A Robust Homogeneous Fluorescence Polarization Immunoassay for Rapid Determination of Erythromycin in Milk
Abstract
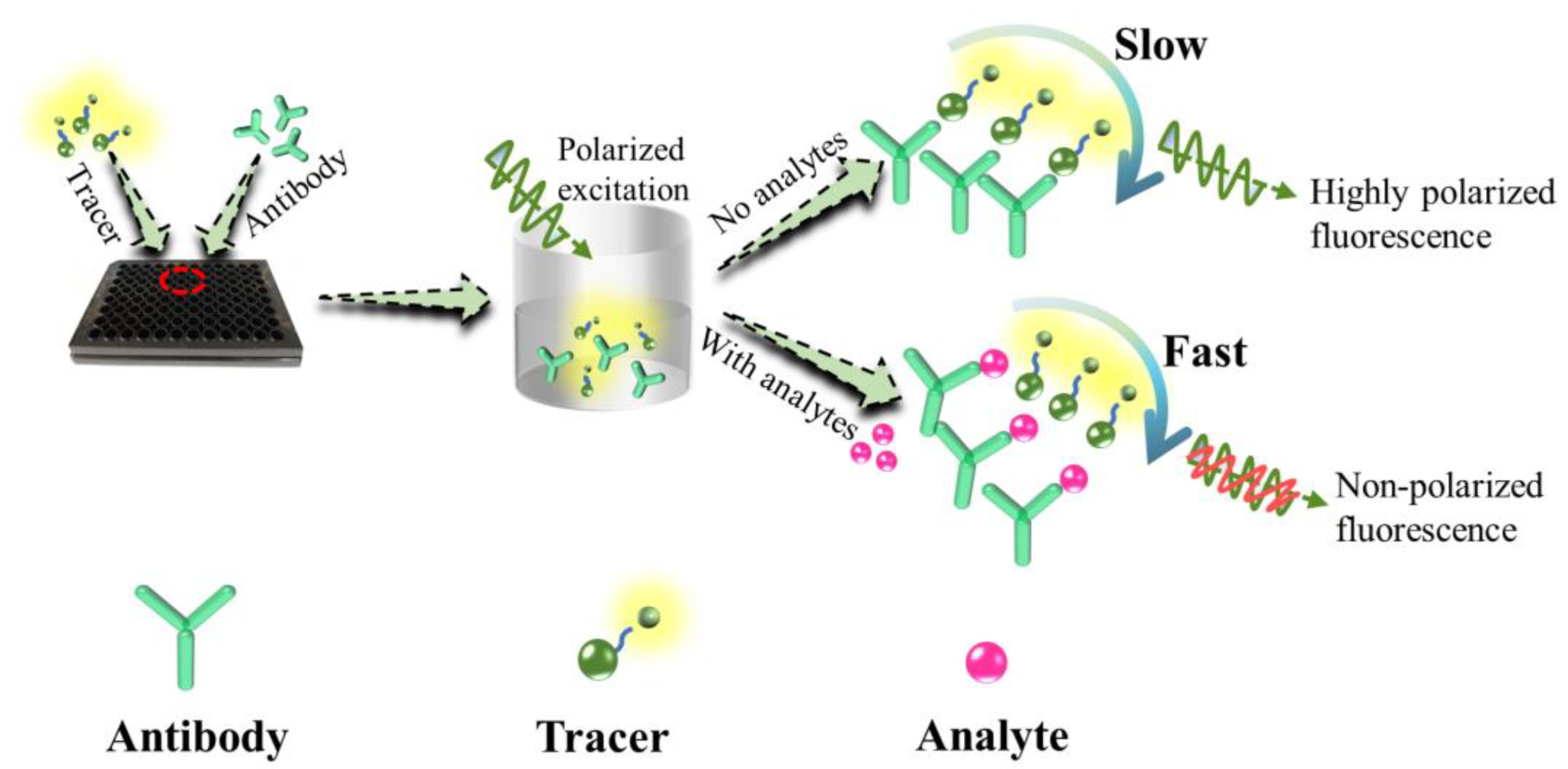
1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
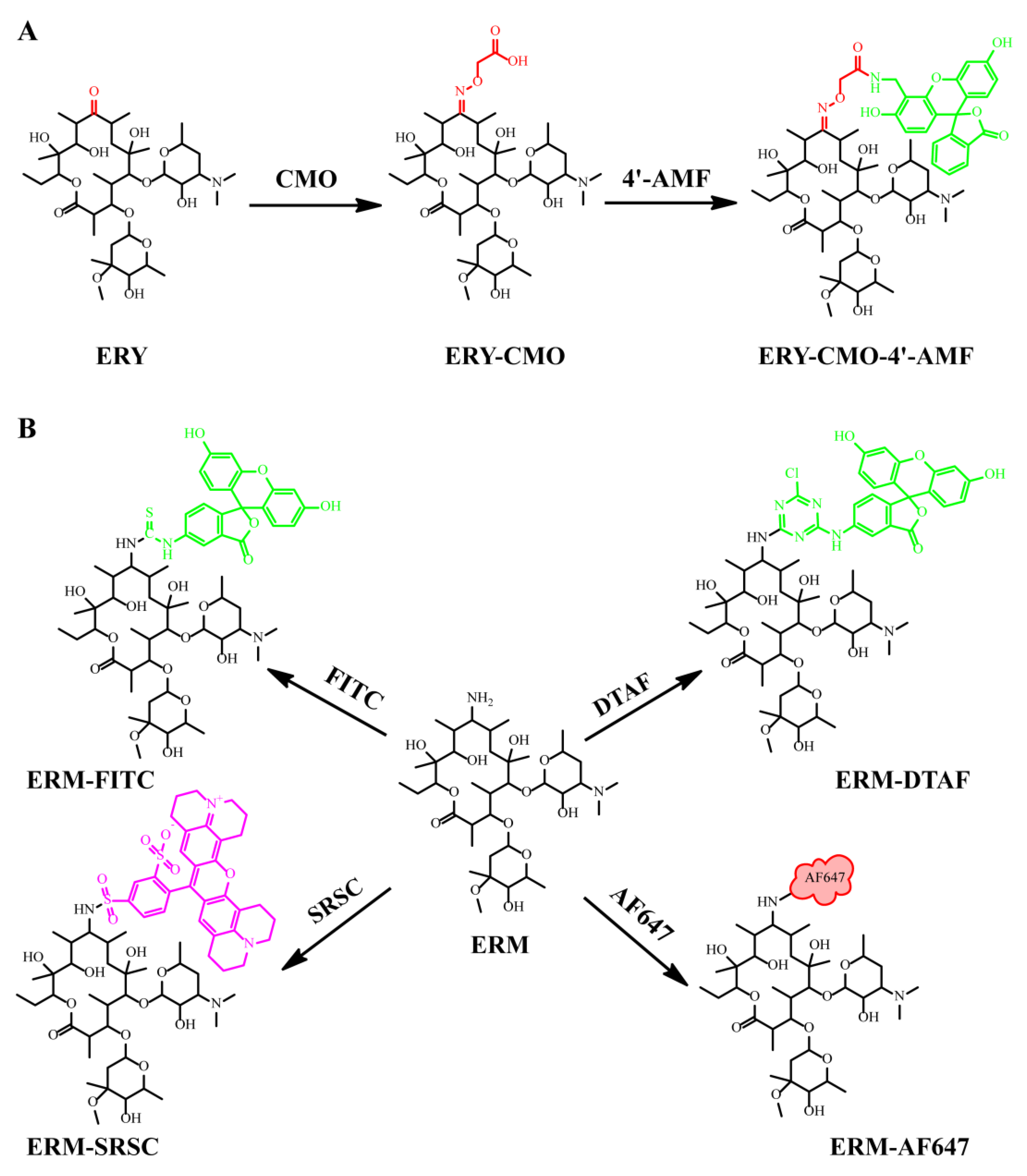
2.2. Synthesis and Characterization of Tracers
2.2.1. Synthesis and Purification of Tracers
2.2.2. Characterization of Tracers
2.3. Development of FPIA for ERM
2.3.1. The mAb Dilution Curve
2.3.2. The Calibration Curves
2.3.3. The FPIA Optimization
2.3.4. Specificity Evaluation of the FPIA
2.3.5. Preparation of the Milk Samples
3. Results and Discussions
3.1. Synthesis and Characterization of Tracers
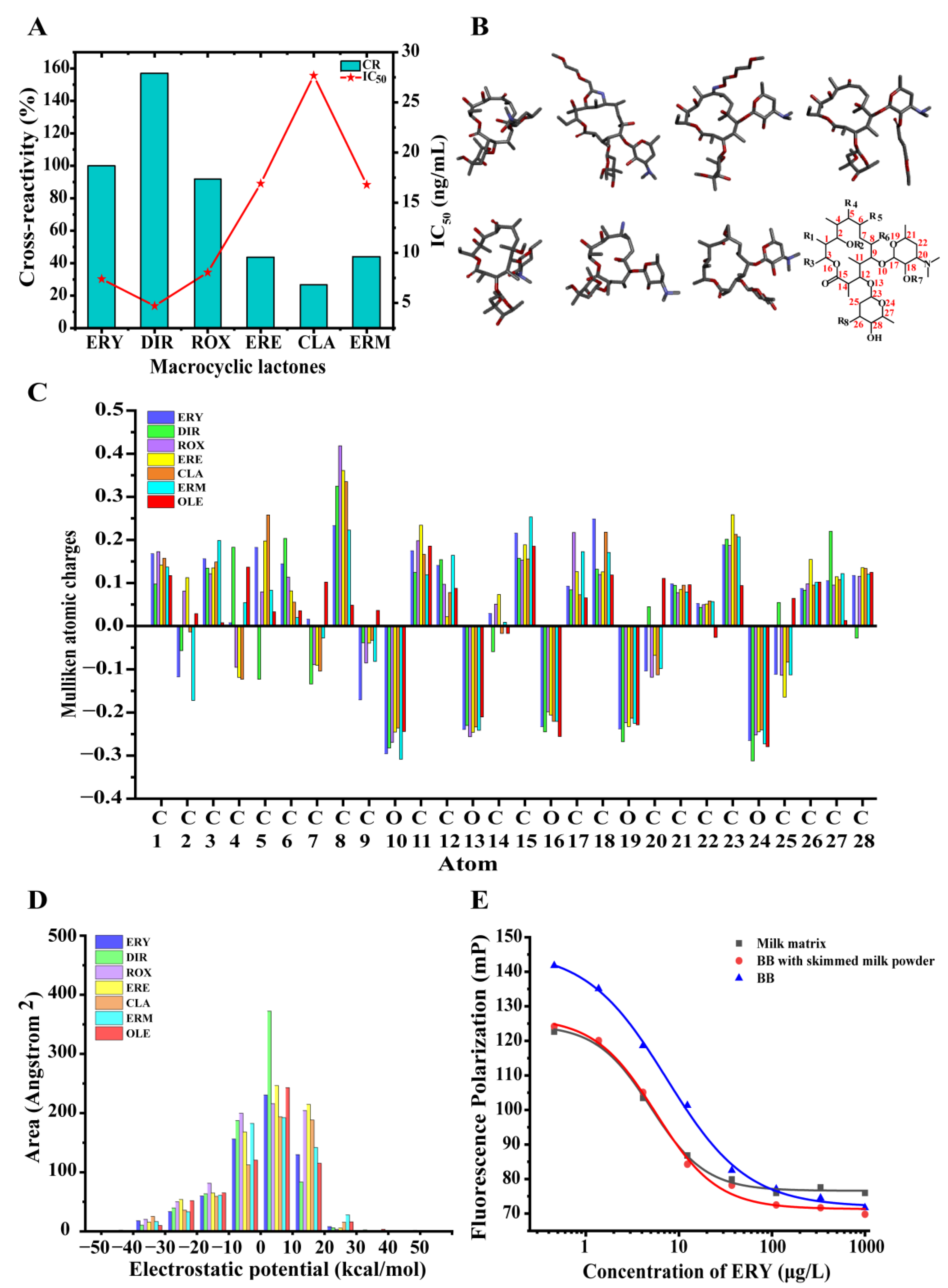
3.2. Selection of the mAb–Tracer Pairs
3.3. Development and Optimization of the FPIA for ERY
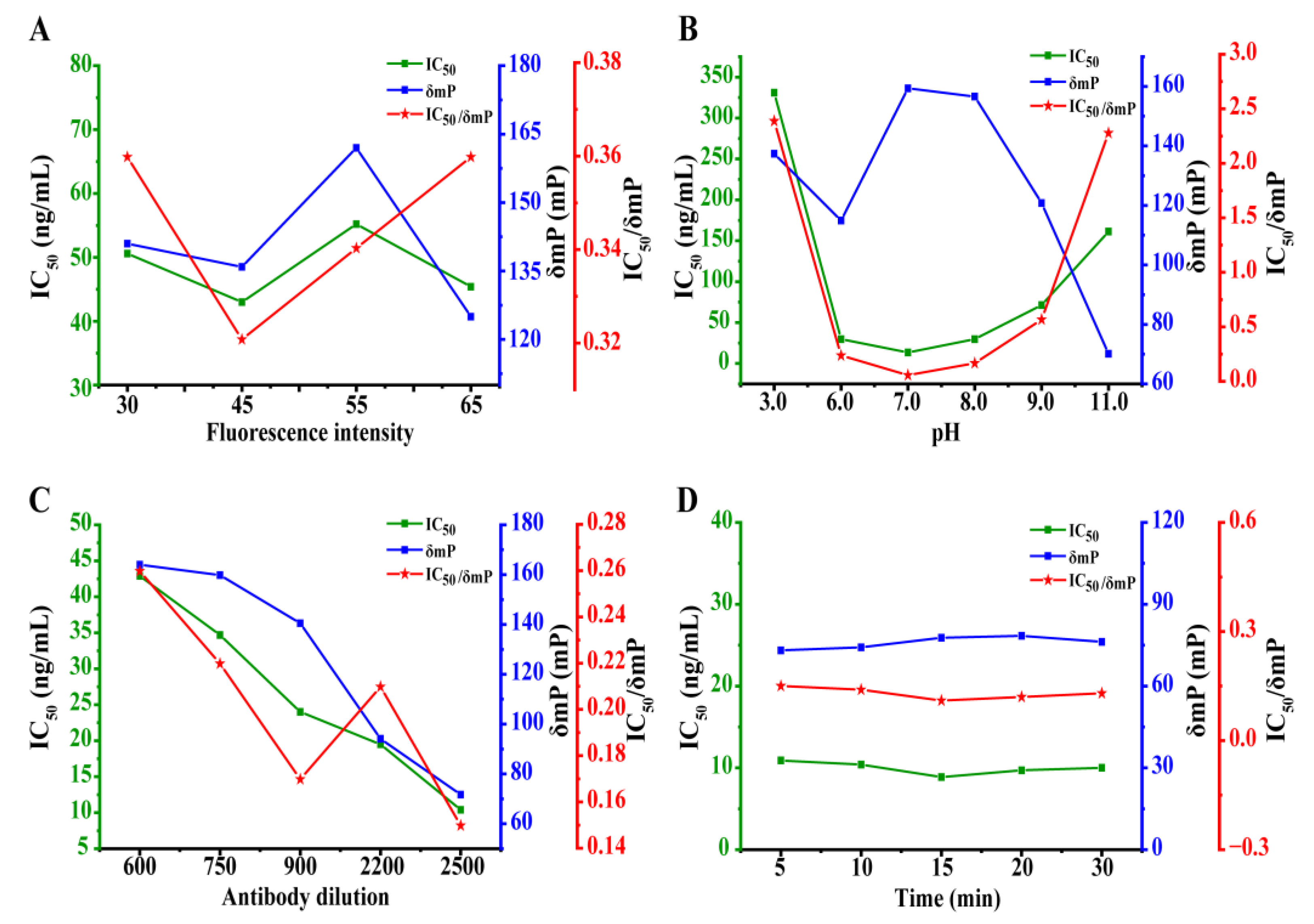
3.3.1. Optimization of the Tracer Concentration
3.3.2. The Effect of the Buffer pH
3.3.3. Optimization of the mAb 5B2 Concentration
3.3.4. The Study of Reaction Kinetic of the Competition
3.3.5. Specificity of the FPIA
3.4. Analysis of Milk Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Edder, P.; Coppex, L.; Cominoli, A.; Corvi, C. Analysis of Erythromycin and Oleandomycin Residues in Food by High-Performance Liquid Chromatography with Fluorometric Detection. Food Addit. Contam. 2002, 19, 232–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Leung, D.; Lenz, S.P. Determination of Five Macrolide Antibiotic Residues in Raw Milk Using Liquid Chromatography−Electrospray Ionization Tandem Mass Spectrometry. J. Agric. Food Chem. 2006, 54, 2873–2880. [Google Scholar] [CrossRef] [PubMed]
