Abstract
Abscisic acid (ABA) plays a crucial role in regulating the ripening of non-climacteric strawberry fruit. In the present study, ABA was confirmed to promote strawberry ripening and induce the down-regulation of FaMADS1. The transient silence of FaMADS1 in strawberries promoted fruit ripening and induced the content of anthocyanin and soluble pectin but reduced firmness and protopectin through a tobacco rattle virus-induced gene silencing technique. In parallel with the accelerated ripening, the genes were significantly induced in the transiently modified fruit, including anthocyanin-related PAL6, C4H, 4CL, DFR, and UFGT, softening-related PL and XTH, and aroma-related QR and AAT2. In addition, the interaction between FaMADS1 and ABA-related transcription factors was researched. Yeast one-hybrid analysis indicated that the FaMADS1 promoter could interact with FaABI5-5, FaTRAB1, and FaABI5. Furthermore, dual-luciferase assay suggested that FaTRAB1 could actively bind with the FaMADS1 promoter, resulting in the decreased expression of FaMADS1. In brief, these results suggest that the ABA-dependent ripening of strawberry fruit was probably inhibited through inhibiting FaMADS1 expression by the active binding of transcript FaTRAB1 with the FaMADS1 promoter.
1. Introduction
Phytohormones strongly dominate transcription factors and genetic regulators to regulate fruit ripening, and the degree of fruit maturity determines fruit quality [1]. As one of the main classical phytohormones, abscisic acid (ABA) plays a crucial role in many physiological processes during plant growth and abiotic stress responses, particularly in fruit ripening [2,3,4]. Studies have revealed that strawberry ripening is controlled by ABA [5,6,7] through the expression of related genes and miRNAs [8,9,10].
ABA functioned through a network of signaling pathways, and numerous signaling components were identified. The most prominent upstream regulators were putative ABA receptors, which perceived ABA signals and then triggered downstream signaling cascades, ultimately inducing physiological responses. In plants, three kinds of ABA receptors have been identified: normal G protein-coupled receptors (GCR) and novel GTP-binding protein receptors (GTG) in the cytoplasmic membrane [11], PYL/PYR/RCAR receptors in the cytosol [12], and ABAR/CHLH receptors in plastid or chloroplast [13]. In normal conditions, the ABA signal was perceived and transferred by central transduction complex PYR/PP2Cs/SnRK2s [14]. Activated SnRK2 switched on ABA signaling by phosphorylating the downstream transcription factors such as AREB/ABF, activating several genes sets [15,16]. Moreover, ABAR could repress the expression of WRKY40 in response to a high level of ABA signal, thereby releasing the expression of the ABI5 gene [17]. A primary region leucine zipper (bZIP) factor, TRAB1, could interact with ABREs and has been identified as a true trans-acting factor involved in ABA-inducible transcription, which controlled maturation and dormancy in plant embryos [18,19]. However, the complexity and coordination of the network of ABA signaling pathways were more widespread than previously believed.
MADS-box genes are present in nearly all major eukaryotic groups and are pivotal regulators in several processes during plant vegetative and reproductive development [20]. Previous studies have reported that MADS-box genes might be the downstream targets of ABA signaling to participate in ABA-associated plant development. In barley, two MADS-box genes, HvOS1 and HvOS2, are induced by exogenous ABA, and differences in DNA methylation difference after the application of ABA were suggestive of epigenetic regulation of these genes [20]. MADS-box genes also participated in the ABA-associated flower and fruit development [21,22,23]. During the transition from inflorescence to floral meristems, some MADS-box genes have been confirmed to influence the meristem identify genes, regulating the flowering time [24,25]. A SEP-like gene MADS7 was cloned in sweet cherry, an indispensable positive regulator of fruit ripening, and exogenous ABA could rescue phenotype defects induced by PaMADS7 silencing [26]. A C-type MADS-box gene FaSHP was induced by ABA in either over-expression or RNAi-mediated down-regulation of strawberry fruit, and over-expression of FaSHP promoted fruit transformation from green to pink, whereas down-regulation of FaSHP could delay the aforementioned transformation [6]. Strawberry FaMADS1 belonged to the SEP1/2 clade based on the amino acid sequence and was suppressed by ABA treatment during accelerated ripening [27,28].
To research the interaction between FaMADS1 and ABA-related transcription factors, strawberry fruit were injected with ABA and a suspension of A. tumefaciens cells carrying a construct for FaMADS1-RNAi to analyze the function of FaMADS1. The activity regulation of the five ABA-related transcription factors (FaABI5-5, FaTRAB1, FaABI5, FaABI5-2, and FaABI5-3) to FaMADS1 was then identified using yeast one-hybrid analysis and dual-luciferase assay.
2. Materials and Methods
2.1. Fruit and ABA Injection
Octaploid strawberry (Fragaria × ananassa Duch. cv. Akihime) plants were grown in suitable environmental conditions. The plastic greenhouse in Hangzhou, Zhejiang Province, China (30°15′ N 120°10′ E), was used for fruit development with a temperature range of 15–25 °C, relative humidity of 70−90%, and no supplemental light. Strawberry fruit at different stages (SG: small green fruit, LG: large green fruit, DG: de-green fruit, W: white fruit, IR: initial red fruit, PR: partial red fruit, and FR: full red fruit) were collected for analysis [7].
Twenty fruit on the plant at DG stage were injected with 0.1 mL of 1 mM ABA (dissolved in 2% ethanol v/v total volume) into the receptacle core through the pedicel using a sterile microsyringe. Another twenty fruit were injected with 2% ethanol as the control [29]. At 0 d, samples of the control and ABA treatment were collected immediately after injection, and subsequently, all samples were collected after 3, 6, and 9 d. After sanitizing with 1% sodium hypochlorite solution, all fruit was washed with ddH2O, cut into small cubes devoid of achenes with a sterilized blade on a sterile bench, and then rapidly frozen in liquid nitrogen [28].
2.2. Determination of Fruit Firmness, Total Anthocyanin, Soluble Solid, Titratable Acid, Protopectin, and Soluble Pectin
The firmness of the fruit was determined using a TA-XT2i Texture Analyzer (Stable Micro Systems Ltd., Surrey, UK). According to the manufacturer, two opposite sides of each fruit were identified as the detection site, and a 5 mm diameter probe was used for testing strawberry fruit. During the experiment, a 5 mm penetration depth and a speed of 1 mm s−1 were established, and the resulting value, N, was recorded.
Ten strawberry fruit were longitudinally cut into four identical portions and distributed into four subgroups to determine the contents of total anthocyanin, soluble solid, titratable acid, protopectin, and soluble pectin. The content of total anthocyanin (TAC) was detected using the pH differential method [30]. The content of soluble solid was directly measured using a Pocket Refractometer (PAL-1, Atago Co. Ltd., Tokyo, Japan). The total titratable acid content was determined using the acid–base titration method, according to Kafkas et al. [31]. The contents of soluble solid and titratable acidity were expressed as %. The contents of protopectin and soluble pectin were measured by coloring carbazole/vitriol according to Lei et al. [32].
