ± 0.08 | 0.88 ± 0.09 | 0.039 | ||
| LYS | 1.46 ± 0.12 | 4.06 ± 0.33 | 0.57 ± 0.06 | 0.74 ± 0.11 | 0.94 ± 0.06 | 0.030 | ||
| MET | 0.24 ± 0.01 | 1.92 ± 0.18 | 0.14 ± 0.01 | 0.04 ± 0.01 | 0.17 ± 0.09 | 0.010 | ||
| PHE | 0.85 ± 0.10 | 3.62 ± 0.28 | 0.34 ± 0.02 | 0.42 ± 0.06 | 0.46 ± 0.02 | Tryo + phyla = 0.025 | ||
| THR | 0.93 ± 0.10 | 2.56 ± 0.22 | 0.37 ± 0.02 | 0.42 ± 0.06 | 0.50 ± 0.03 | 0.015 | ||
| Nonessential amino acids (NEAAs) | ||||||||
| CYS | 0 | 0.62 ± 0.06 | 0 | 0 | 0 | 0.004 | ||
| GLU | 3.36 ± 0.22 | 7.61 ± 0.63 | 1.38 ± 0.11 | 1.52 ± 0.08 | 1.84 ± 0.11 | - | ||
| GLY | 1.04 ± 0.09 | 3.10 ± 0.28 | 0.52 ± 0.04 | 0.54 ± 0.03 | 0.48 ± 0.03 | - | ||
| PRO | 0.74 ± 0.08 | 2.35 ± 0.22 | 0.34 ± 0.02 | 0.46 ± 0.04 | 0.38 ± 0.02 | - | ||
| TYR | 0.68 ± 0.07 | 2.92 ± 0.24 | 0.24 ± 0.01 | 0.28 ± 0.01 | 0.42 ± 0.02 | Tryo + phyla = 0.025 | ||
| ALA | 1.25 ± 0.16 | 3.18 ± 0.32 | 0.43 ± 0.01 | 0.53 ± 0.08 | 0.57 ± 0.05 | - | ||
| ASP | 1.99 ± 0.08 | 4.55 ± 0.045 | 0.84 ± 0.06 | 0.91 ± 0.06 | 1.04 ± 0.11 | - | ||
| SER | 0.86 ± 0.06 | 2.51 ± 0.22 | 0.36 ± 0.01 | 0.41 ± 0.02 | 0.47 ± 0.02 | - | ||
| Amino Acid (g/100 g) | C. nama | C. latius | N. nandus | S. gora | E. vacha * | C. reba | G. giuris | Requirement for Adult Human (g/kg body wt./day) [12] |
| Crude protein (%) | 14.99 ± 0.32 | 13.19 ± 0.07 | 15.07 ± 0.11 | 16.8 ± 0.05 | 19.7 ± 0.08 | 16.5 ± 0.03 | 17.5 ± 0.14 | |
| Essential amino acids (EAAs) | ||||||||
| ARG | 1.07 ± 0.05 | 0.13 ± 0.10 | 0.15 ± 0.01 | 3.17 ± 0.34 | 0.64 ± 0.08 | 1.03 ± 0.04 | 0.93 ± 0.06 | - |
| VAL | 0.69 ± 0.05 | 0.41 ± 0.02 | 0.35 ± 0.02 | 2.14 ± 0.14 | 0.32 ± 0.03 | 0.57 ± 0.06 | 0.48 ± 0.04 | 0.026 |
| HIS | 0.27 ± 0.01 | 0.95 ± 0.01 | 0.18 ± 0.02 | 2.57 ± 0.32 | 0.15 ± 0.01 | 0.31 ± 0.02 | 0.25 ± 0.01 | 0.01 |
| ILE | 0.55 ± 0.07 | 0.34 ± 0.02 | 0.01 ± 0.0 | 2.04 ± 0.14 | 0.86 ± 0.08 | 0.53 ± 0.08 | 0.45 ± 0.03 | 0.02 |
| LEU | 0.96 ± 0.11 | 0.74 ± 0.07 | 0.66 ± 0.05 | 3.36 ± 0.41 | 0.57 ± 0.05 | 0.91 ± 0.07 | 0.81 ± 0. 06 | 0.039 |
| LYS | 1.08 ± 0.09 | 0.61 ± 0.05 | 0.80 ± 0.07 | 2.48 ± 0.18 | 0.26 ± 0.01 | 1.26 ± 0.14 | 1.13 ± 0.11 | 0.030 |
| MET | 0.16 ± 0.01 | 0.05 ± 0.01 | 0.15 ± 0.01 | 1.45 ± 0.11 | 0.09 ± 0.01 | 0.16 ± 0.01 | 0.16 ± 0.01 | 0.010 |
| PHE | 0.50 ± 0.05 | 0.45 ± 0.04 | 0.35 ± 0.01 | 2.13 ± 0.22 | 0.29 ± 0.01 | 0.50 ± 0.04 | 0.46 ± 0.02 | Tryo + phyla = 0.025 |
| THR | 0.49 ± 0.02 | 0.64 ± 0.06 | 0.35 ± 0.01 | 2.69 ± 0.21 | 0.30 ± 0.02 | 0.50 ± 0.04 | 0.44 ± 0.03 | 0.015 |
| Nonessential amino acids (NEAAs) | ||||||||
| CYS | nd | nd | nd | 0.29 ± 0.02 | nd | nd | nd | 0.004 |
| GLU | 1.97 ± 0.13 | 1.50 ± 0.11 | 1.43 ± 0.11 | 1.63 ± 0.14 | 1.25 ± 0.14 | 2.01 ± 0.18 | 1.81 ± 0.18 | - |
| GLY | 0.02 ± 0.0 | 0.80 ± 0.07 | 0.37 ± 0.04 | 0.83 ± 0.10 | 0.28 ± 0.04 | 0.51 ± 0.04 | 0.47 ± 0.03 | - |
| PRO | 0.41 ± 0.02 | 0.43 ± 0.01 | 0.28 ± 0.02 | 2.33 ± 0.22 | 0.24 ± 0.01 | 1.57 ± 0.14 | 1.64 ± 0.12 | - |
| TYR | 0.44 ± 0.03 | 0.32 ± 0.01 | 0.30 ± 0.01 | 1.81 ± 0.11 | 0.25 ± 0.04 | 0.42 ± 0.02 | 0.37 ± 0.03 | Tryo + phyla = 0.025 |