- McGlinchey, T.A.; Rafter, P.A.; Regan, F.; McMahon, G.P. A Review of Analytical Methods for the Determination of Aminoglycoside and Macrolide Residues in Food Matrices. Anal. Chim. Acta 2008, 624, 1–15. [Google Scholar] [CrossRef]
- Varaldo, P.E.; Montanari, M.P.; Giovanetti, E. Genetic Elements Responsible for Erythromycin Resistance in Streptococci. Antimicrob. Agents Chemother. 2009, 53, 343–353. [Google Scholar] [CrossRef]
- Zhang, A.; Song, L.; Liang, H.; Gu, Y.; Zhang, C.; Liu, X.; Zhang, J.; Zhang, M. Molecular Subtyping and Erythromycin Resistance of Campylobacter in China. J. Appl. Microbiol. 2016, 121, 287–293. [Google Scholar] [CrossRef]
- Desjardins, M.; Delgaty, K.L.; Ramotar, K.; Seetaram, C.; Toye, B. Prevalence and Mechanisms of Erythromycin Resistance in Group A and Group B Streptococcus: Implications for Reporting Susceptibility Results. J. Clin. Microbiol. 2004, 42, 5620–5623. [Google Scholar] [CrossRef]
- Martin, J.M.; Green, M.; Barbadora, K.A.; Wald, E.R. Erythromycin-Resistant Group A Streptococci in School Children in Pittsburgh. N. Engl. J. Med. 2002, 346, 1200–1206. [Google Scholar] [CrossRef]
- Ray, W.A.; Murray, K.T.; Meredith, S.; Narasimhulu, S.S.; Hall, K.; Michael Stein, C. Oral Erythromycin and the Risk of Sudden Death from Cardiac Causes. N. Engl. J. Med. 2004, 351, 1089–1096. [Google Scholar] [CrossRef]
- The Ministry of Agriculture and Rural Affairs of the People’s Republic of China. GB 31650-2019; National Food Safety Standard-Maximum Residue Limits for Veterinary Drugs in Foods. Standards Press of China: Beijing, China, 2019.