2.3. RNA Extraction and RT-qPCR Analysis
Strawberry RNA was extracted using the cetyltrimethylammonium bromide (CTAB) method [33]. The extraction buffer was prepared before RNA extraction, including 2% CTAB, 2% PVP, 2 mol L−1 NaCl, 30 mmol L−1 spermidine, 100 mmol L−1 Tris-HCl (pH 8.0), 25 mmol L−1 EDTA (pH 8.0), chloroform:isoamyl alcohol (24:1), 10 mol L−1 LiCl, and 2% β-mercaptoethanol, which was added just before use. Furthermore, SSTE was prepared for RNA extraction. The RNA was reverse transcribed using the PrimeScriptTM RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Biotech, Dalian, China). SYBR®PremixEx Taq™ (TliRNaseH Plus) (Takara Biotech, Dalian, China) was used for quantitative PCR. FaActin gene (GenBank No.: AB116565.1) was considered a loading control gene. The 2−ΔΔCT method was used to calculate the relative gene expressions.
2.4. RNAi Plasmid Construction
VIGS vectors of pTRV1 and pTRV2 were selected as described by Liu et al. [34]. The FaMADS1 coding sequence (750 bp) was cloned in the vector pTRV2 to obtain the FaMADS1 down-expression construct. The cDNA was inserted into the vector pTRV2 via XbaI and EcoRI sites. The recombinant plasmid pTRV2-FaMADS1 was introduced into Agrobacterium tumefaciens (strain EHA105). Agrobacterium cells were harvested when the culture reached an OD600 of 0.8. The infection method was performed according to Jia et al. [5]. The fruit was gathered when either one of the two groups turned red. The expression of FaMADS1 in FaMADS1-RNAi fruit was examined using the primers FaMADS1-F and FaMADS1-R (Table S1).
2.5. Determination of Fruit Aroma Components
Headspace solid-phase microextraction (HS-SPME) was adopted to prepare the samples, and gas chromatography-mass spectrometry (GC-MS) was used to analyze fruit aroma production as described by Vandendriessche et al. [35]. Pure 2-Octanone was added to each sample at a concentration of 0.00524 g L−1 to serve as an internal standard composition. The peak areas of components relating to the internal standard were considered for each peak. All extractions were performed manually using a Supelco fiber holder (Bellefonte, PA, USA), and volatile flavor compounds were extracted using a 50/30 μm DVB/CAR/PDMS fiber. Five grams of fruit puree mixed with 5 mL saturated salt water and 50 μL of internal standard (0.00524 g L−1, 2-octanone) were added into a 20 mL headspace vial. Semi-quantitative results are expressed as μg kg−1 FW compared with the standard.
2.6. Determination of Enzyme Activity
The activities of PAL (phenylalanine ammonia lyase), 4CL (ρ-coumarate ligase), C4H (cinnamicacid-4-hydroxylase), DFR (dihydroflavonol 4-reductase), PG (polygalacturonase), PL (pectate lyase), and AAT (alcohol acyltransferase) were detected in the control and FaMADS1-RNAi fruit. PAL, C4H, and DFR enzyme activities were measured according to Chen et al. [36]. PG enzyme activity was measured using the protocol of Andrews and Li [37]. PL enzyme activity was detected using the protocol of Mao et al. [38]. AAT enzyme activity was determined using the protocol of Pérez et al. [39].
2.7. Subcellular Localization Assay
The open reading frame of FaMADS1 was cloned as C-terminal fusion in frame with the green fluorescent protein (GFP) gene into the pBI121 vector. The fusion construction and infection methods were performed according to Jia et al. [5] and Lu et al. [40]. The Agrobacterium suspension was evenly injected into the back of the leaf attached to the BY-2 tobacco (Nicotiana tabacum). After 48 h, at least six replicates of transfected leaves were analyzed using the laser scanning confocal imaging system (TCS SP8, Leica, Wetzlar, Germany).
2.8. Analysis of DNA Sequence and Yeast One-Hybrid Analysis
The PLANTCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) and MatInspector (http://www.genomatix.de/online_help/help_matinspector/matinspector_help.html) software were used to analyze the identification and characterization of putative cis-acting elements of the FaMADS1 promoter.
Yeast one-hybrid assay was performed using the Matchmaker® Gold Yeast One-Hybrid Library Screening System (Clontech, Mountain View, CA, USA). The promoters of FaMADS1 (FaMADS1-pro1 and FaMADS1-pro2) were constructed into the pAbAi vector. FaMADS1-pro1 and FaMADS1-pro2 lack auto-activation activities, so they were chosen for interaction tests. The test was conducted according to the instructions of Wei et al. [41].
2.9. Dual-Luciferase Assay
The dual-luciferase assay was performed according to Min et al. [42] and Wei et al. [41]. The vectors pGreen II 0029 62-SK (SK) and pGreen II 0800-LUC were used for building fusion vectors with FaABI5-5, TRAB1, ABI5, and FaMADS1. The proteins were expressed in tobacco (Nicotiana benthamiana) by agrobacterium transformation of Agrobacterium tumefaciens GV3101. The activities of the enzymes (FLUC and RLUC) were analyzed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA, E1910) with Modulus Luminometers (Promega).
2.10. Statistical Assay
All determinations were conducted in triplicate, and the data are presented as means ± standard deviations. All data were analyzed for significance using SPSS software (p < 0.05, significance) and Origin 9.0. The error bars represent the standard deviation (SD, n = 3). Asterisks indicate values significantly different from the control samples (* p < 0.05; ** p < 0.01) through the Student’s t test.
3. Results
3.1. ABA Induced the Ripening of Strawberry Fruit
ABA induced fruit ripening by shortening the time to attain the full red stage (Figure 1A). Anthocyanin and soluble solid accumulated rapidly with significant differences between the control and ABA-treated fruit (Figure 1B,C). However, ABA treatment reduced total titratable acid content and fruit firmness (Figure 1D,E). These results indicated that ABA stimulated the ripening of strawberry fruit.
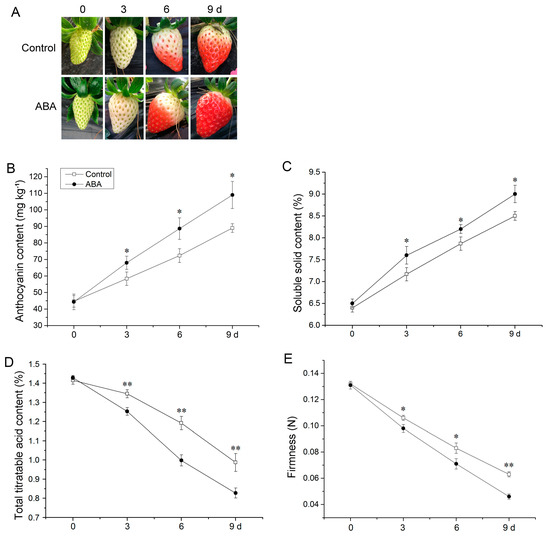
Figure 1.