| ALA | 0.68 ± 0.05 | 0.52 ± 0.04 | 0.50 ± 0.04 | 2.48 ± 0.25 | 0.39 ± 0.04 | 0.69 ± 0.05 | 0.62 ± 0.04 | - |
| ASP | 1.45 ± 0.14 | 0.92 ± 0.07 | 0.85 ± 0.05 | 0.87 ± 0.07 | 0.75 ± 0.08 | 1.26 ± 0.11 | 1.13 ± 0.11 | - |
| SER | 0.53 ± 0.02 | 0.45 ± 0.03 | 0.36 ± 0.03 | 2.49 ± 0.22 | 0.31 ± 0.02 | 0.51 ± 0.06 | 0.47 ± 0.02 | - |
| Essential amino acids (EAAs) | |||||
| ARG c | E. fusca | S. gora | M. cavasius | A. botia | S. silondia |
| VAL | E. fusca | M. malcomsonii | M. cavasius | C. cynoglossus | A. botia |
| HIS | M. malcomsonii | E. fusca | C. morar | S. ruconius | A. bato |
| ILE | E. fusca | M. malcomsonii | M. cavasius | C. cynoglossus | S. silondia |
| LEU c | E. fusca | M. malcomsonii | M. cavasius | S. silondia | C. cynoglossus |
| LYS | E. fusca | M. cavasius | M. malcomsonii | C. cynoglossus | S. silondia |
| MET c | E. fusca | S. gora | M. malcomsonii | M. cavasius | C. cynoglossus |
| PHE | E. fusca | M. malcomsonii | M. cavasius | C. cynoglossus | A. botia |
| THR | M. malcomsonii | E. fusca | M. cavasius | A. botia | S. silondia |
| Nonessential amino acids (NEAAs) | |||||
| CYS | G. cenia | E. fusca | A. bato | S. bacaila | |
| GLU ac | E. fusca | M. cavasius | A. botia | C. cynoglossus | M. panculus |
| GLY ac | E. fusca | M. malcomsonii | M. panculus | M. cavasius | C. cynoglossus |
| PRO ac | C. cynoglossus | E. fusca | S. gora | G. giuris | C. reba |
| TYR c | M. malcomsonii | E. fusca | M. cavasius | S. silondia | A. botia |
| ALA | E. fusca | S. gora | M. malcomsonii | M. cavasius | M. panculus |
| ASP | E. fusca | M. cavasius | C. cynoglossus | A. botia | M. panculus |
| SER | E. fusca | S. gora | M. malcomsonii | M. cavasius | C. morar |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, B.K.; Ganguly, S.; Bayen, S.; Talukder, A.K.; Ray, A.; Das Gupta, S.; Kumari, K. Amino Acid Composition of Thirty Food Fishes of the Ganga Riverine Environment for Addressing Amino Acid Requirement through Fish Supplementation. Foods 2024, 13, 2124. https://doi.org/10.3390/foods13132124
Das BK, Ganguly S, Bayen S, Talukder AK, Ray A, Das Gupta S, Kumari K. Amino Acid Composition of Thirty Food Fishes of the Ganga Riverine Environment for Addressing Amino Acid Requirement through Fish Supplementation. Foods. 2024; 13(13):2124. https://doi.org/10.3390/foods13132124
Chicago/Turabian StyleDas, Basanta Kumar, Satabdi Ganguly, Supriti Bayen, Anjon Kumar Talukder, Archisman Ray, Subhadeep Das Gupta, and Kajal Kumari. 2024. "Amino Acid Composition of Thirty Food Fishes of the Ganga Riverine Environment for Addressing Amino Acid Requirement through Fish Supplementation" Foods 13, no. 13: 2124. https://doi.org/10.3390/foods13132124
APA StyleDas, B. K., Ganguly, S., Bayen, S., Talukder, A. K., Ray, A., Das Gupta, S., & Kumari, K. (2024). Amino Acid Composition of Thirty Food Fishes of the Ganga Riverine Environment for Addressing Amino Acid Requirement through Fish Supplementation. Foods, 13(13), 2124. https://doi.org/10.3390/foods13132124