- The European Union. Commission Regulation (EU) No 37/2010 of 22 December 2009 on Pharmacologically Active Substances and Their Classification Regarding Maximum Residue Limits in Foodstuffs of Animal Origin; Official Journal of the European Union: Brussels, Belgium, 2010. [Google Scholar]
- The, U.S. Food and Drug Administration. 21CFR556.230. Specific Tolerances for Residues of Approved and Conditionally Approved New Animal Drugs, Sec. 556.230 Erythromycin. In The Code of Federal Regulations; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2023. [Google Scholar]
- Song, X.; Zhou, T.; Liu, Q.; Zhang, M.; Meng, C.; Li, J.; He, L. Molecularly Imprinted Solid-Phase Extraction for the Determination of Ten Macrolide Drugs Residues in Animal Muscles by Liquid Chromatography-tandem Mass Spectrometry. Food Chem. 2016, 208, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Hu, Q.; Zhao, M.; Cheng, Y.; Guo, Y.; Qian, H.; Yao, W. Simultaneous Determination of Erythromycin, Tetracycline, and Chloramphenicol Residue in Raw Milk by Molecularly Imprinted Polymer Mixed with Solid-Phase Extraction. Food Anal. Methods 2017, 11, 374–381. [Google Scholar] [CrossRef]
- Zhao, L.; Cao, W.; Xue, X.; Wang, M.; Wu, L.; Yu, L. Occurrence of Erythromycin and its Degradation Products Residues in Honey. Validation of an analytical method. J. Sep. Sci. 2017, 40, 1353–1360. [Google Scholar] [CrossRef]
- Park, M.J.; Park, M.S.; Lee, T.S.; Shin, I.S. A New Analytical Method for Erythromycin in Fish by Liquid Chromatography/Tandem Mass Spectrometry. Food Sci. Biotechnol. 2008, 17, 508–513. [Google Scholar]