Influence of ABA on strawberry fruit phenotype (A), anthocyanin (B), soluble solid (C), total titratable acid (D), and firmness (E). Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (* p < 0.05; ** p < 0.01).
3.2. Down-Regulation of FaMADS1 and Up-Regulation of Ripening-Related Gene Expressions by ABA
ABA significantly reduced the expression of FaMADS1 and a rapid decrease trend was observed during strawberry ripening (Figure 2A). Furthermore, there was no significant difference between the control and ABA-treated fruit on the expression levels of FaC4H, Fa4CL, FaANS, FaUFGT, FaPG, and FaQR, whereas the expression of several anthocyanin biosynthesis genes (PAL6, CHS, and DFR), softening-related gene (PL), and aroma biosynthesis gene (AAT2) were significantly up-regulated by ABA treatment (Figure 2B).
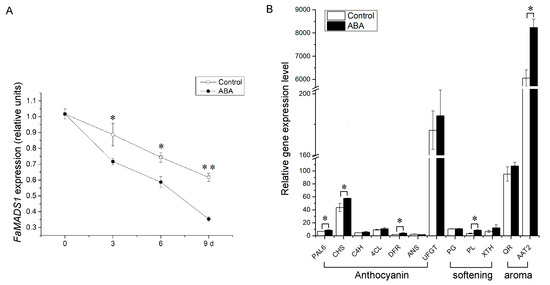
Figure 2.
Relative expression levels of FaMADS1 (A) in strawberry fruit at 0, 3, 6, and 9 d after ABA treatment and other ripening-related genes (B) in strawberry fruit at 3 d after ABA treatment. Anthocyanin-related genes: FaANS, anthocyanidin synthase; Fa4CL, ρ-coumarate ligase; FaC4H, cinnamicacid-4-hydroxylase; FaCHS, chalcone synthase; FaDFR, dihydroflavonol 4-reductase; FaPAL6, phenylalanine ammonia lyase; FaUFGT, flavonoid 3-O-glucosyltransferase. Softening-related genes: FaPG, Polygalacturonase; FaPL, pectate lyase; FaXTH, xyloglucan endotransglycosylase/hydrolase. Aroma-related genes: FaAAT2, alcohol acyltransferase; FaQR, quinone oxidoreductase. Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (* p < 0.05; ** p < 0.01).
3.3. Down-Regulation of FaMADS1 in FaMADS1-RNAi Strawberry Fruit
In this study, the fruit at the DG stage was injected with a suspension of A. tumefaciens cells carrying a construct of FaMADS1-RNAi (Figure 3A,B). FaMASD1-RNAi decreased the FaMADS1 transcript level by more than three folds, suggesting that the transient RNAi technique successfully manipulated FaMADS1 expression in the strawberry fruit (Figure 3C).
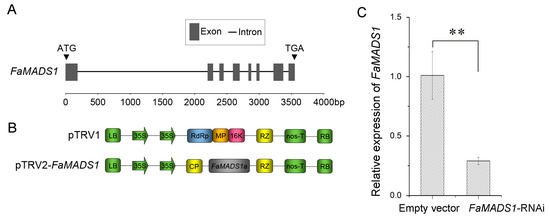
Figure 3.
Construction of FaMADS1-RNAi strawberry fruit. Genomic DNA sequence analysis of FaMADS1 (A). Dark gray boxes represent exons and lines indicate introns. The scale indicates the length of the sequence. Schematic representation of the pTRV1 and pTRV2-FaMADS1 VIGS constructs (B). CP: viral coat protein; RdRp: RNA-dependent RNA polymerase; MP: movement protein; 16 K: 16 kDa protein; RZ: ribozyme. Relative FaMADS1 transcript levels in TRV (empty vector) and TRV-FaMADS1 (FaMADS1-RNAi) strawberry fruit were measured using qRT-PCR at 7 d after infiltration (C). Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (** p < 0.01).
3.4. Promoted Ripening in FaMADS1-RNAi Strawberry
As shown in Figure 4A, FaMASD1-RNAi greatly promoted the ripening progression of strawberry fruit. The fruit on FaMADS1-RNAi plants became partially red (nearly 75%), whereas the control fruit remained in the white stage at 7 d after injection. FaMADS1-RNAi fruit displayed a substantial increase in anthocyanin content and a decrease in firmness (Figure 4B,C). Furthermore, FaMADS1-RNAi led to a decrease in protopectin and an increase in soluble pectin (Figure 4D,E).
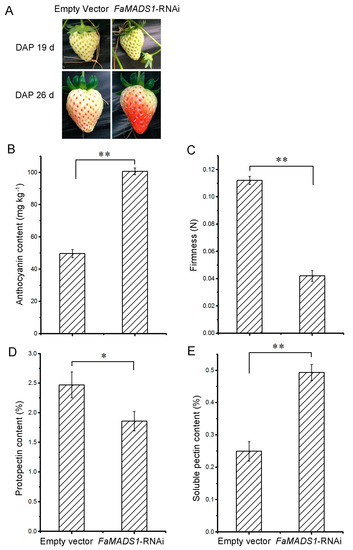
Figure 4.
Fruit properties of FaMADS1-RNAi strawberry. RNAi fruit was already red when the control fruit was white at 7 d after injection (A). Anthocyanin content (B), firmness (C), protopectin (D), and soluble pectin content (E) were detected in control (empty vector) and FaMADS1-RNAi strawberry fruit at 7 d after injection. Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (* p < 0.05; ** p < 0.01).
Furthermore, FaMADS1-RNAi increased the levels of esters (methyl hexanoate; 1-methyl hexanoate; methyl 2-octynoate; octyl butyrate; octyl 3-methylbutyrate; (3,7,11-trimethyldodeca-1,6,10-trien-3-yl) formate; 3,7,11-trimethyl-1,6,10-dodecatrien-3-olacetate and 5-octyloxolan-2-one), ketones [DMMF and 1-(2,6,6-trimethyl-1-cyclohexen-1-yl)-1-penten-3-one], acids (octanoic acid and 3-hydroxydodecanoic acid), terpenes (2,3-dihydro-1H-indene), and benzene [(4aS-cis)-2,4a,5,6,7,8,9,9a-octahydro-3,5,5-trimethyl-9-methylene-1H-Benzocycloheptene]. Meanwhile, we observed increased levels of some alcohol, aldehyde, and olefin in FaMADS1-RNAi strawberry fruit (Table 1). This increase in aroma indicated that a metabolic boost facilitated fruit ripening in FaMADS1-RNAi lines.

Table 1.
Main aroma compounds in FaMADS1-RNAi strawberry fruit.
3.5. Increased Ripening Gene Expressions and Enzyme Activity in FaMADS1-RNAi Strawberry Fruit
The expression of genes related to fruit color, firmness, and aroma metabolism was significantly regulated by FaMADS1-RNAi (Figure 5). FaMADS1-RNAi greatly promoted the expression levels of FaPAL6, FaC4H, Fa4CL, FaDFR, and FaUFGT, which are involved in anthocyanin biosynthesis [43]. FaMADS1-RNAi also induced the transcription of fruit softening-related genes, such as FaPL and FaXTH [44]. Notably, the expression level of FaQR in FaMADS1-RNAi fruit was 4-fold higher than the controls, and FaMADS1-RNAi led to an almost 3-fold increase in the expression level of FaAAT2.
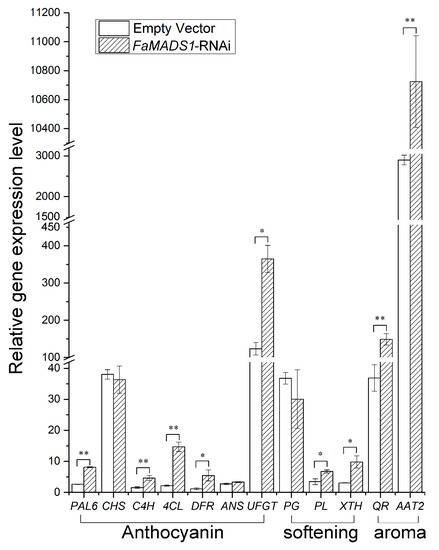
Figure 5.
Relative expression of ripening-related genes in the control fruit (empty vector) and FaMADS1-RNAi fruit at 7 d after injection. Each value represents the mean of six independent fruit. Anthocyanin-related genes: FaANS, anthocyanidin synthase; Fa4CL, ρ-coumarate ligase; FaC4H, cinnamicacid-4-hydroxylase; FaCHS, chalcone synthase; FaDFR, dihydroflavonol 4-reductase; FaPAL6, phenylalanine ammonia lyase; FaUFGT, flavonoid 3-O-glucosyltransferase. Softening-related genes: FaPG, Polygalacturonase; FaPL, pectate lyase; FaXTH, xyloglucan endotransglycosylase/hydrolase. Aroma-related genes: FaAAT2, alcohol acyltransferase; FaQR, quinone oxidoreductase. Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (* p < 0.05; ** p < 0.01).
FaMADS1-RNAi significantly promoted the activities of PAL, 4CL, C4H, and DFR (Figure 6A–D), which are involved in the phenylpropanoid pathway of anthocyanin biosynthesis [45]. There was no significant difference in PG activity between the control and FaMADS1-RNAi fruit, but there was a significant increase in PL activity in FaMADS1-RNAi fruit (Figure 6E). Furthermore, the activity of AAT in FaMADS1-RNAi fruit was higher than that in the control group (Figure 6F).
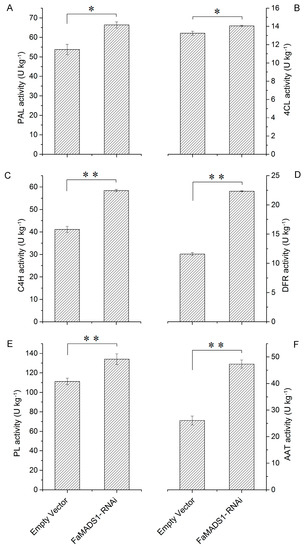
Figure 6.
PAL (A), 4CL (B), C4H (C), DFR (D), PL (E), and AAT (F) activities in the control (empty vector) and FaMADS1-RNAi fruit at 7 d after injection. PAL, phenylalanine ammonia lyase; 4CL, ρ-coumarate ligase; C4H, cinnamicacid-4-hydroxylase; DFR, dihydroflavonol-4-reductase; PL, pectate lyase; AAT, alcohol acyltransferase. Error bars represent standard deviation (SD, n = 3). Asterisks indicate values significantly different by Student’s t test from the control samples (* p < 0.05; ** p < 0.01).
3.6. Localization and Interaction of FaMADS1 with ABI5-5, TRAB1, and ABI5
The coding sequence of FaMADS1 without stop codes was constructed in pBI221. The leaf epidermal cells with 35S::GFP demonstrated green fluorescence throughout the cell. By contrast, green fluorescence was especially detected in the nuclei of leaf epidermal cells expressing the 35S::FaMADS1-GFP fusion plasmids (Figure 7), suggesting that FaMADS1 was a nuclear protein.
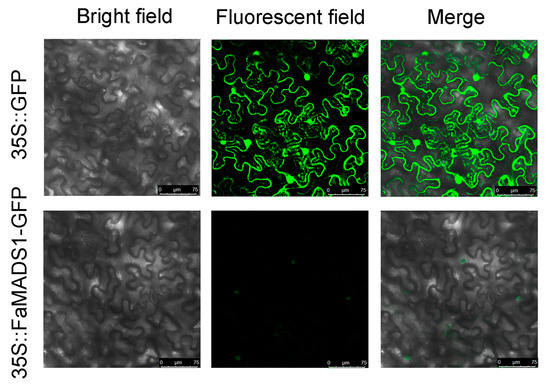
Figure 7.
Sub-cellular localization of 35S::FaMADS1-GFP fusion protein in leaf epidermal cells of tobacco. Plasmids of 35S::GFP and 35S::FaMADS1-GFP were transformed into leaf epidermal cells by agro-infiltration, respectively. Bright field images, fluorescent field images, and the merged images of leaf epidermal cells expressing 35S::GFP or 35S::FaMADS1-GFP fusion protein.
An online informatics survey was implemented to analyze a fragment of the FaMADS1 promoter sequence (1095 bp upstream from the ATG start codon). A putative domain (ABRE) for responding to ABA was found (Table S2). Therefore, two sequences (FaMADS1-pro1 and FaMADS1-pro2) of FaMADS1 promoter were cloned to study the interaction between FaMADS1 and ABA-related transcription factors (FaABI5-5, FaTRAB1, FaABI5, FaABI5-2, and FaABI5-3). FaABI5-5, FaTRAB1, and FaABI5 interacted with FaMADS1-pro1, whereas ABI5-2 and ABI5-3 did not interact with FaMADS1-pro1 (Figure 8A). FaABI5-5, FaTRAB1, FaABI5, FaABI5-2, and FaABI5-3 also did not interact with FaMADS1-pro2 (Figure 8B). These results revealed that FaMADS1-pro1 contained the active cis-acting elements, which could interact with FaABI5-5, FaTRAB1, and FaABI5.
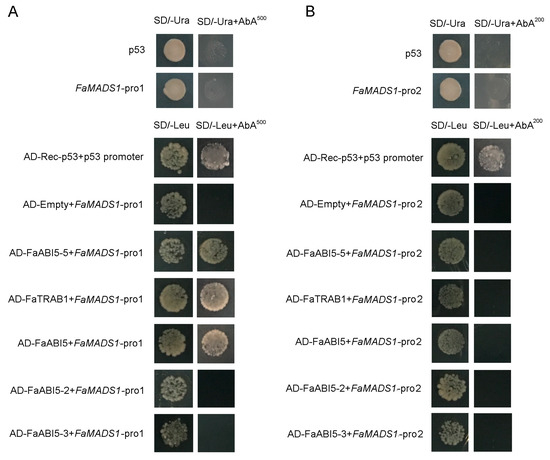
Figure 8.