- Lucchetti, D.; Fabrizi, L.; Esposito, A.; Guandalini, E.; Pasquale, M.D.; Coni, E. Simple Confirmatory Method for the Determination of Erythromycin Residues in Trout: A Fast Liquid−Liquid Extraction Followed by Liquid Chromatography−Tandem Mass Spectrometry. J. Agric. Food Chem. 2005, 53, 9689–9694. [Google Scholar] [CrossRef]
- Smith, D.S.; Eremin, S.A. Fluorescence Polarization Immunoassays and Related Methods for Simple, High-Throughput Screening of Small Molecules. Anal. Bioanal. Chem. 2008, 391, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Liu, J.; Wang, J.P. Development of a Dihydropteroate Synthase-Based Fluorescence Polarization Assay for Detection of Sulfonamides and Studying Its Recognition Mechanism. J. Agric. Food Chem. 2021, 69, 13953–13963. [Google Scholar] [CrossRef]
- Wang, Z.; Liang, X.; Wen, K.; Zhang, S.; Li, C.; Shen, J. A highly Sensitive and Class-Specific Fluorescence Polarisation Assay for Sulphonamides Based on Dihydropteroate Synthase. Biosens. Bioelectron. 2015, 70, 1–4. [Google Scholar] [CrossRef]
- Wang, Q.; Haughey, S.A.; Sun, Y.; Eremin, S.A.; Li, Z.; Liu, H.; Xu, Z.; Shen, Y.; Lei, H. Development of a Fluorescence Polarization Immunoassay for the Detection of Melamine in Milk and Milk Powder. Anal. Bioanal. Chem. 2011, 399, 2275–2284. [Google Scholar] [CrossRef]
- Zhang, Q.; Zou, M.; Wang, W.; Li, J.; Liang, X. Design, Synthesis, and Characterization of Tracers and Development of a Fluorescence Polarization Immunoassay for Rapid Screening of 4,4’-Dinitrocarbanilide in Chicken Muscle. Foods 2021, 10, 1822. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, S.; Ding, S.; Eremin, S.A.; Shen, J. Simultaneous Determination of Sulphamerazine, Sulphamethazine and Sulphadiazine in Honey and Chicken Muscle by a New Monoclonal Antibody-based Fluorescence Polarisation Immunoassay. Food Addit. Contam. 2008, 25, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Wang, Z.; Nesterenko, I.S.; Eremin, S.A.; Shen, J. Fluorescence Polarisation Immunoassay Based on a Monoclonal Antibody for the Detection of Sulphamethazine in Chicken Muscle. Int. J. Food Sci. Technol. 2007, 42, 36–44. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, J.; Tao, Z.; Eremin, S.A.; Hua, X.; Wang, M. Development of Fluorescence Polarization Immunoassay for Imidacloprid in Environmental and Agricultural Samples. Front. Chem. 2020, 8, 615594. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Cai, J.; Eremin, S.A.; Xiao, Z.; Shen, Y.; Tian, Y.; Xu, Z.; Yang, J.; Lei, H.; Sun, Y.; et al. Fluorescence Polarization Immunoassay for Alternaria Mycotoxin Tenuazonic Acid Detection and Molecular Modeling Studies of Antibody Recognition. Food Anal. Methods 2018, 11, 2455–2462. [Google Scholar] [CrossRef]
- Zhang, X.; Tang, Q.; Mi, T.; Zhao, S.; Wen, K.; Guo, L.; Mi, J.; Zhang, S.; Shi, W.; Shen, J.; et al. Dual-Wavelength Fluorescence Polarization Immunoassay to Increase Information Content Per Screen: Applications for Simultaneous Detection of Total Aflatoxins and Family Zearalenones in Maize. Food Control 2018, 87, 100–108. [Google Scholar] [CrossRef]
- Li, C.; Mi, T.; Conti, G.O.; Yu, Q.; Wen, K.; Shen, J.; Ferrante, M.; Wang, Z. Development of a Screening Fluorescence Polarization Immunoassay for the Simultaneous Detection of Fumonisins B1 and B2 in Maize. J. Agric. Food Chem. 2015, 63, 4940–4946. [Google Scholar] [CrossRef]