Yeast one-hybrid analysis of ABI5-5, TRAB1, ABI5, ABI5-2, and ABI5-3 binding to the FaMADS1 promoter. Auto-activation tests of p53, FaMADS1-pro1, and FaMADS1-pro2 were experimented on SD medium lacking Ura in the presence of aureobasidin A. Interaction tests between FaMADS1 promoters with transcription factors were carried out on SD medium lacking Leu in the presence of aureobasidin A. (A) The interaction between ABI5-5, TRAB1, ABI5, ABI5-2, ABI5-3, and FaMADS1-pro1. (B) The interaction between ABI5-5, TRAB1, ABI5, ABI5-2, ABI5-3, and FaMADS1-pro2.
To further verify the regulatory roles of FaABI5-5, FaTRAB1, and FaABI5 in FaMADS1, a dual-luciferase assay was performed in tobacco leaves (Figure 9). FaTRAB1 could significantly inhibit the promoter of FaMADS1, while FaABI5-5 and FaABI5 had little influence on the FaMADS1 promoter.
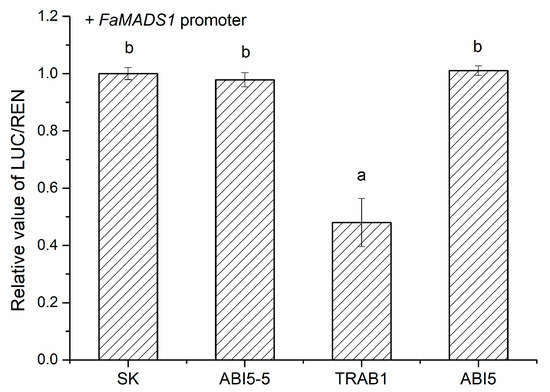
Figure 9.
Regulatory effects of FaABI5-5, FaTRAB1, and FaABI5 on the promoters of the FaMADS1 gene using the dual-luciferase assay. The LUC/REN ratio of the empty vector (SK) plus promoter was set as 1. Different letters above the points indicate significantly different values (p < 0.05) through the LSD test.
4. Conclusions and Discussion
In this study, ABA significantly promoted the accumulation of anthocyanin and soluble solid in strawberry fruit, but reduced fruit firmness and total titratable acid; thus, ABA regulated almost the entire fruit ripening process [46]. This significant acceleration of fruit ripening by ABA was accompanied by increased expression levels of series genes involved in anthocyanin biosynthesis (PAL, CHS, and DFR), softening (PL), and easier biosynthesis (AAT). PAL was discovered to trigger the production of anthocyanin [47]. CHS was indicated to increase the attractive red color of the strawberry fruit [48], and DFR was one of the key anthocyanin structural enzymes [49]. PL was responsible for the eliminative cleavage of pectate, and then oligosaccharides were produced with 4-deoxy-α-D-mann-4-enuronosyl groups at their non-reducing ends, resulting in fruit softening [50]. During ripening, strawberry fruit produced lots of secondary metabolites contributing to flavor and organoleptic peculiarities. Zabetakis and Holden [51] discovered more than 350 compounds from strawberries, including alcohols, esters, aldehydes, furanone derivative ketones, sulfur compounds, and terpenes; however, there were only 20 compounds actually contributing to the aroma and flavor of strawberries [52]. Alcohol acyltransferase (AAT) was involved in the biosynthesis of short-chain esters, contributing to the blend of volatiles and defining the fruit aroma [53]. These results demonstrated that ABA regulated ripening by promoting softening metabolism and biosynthesis of anthocyanin and aroma in strawberries.
Studies have suggested that some MADS members contributed to the fruit ripening of tomatoes [54], grape [55], strawberries [56,57], and bananas [58]. The MADS-box gene FaMADS1 was suppressed with an accelerated ripening process following ABA treatment [28]. These results confirmed that FaMADS1 might play a negative role in fruit ripening via the downstream ABA signal transduction pathway connected to related metabolic processes.
Previous studies have reported that tobacco rattle virus (TRV)-mediated VIGS was a rapid and feasible technique for analyzing fruit ripening of tomato [59] and strawberry [60,61]. In the present study, the negative role of FaMADS1 in the ripening of strawberry fruit was verified through a gene silencing technique. Compared with the control (empty vector), FaMADS1-RNAi fruit exhibited accelerated ripening with high levels of anthocyanin and soluble pectin, low firmness, and protopectin content. In addition, an increase in the content levels of ester, ketone, acid, terpenes, and benzene was observed in the FaMADS1-RNAi fruit. The down-regulation of FaMADS1 resulted in increased expression levels of anthocyanin-related genes (PAL6, C4H, 4CL, DFR, and UFGT), softening-related genes (PL and XTH), and aroma-related genes (QR and AAT2). In tomato, the silence experiment also caused changes in fruit ripening. Mekontso et al. [62] found that postharvest ripening could be delayed by knocking down miR160a, and some related genes, such as ARF10A, ARF10B, and ZRF17, were up-regulated. Meanwhile, FaMADS1-RNAi significantly promoted the activities of PAL, 4CL, C4H, DFR, PL, and AAT enzymes. These results collectively suggested that the FaMADS1 gene had a strong negative function in strawberry ripening.
The gene ABI encoded protein PP2C in the ABA signal pathway. FaABI1 and FaABI2 negatively regulated strawberry fruit ripening, whereas FaABI4 and FaABI5 positively regulated fruit ripening [5,63]. Previous studies have confirmed that ABI and TRAB are the major transcription factors in the ABA signaling pathway [18,63,64,65,66]. In the present study, yeast one-hybrid analysis revealed that FaABI5-5, FaTRAB1, and FaABI5 could interact with FaMADS1-pro1. Furthermore, the dual-luciferase assay confirmed that the promoter of FaMADS1 could be significantly inhibited by FaTRAB1, whereas FaABI5-5 and FaABI5 had little influence on the promoter of FaMADS1. These results suggested that the ABA signal was transmitted to the FaMADS1 promoter with the interaction of FaTRAB1. FaMADS1 regulated the transcriptional level of the downstream anthocyanin-related genes, softening-related genes, and aroma-related genes, eventually initiating fruit ripening.
In conclusion, FaMADS1 expression negatively depended on ABA to regulate strawberry fruit ripening. The yeast one-hybrid analysis and dual-luciferase assay suggested that the FaMADS1 promoter was inhibited by the interaction with FaTRAB1, resulting in the blockage of FaMADS1 translation. ABA application and FaMADS1-RNAi data reflected a negative regulation of FaMADS1 to the downstream anthocyanin-related genes (PAL6, C4H, 4CL, DFR, and UFGT), softening-related genes (PL and XTH), and aroma-related genes (QR and AAT2), as well as the enzyme activities (Figure 10). This observation enhanced our understanding of the molecular mechanism of ABA signaling during the non-climacteric fruit ripening.
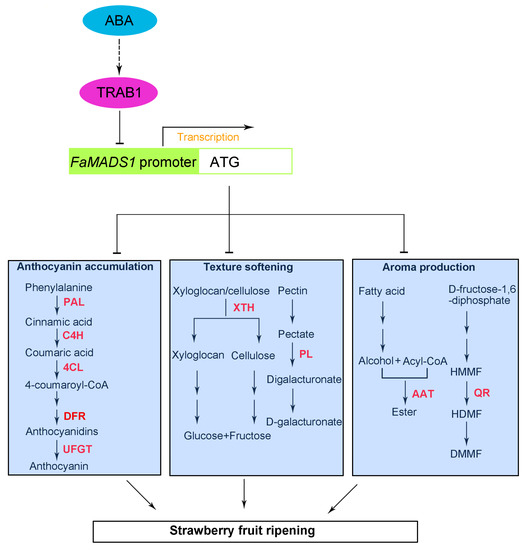
Figure 10.
A model for ABA receptors-mediated signaling pathway connected to FaMADS1 regulating strawberry fruit ripening. In the presence of ABA, ABA signal activated ABI and TRAB transcription factors, and then FaMADS1 was inhibited. Depression of FaMADS1 induced the expression of anthocyanin-related FaPAL, FaC4H, Fa4CL, and FaUFGT, softening-related FaPL and FaXTH, and aroma-related FaQR and FaAAT. Abbreviations: AAT2, alcohol acyltransferase; ABA, abscisic acid; 4CL, ρ-coumarate ligase; C4H, cinnamicacid-4-hydroxylase; PAL6, phenylalanine ammonia lyase; PL, pectate lyase; QR, quinone oxidoreductase; SnRK2s, phosphorylation of ABA-activated sucrose non-fermenting 1-related protein kinases subfamily 2; TRAB, transcription factor responsible for ABA regulation; UFGT, flavonoid 3-O-glucosyltransferase; XTH, xyloglucan endotransglycosylase/hydrolase. The solid arrows indicate direct action and the dashed arrow indicates indirect action.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods12091802/s1, Table S1: All primers used in the study for PCR analysis; Table S2: Bioinformatics analysis of the FaMADS1 gene promoter.
Author Contributions
Conceptualization, L.M. and W.L.; methodology, W.L.; software, X.W.; validation, X.W., X.H. and R.C.; formal analysis, W.L.; investigation, X.H.; resources, X.W.; data curation, W.L.; original draft preparation, W.L.; review and editing, X.Z. and L.M.; visualization, W.L.; supervision, L.M.; project administration, C.X. and X.W.; funding acquisition, L.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (31972468).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Forlani, S.; Masiero, S.; Mizzotti, C. Fruit ripening: The role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 2019, 70, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Park, Y.; Hwang, I. Abscisic acid: Biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 2015, 58, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Ju, Y.L.; Liu, M.; Zhao, H.; Meng, J.F.; Fang, Y.L. Effect of exogenous abscisic acid and methyl jasmonate on anthocyanin composition, fatty acids, and volatile compounds of cabernet sauvignon (Vitis vinifera L.) grape berries. Molecules 2016, 21, 1354. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.P.; Mao, L.C.; Wei, X.B.; Guan, W.L.; Lu, W.J.; Xu, C.J. ABA stimulates wound suberization through antagonizing the MYB4-mediated transcriptional repression of CYP86A1 and FAR in postharvest kiwifruit. Postharvest Biol. Technol. 2021, 172, 111354. [Google Scholar] [CrossRef]
- Jia, H.F.; Lu, D.; Sun, J.H.; Li, C.L.; Xing, Y.; Qin, L.; Shen, Y.Y. Type 2C protein phosphatase ABI1 is a negative regulator of strawberry fruit ripening. J. Exp. Bot. 2013, 64, 1677–1687. [Google Scholar] [CrossRef]
- Daminato, M.; Guzzo, F.; Casadoro, G. A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 2013, 64, 3775–3786. [Google Scholar] [CrossRef]
- Han, Y.; Dang, R.; Li, J.; Jiang, J.; Zhang, N.; Jia, M.; Wei, L.; Li, Z.; Li, B.; Jia, W. SUCROSE NONFERMENTING1-RELATED PROTEIN KINASE2.6, an ortholog of OPEN STOMATA1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 2015, 167, 915–930. [Google Scholar] [CrossRef]
- Hou, B.Z.; Xu, C.; Shen, Y.Y. A leu-rich repeat receptor-like protein kinase, FaRIPK1, interacts with the ABA receptor, FaABAR, to regulate strawberry (Fragaria × ananassa) fruit ripening. J. Exp. Bot. 2018, 69, 1569–1582. [Google Scholar] [CrossRef]
- Li, D.D.; Mou, W.S.; Xia, R.; Li, L.; Zawora, C.; Ying, T.J.; Mao, L.C.; Liu, Z.C.; Luo, Z.S. Integrated analysis of high-throughput sequencing data shows abscisic acid-responsive genes and miRNAs in strawberry receptacle fruit ripening. Hortic. Res. 2019, 6, 26. [Google Scholar] [CrossRef]
- Xie, Y.G.; Ma, Y.Y.; Bi, P.P.; Wei, W.; Liu, J.; Hu, Y.; Gou, Y.J.; Zhu, D.; Wen, Y.Q.; Feng, J.Y. Transcription factor FvTCP9 promotes strawberry fruit ripening by regulating the biosynthesis of abscisic acid and anthocyanins. Plant Physiol. Biochem. 2020, 146, 374–383. [Google Scholar] [CrossRef]
- Liu, X.; Yue, Y.; Li, B.; Nie, Y.; Li, W.; Wu, W.H.; Ma, L.A.G. A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 2007, 315, 1712–1716. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Q.; Xin, Q.; Can, Z.; Liu, Z.Q.; Du, S.Y.; Mei, C.; Zhao, C.X.; Wang, X.F.; Shang, Y.; Jiang, T.; et al. The Magnesium-chelatase H Subunit Binds Abscisic Acid and Functions in Abscisic Acid Signaling: Newevidence in Arabidopsis. Plant Physiol. 2009, 150, 1940–1954. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.P.; Chen, P.; Ren, J.; Ji, K.; Li, Q.; Li, P.; Dai, S.J.; Leng, P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J. Exp. Bot. 2011, 62, 5659–5669. [Google Scholar] [CrossRef] [PubMed]
- Bastías, A.; Yañez, M.; Osorio, S.; Arbona, V.; Gómez-Cadenas, A.; Fernie, A.R.; Casaretto, J.A. The transcription factor AREB1 regulates primary metabolic pathways in tomato fruit. J. Exp. Bot. 2014, 65, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, P.; Lecourieux, D.; Kappel, C.; Cluzet, S.; Cramer, G.; Delrot, S.; Lecourieux, F. The basic leucine zipper transcription factor Abscisic Acid Response Element-Binding Factor2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol. 2014, 164, 365–383. [Google Scholar] [CrossRef]
- Shang, Y.; Yan, L.; Liu, Z.Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.Q.; Wang, X.F.; Du, S.Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef]
- Hobo, T.; Kowyama, Y.; Hattori, T. A bzip factor, trab1, interacts with vp1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 1999, 96, 15348–15353. [Google Scholar] [CrossRef]