- Wang, Z.; Mi, T.; Beier, R.C.; Zhang, H.; Sheng, Y.; Shi, W.; Zhang, S.; Shen, J. Hapten Synthesis, Monoclonal Antibody Production and Development of a Competitive Indirect Enzyme-Linked Immunosorbent Assay for Erythromycin in Milk. Food Chem. 2015, 171, 98–107. [Google Scholar] [CrossRef]
- Guo, L.; Liu, M.; Li, Q.; Dong, B.; Li, H.; Mari, G.M.; Liu, R.; Yu, W.; Yu, X.; Wang, Z.; et al. Synthesis and Characterization of Tracers and Development of a Fluorescence Polarization Immunoassay for Amantadine with High Sensitivity in Chicken. J. Food Sci. 2021, 86, 4754–4767. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Zhao, S.; Li, H.; Wen, K.; Ke, Y.; Shen, J.; Zhang, S.; Shi, W.; Wang, Z. Design, Synthesis and Characterization of Tracers and Development of a Fluorescence Polarization Immunoassay for the Rapid Detection of Ractopamine in Pork. Food Chem. 2019, 271, 9–17. [Google Scholar] [CrossRef]
- Mi, T.; Wang, Z.; Eremin, S.A.; Shen, J.; Zhang, S. Simultaneous Determination of Multiple (Fluoro)quinolone Antibiotics in Food Samples by a One-Step Fluorescence Polarization Immunoassay. J. Agric. Food Chem. 2013, 61, 9347–9355. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y.; Wang, H.; Xu, Z.; Eremin, S.A.; Shen, Y.; Wu, Q.; Lei, H.; Sun, Y. Development of Fluorescence Polarization Immunoassay for Carbofuran in Food and Environmental Water Samples. Food Agric. Immunol. 2014, 26, 340–355. [Google Scholar] [CrossRef]
- Mi, T.; Liang, X.; Ding, L.; Zhang, S.; Eremin, S.A.; Beier, R.C.; Shen, J.; Wang, Z. Development and Optimization of a Fluorescence Polarization Immunoassay for Orbifloxacin in Milk. Anal. Methods 2014, 6, 3849–3857. [Google Scholar] [CrossRef]