- Kagaya, Y.; Hobo, T.; Murata, M.; Ban, A.; Hattori, T. Abscisic acid-induced transcription is mediated by phosphorylation of an abscisic acid response element binding factor, TRAB1. Plant Cell 2002, 14, 3177–3189. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Engineer, C.; Drosou, V.; Kalloniati, C.; Tani, E.; Tsaballa, A.; Kouri, E.D.; Ganopoulos, I.; Flemetakis, E.; Tsaftaris, A.S. The study of two barley type I-like MADS-box genes as potential targets of epigenetic regulation during seed development. BMC Plant Biol. 2012, 12, 166. [Google Scholar] [CrossRef]
- Lovisetto, A.; Guzzo, F.; Tadiello, A.; Toffali, K.; Favretto, A.; Casadoro, G. Molecular analyses of MADS-box genes trace back to gymnosperms the invention of fleshy fruits. Mol. Biol. Evol. 2012, 29, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.H.; Wu, J.; Zhang, Z.S.; Miao, Z.Q.; Zhao, P.X.; Wang, Z.; Xiang, C.B. Arabidopsis MADS-box transcription factor AGL21 acts as environment surveillance of seed germination by regulating ABI5 expression. Mol. Plant 2017, 10, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Tuan, P.A.; Bai, S.; Saito, T.; Ito, A.; Moriguchi, T. Dormancy-associated MADS-box (DAM) and Abscisic Acid pathway regulate pear endodormancy through a feedback mechanism. Plant Cell Physiol. 2017, 58, 1378–1390. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, C.C.; Burn, J.E.; Perez, P.P.; Metzger, J.; Edwards, W.J.; Peacock, W.J.; Dennis, E.S. The FLF MADS-box gene: Arepressor of flowering in Arabidopsis regulated by vernalization andmethylation. Plant Cell 1999, 11, 444–458. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Rouse, D.T.; Finnegan, E.J.; Peacock, W.J.; Dennis, E.S. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC). Proc. Natl. Acad. Sci. USA 2000, 97, 3753–3758. [Google Scholar] [CrossRef]
- Qi, X.L.; Liu, C.L.; Song, L.L.; Li, M. PaMADS7, a MADS-box transcription factor, regulates sweet cherry fruit ripening and softening. Plant Sci. 2020, 301, 110634. [Google Scholar] [CrossRef]
- Chen, R.C.; Mao, L.C.; Guan, W.L.; Wei, X.B.; Huang, Z.H.; Wu, Y.Y. ABA-mediated miR5290 promotes anthocyanin biosynthesis by inhibiting the expression of FaMADS1 in postharvest strawberry fruit. Postharvest Biol. Technol. 2022, 189, 111934. [Google Scholar] [CrossRef]
- Lu, W.J.; Chen, J.X.; Ren, X.C.; Yuan, J.J.; Han, X.Y.; Mao, L.C.; Ying, T.J.; Luo, Z.S. One novel strawberry MADS-box transcription factor FaMADS1a acts as a negative regulator in fruit ripening. Sci. Hortic. 2018, 227, 124–131. [Google Scholar] [CrossRef]
- Li, D.D.; Mou, W.S.; Luo, Z.S.; Li, L.; Limwachiranon, J.; Mao, L.C.; Ying, T.J. Developmental and stress regulation on expression of a novel miRNA, Fan-miR73, and its target ABI5 in strawberry. Sci. Rep. 2016, 6, 28385. [Google Scholar] [CrossRef]
- Ornelas-Paz, J.J.; Yahia, E.M.; Ramírez-Bustamante, N.; Pérez-Martínez, J.D.; Escalante-Minakata, M.P.; Ibarra-Junquera, V.; Acosta-Muñiz, C.; Guerrero-Prieto, V.; Ochoa-Reyes, E. Physical attributes and chemical composition of organic strawberry fruit (Fragaria ×ananassa Duch., cv. Albion) at six stages of ripening. Food Chem. 2013, 138, 372–381. [Google Scholar] [CrossRef]
- Kafkas, E.; Koşar, M.; Paydaş, S.; Kafkas, S.; Başer, K.H.C. Quality characteristics of strawberry genotypes at different maturation stages. Food Chem. 2007, 100, 1229–1236. [Google Scholar] [CrossRef]
- Lei, Y.; Liu, Y.Z.; Gu, Q.Q.; Yang, X.Y.; Deng, X.X.; Chen, J.Y. Comparison of cell wall metabolism in the pulp of three cultivars of ‘Nanfeng’ tangerine differing in mastication trait. J. Sci. Food Agric. 2012, 92, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Puryear, J.; Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Report. 1993, 11, 113–116. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Marathe, R.; Dinesh-Kumar, S.P. Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 2002, 30, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Vandendriessche, T.; Nicolai, B.M.; Hertog, M.L.A.T.M. Optimization of HS SPME Fast GC-MS for high-throughput analysis of strawberry aroma. Food Anal. Methods 2013, 6, 512–520. [Google Scholar] [CrossRef]
- Chen, J.X.; Mao, L.C.; Mi, H.B.; Lu, W.J.; Ying, T.J.; Luo, Z.S. Involvement of abscisic acid in postharvest water-deficit stress associated with the accumulation of anthocyanins in strawberry fruit. Postharvest Biol. Technol. 2016, 111, 99–105. [Google Scholar] [CrossRef]
- Andrews, P.K.; Shulin, L. Cell wall hydrolytic enzyme activity during development of nonclimacteric sweet cherry (Prunus avium L.) fruit. J. Hortic. Sci. 1995, 70, 561–567. [Google Scholar] [CrossRef]
- Mao, H.W.; Jin, G.K.; Sun, E.A.; Lee, A.Y.; Bae, T.M.; Kim, D.R.; Hwang, Y.S. Potential role of pectate lyase and Ca2+, in the increase in strawberry fruit firmness induced by short-term treatment with high-pressure CO2. J. Food Sci. 2014, 79, 685–692. [Google Scholar] [CrossRef]
- Pérez, A.G.; Sanz, C.; Olías, R.; Ríos, J.J.; Olías, J.M. Evolution of strawberry alcohol acyltransferase activity during fruit development and storage. J. Agric. Food Chem. 1996, 44, 3286–3290. [Google Scholar] [CrossRef]
- Lu, W.J.; Chu, X.Q.; Li, Y.Z.; Wang, C.; Guo, X.Q. Cotton GhMKK1 induces the tolerance of salt and drought stress, and mediates defence responses to pathogen infection in transgenic Nicotiana benthamiana. PLoS ONE 2013, 8, e68503. [Google Scholar] [CrossRef]