- Gasilova, N.V.; Eremin, S.A. Determination of Chloramphenicol in Milk by a Fluorescence Polarization Immunoassay. J. Anal. Chem. 2010, 65, 255–259. [Google Scholar] [CrossRef]
- Thompson, T.S.; van den Heever, J.P. Degradation of Erythromycin in Honey and Selection of Suitable Marker Residues for Food Safety Analysis. Food Chem. 2012, 133, 1510–1520. [Google Scholar] [CrossRef]
- Galvidis, I.; Lapa, G.; Burkin, M. Group Determination of 14-Membered Macrolide Antibiotics and Azithromycin Using Antibodies Against Common Epitopes. Anal. Biochem. 2015, 468, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wen, K.; Chen, Y.; Wu, X.; Pei, X.; Wang, Q.; Liu, A.; Shen, J. Multiplex Immunogold Chromatographic Assay for Simultaneous Determination of Macrolide Antibiotics in Raw Milk. Food Anal. Methods 2015, 8, 2368–2375. [Google Scholar] [CrossRef]
- Tao, Y.; Yu, G.; Chen, D.; Pan, Y.; Liu, Z.; Wei, H.; Peng, D.; Huang, L.; Wang, Y.; Yuan, Z. Determination of 17 Macrolide Antibiotics and Avermectins Residues in Meat with Accelerated Solvent Extraction by Liquid Chromatography-Tandem Mass Spectrometry. J. Chromatogr. B 2012, 897, 64–71. [Google Scholar] [CrossRef] [PubMed]
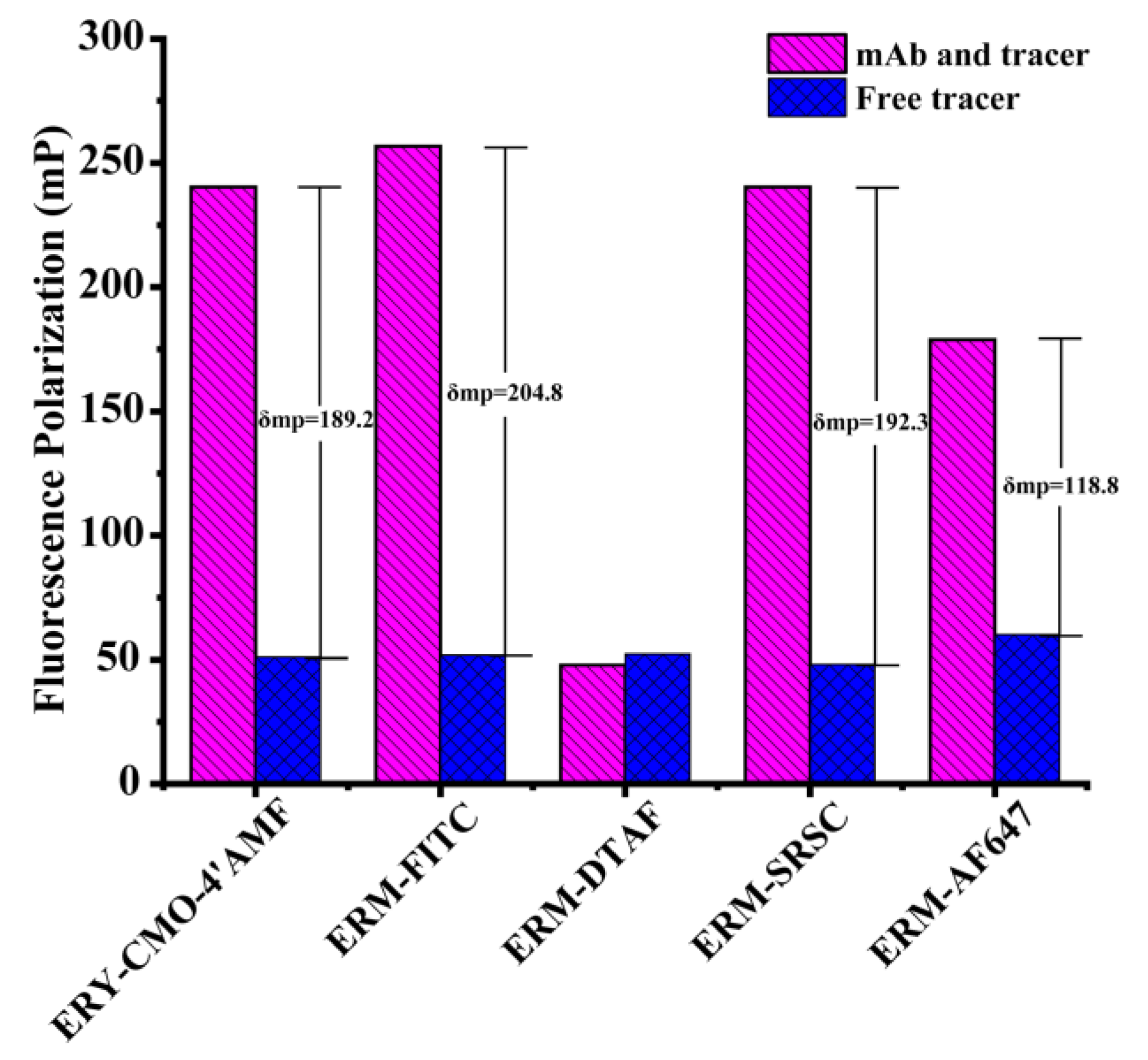
| Tracers | mAbs | δmP (mP) | Antibody Dilution | IC50 (ng/mL) |
|---|---|---|---|---|
| ERY-CMO-4′-AMF | mAb 5B2 | 145 | 1/1500 | 80.3 |
| mAb 6C2 | 164 | 1/2500 | 107.4 | |
| mAb 6D9 | 183 | 1/2000 | 92.0 | |
| ERM-FITC | mAb 5B2 | 210 | 1/1000 | 24.0 |
| mAb 6C2 | 109 | 1/1200 | 154.3 | |
| mAb 6D9 | 225 | 1/1700 | 34.7 | |
| ERM-SRSC | mAb 5B2 | 97 | 1/600 | 62.1 |
| mAb 6C2 | 90 | 1/1600 | 390.0 | |
| mAb 6D9 | 76 | 1/500 | 159.5 | |
| ERM-AF647 | mAb 5B2 | 156 | 1/3000 | 13.0 |
| mAb 6C2 | 177 | 1/10,000 | 85.0 | |
| mAb 6D9 | 142 | 1/3000 | 65.9 |
| Sample | LOD (μg/L) | Spiked Level (μg/L) | Recovery (%) | CVs (%) |
|---|---|---|---|---|
| milk | 14.08 | 50 | 107.77 | 6.15 |
| 100 | 97.33 | 3.41 | ||
| 150 | 96.08 | 10.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, C.; Zhang, H.; Zhang, Y.; Li, Q.; Li, P.; Mari, G.M.; Eremin, S.A.; Shen, J.; Wang, Z. A Robust Homogeneous Fluorescence Polarization Immunoassay for Rapid Determination of Erythromycin in Milk. Foods 2023, 12, 1581. https://doi.org/10.3390/foods12081581
Duan C, Zhang H, Zhang Y, Li Q, Li P, Mari GM, Eremin SA, Shen J, Wang Z. A Robust Homogeneous Fluorescence Polarization Immunoassay for Rapid Determination of Erythromycin in Milk. Foods. 2023; 12(8):1581. https://doi.org/10.3390/foods12081581
Chicago/Turabian StyleDuan, Changfei, Huiyan Zhang, Yingjie Zhang, Qiang Li, Peipei Li, Ghulam Mujtaba Mari, Sergei A. Eremin, Jianzhong Shen, and Zhanhui Wang. 2023. "A Robust Homogeneous Fluorescence Polarization Immunoassay for Rapid Determination of Erythromycin in Milk" Foods 12, no. 8: 1581. https://doi.org/10.3390/foods12081581
APA StyleDuan, C., Zhang, H., Zhang, Y., Li, Q., Li, P., Mari, G. M., Eremin, S. A., Shen, J., & Wang, Z. (2023). A Robust Homogeneous Fluorescence Polarization Immunoassay for Rapid Determination of Erythromycin in Milk. Foods, 12(8), 1581. https://doi.org/10.3390/foods12081581