- Wei, X.P.; Lu, W.J.; Mao, L.C.; Han, X.Y.; Wei, X.B.; Zhao, X.X.; Xia, M.; Xu, C.J. ABF2 an MYB transcription factors regu;ate feruloyl transferase FHT involved in ABA-mediate wound suberization of kiwifruit. J. Exp. Bot. 2020, 71, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Min, T.; Yin, X.R.; Shi, Y.N.; Luo, Z.R.; Yao, Y.C.; Grierson, D.; Ferguson, I.B.; Chen, K.S. Ethylene-responsive transcription factors interact with promoters of ADH and PDC involved in persimmon (Diospyros kaki) fruit de-astringency. J. Exp. Bot. 2012, 63, 6393–6405. [Google Scholar] [CrossRef] [PubMed]
- Goto-Yamamoto, N.; Wan, G.H.; Masaki, K.; Kobayashi, S. Structure and transcription of three chalconesynthase genes of grapevine (Vilis vinifera). Plant Sci. 2002, 162, 867–872. [Google Scholar] [CrossRef]
- Brummell, D.A.; Harpster, M.H. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant Mol. Biol. 2001, 47, 311–339. [Google Scholar] [CrossRef]
- Cao, S.F.; Hu, Z.C.; Zheng, Y.H.; Lu, B.H. Effect of BTH on anthocyanin content and activities of related enzymes in strawberry after harvest. J. Agric. Food Chem. 2010, 58, 5801–5805. [Google Scholar] [CrossRef]
- Kou, X.H.; Yang, S.; Chai, L.P.; Zhou, J.Q.; Liu, Y.F.; Xue, Z.H. Abscisic acid and fruit ripening: Multifaceted analysis of the effect of abscisic acid on fleshy fruit ripening. Sci. Hortic. 2021, 281, 109999. [Google Scholar] [CrossRef]
- Pombo, M.A.; Martìnez, G.A.; Civello, P.M. Cloning of FaPAL6 gene from strawberry fruit and characterization of its expression and enzymatic activity in two cultivars with different anthocyanin accumulation. Plant Sci. 2011, 181, 111–118. [Google Scholar] [CrossRef]
- Lunkenbein, S.; Coiner, H.A.; Ric de Vos, C.H.; Schaart, J.G.; Boone, M.J.; Krens, F.A.; Schwab, W.; Salentijn, E.M.J. Molecular characterization of a stable antisense chalcone synthase phenotype in strawberry (Fragaria×ananassa). J. Agric. Food Chem. 2006, 54, 2145–2153. [Google Scholar] [CrossRef]
- Xu, W.; Peng, H.; Yang, T.; Whitaker, B.; Huang, L.; Sun, J.; Chen, P. Effect of calcium on strawberry fruit flavonoid pathway gene expression and anthocyanin accumulation. Plant Physiol. Biochem. 2014, 82, 289–298. [Google Scholar] [CrossRef]
- Cao, J. The pectin lyases in Arabidopsis thaliana: Evolution, selection and expression profiles. PLoS ONE 2012, 7, e46944. [Google Scholar] [CrossRef]
- Zabetakis, I.; Holden, M.A. Strawberry flavor: Analysis and biosynthesis. J. Sci. Food Agric. 1997, 74, 421–434. [Google Scholar] [CrossRef]
- Forney, C.F.; Kalt, W.; Jordan, M.A. The composition of strawberry aroma is influenced by cultivar, maturity and storage. Hortic. Sci. 2000, 35, 1022–1026. [Google Scholar] [CrossRef]
- Cumplido-Laso, G.; Medina-Puche, L.; Moyano, E.; Hoffmann, T.; Sinz, Q.; Ring, L.; Studart-Wittkowski, C.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J.; et al. The fruit ripening-related gene FaAAT2 encodes an acyl transferase involved in strawberry aroma biogenesis. J. Exp. Bot. 2012, 63, 4275–4290. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Hu, Z.; Deng, L.; Wang, Y.; Zhu, M.; Zhang, J.; Chen, G. A tomato MADS-box transcription factor, SlMADS1, acts as a negative regulator of fruit ripening. Plant Physiol. 2013, 163, 1026–1036. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Vivier, M.; Matsumoto, S.; Dry, I.B.; Thomas, M.R. A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Mol. Biol. 2001, 45, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Seymour, G.B.; Ryder, C.D.; Cevik, V.; Hammond, J.P.; Popovich, A.; King, G.J.; Vrebalov, J.; Giovannoni, J.J.; Manning, K. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 2011, 62, 1179–1188. [Google Scholar] [CrossRef]
- Pi, M.T.; Hu, S.Q.; Cheng, L.C.; Zhong, R.H.; Cai, Z.Y.; Liu, Z.C.; Yao, J.L.; Kang, C.Y. The MADS-box gene FveSEP3 plays essential roles in flower organogenesis and fruit development in woodland strawberry. Hortic. Res. 2021, 8, 247. [Google Scholar] [CrossRef]
- Elitzur, T.; Yakir, E.; Quansah, L.; Zhangjun, F.; Vrebalov, J.; Khayat, E.; Giovannoni, J.J.; Friedman, H. Banana MaMADS Transcription Factors Are Necessary for Fruit Ripening and Molecular Tools to Promote Shelf-Life and Food Security. Plant Physiol. 2016, 171, 380–391. [Google Scholar] [CrossRef]
- Fu, D.Q.; Zhu, B.Z.; Zhu, H.L.; Jiang, W.B.; Luo, Y.B. Virus-induced gene silencing in tomato fruit. Plant J. 2005, 43, 299–308. [Google Scholar] [CrossRef]
- Jia, H.F.; Chai, Y.M.; Li, C.L.; Lu, D.; Luo, J.J.; Qin, L.; Shen, Y.Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef]
- Hoffmann, T.; Kalinowski, G.; Schwab, W. RNAi-induced silencing of gene expression in strawberry fruit (Fragaria × ananassa) byagro-infiltration: A rapid assay for gene function analysis. Plant J. 2006, 48, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Mekontso, F.N.; Wu, S.H.; Fu, R.Z.; Li, W.; Meng, L.H.; Wang, Q.; Li, J.K.; Song, H.M.; Xu, X.B. Konckdown of Sly-miR160a using short tandem target mimic (STTM) enhanced expression of auxin signaling genes and delayed postharvest ripening of tomato fruit. Postharvest Biol. Technol. 2023, 198, 112271. [Google Scholar] [CrossRef]
- Jia, M.R.; Ding, N.; Zhang, Q.; Xing, S.N.; Wei, L.Z.; Zhao, Y.Y.; Du, P.; Mao, W.W.; Li, J.Z.; Li, B.B.; et al. A FERONIA-Like Receptor Kinase regulates strawberry (Fragaria × ananassa) fruit ripening and quality formation. Front. Plant Sci. 2017, 8, 01099. [Google Scholar] [CrossRef]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the Arabidopsis Abscisic Acid-Insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.F.; Wu, Y.R.; Xie, Q. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 2015, 20, 569. [Google Scholar] [CrossRef]
- Utsugi, S.; Ashikawa, I.; Nakamura, S.; Shibasaka, M. TaABI5, a wheat homolog of Arabidopsis thaliana ABA insensitive 5, controls seed germination. J. Plant Res. 2020, 133, 245–256. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).