Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Cheese and Related Food Processing Surfaces
2.2. L. monocytogenes Isolation
2.3. Antibiotic Resistance Profiles of L. monocytogenes Strains
2.4. Whole-Genome Sequencing of Bacterial Isolates
2.4.1. DNA Extraction and Sequencing
2.4.2. Bioinformatics Analyses
3. Results
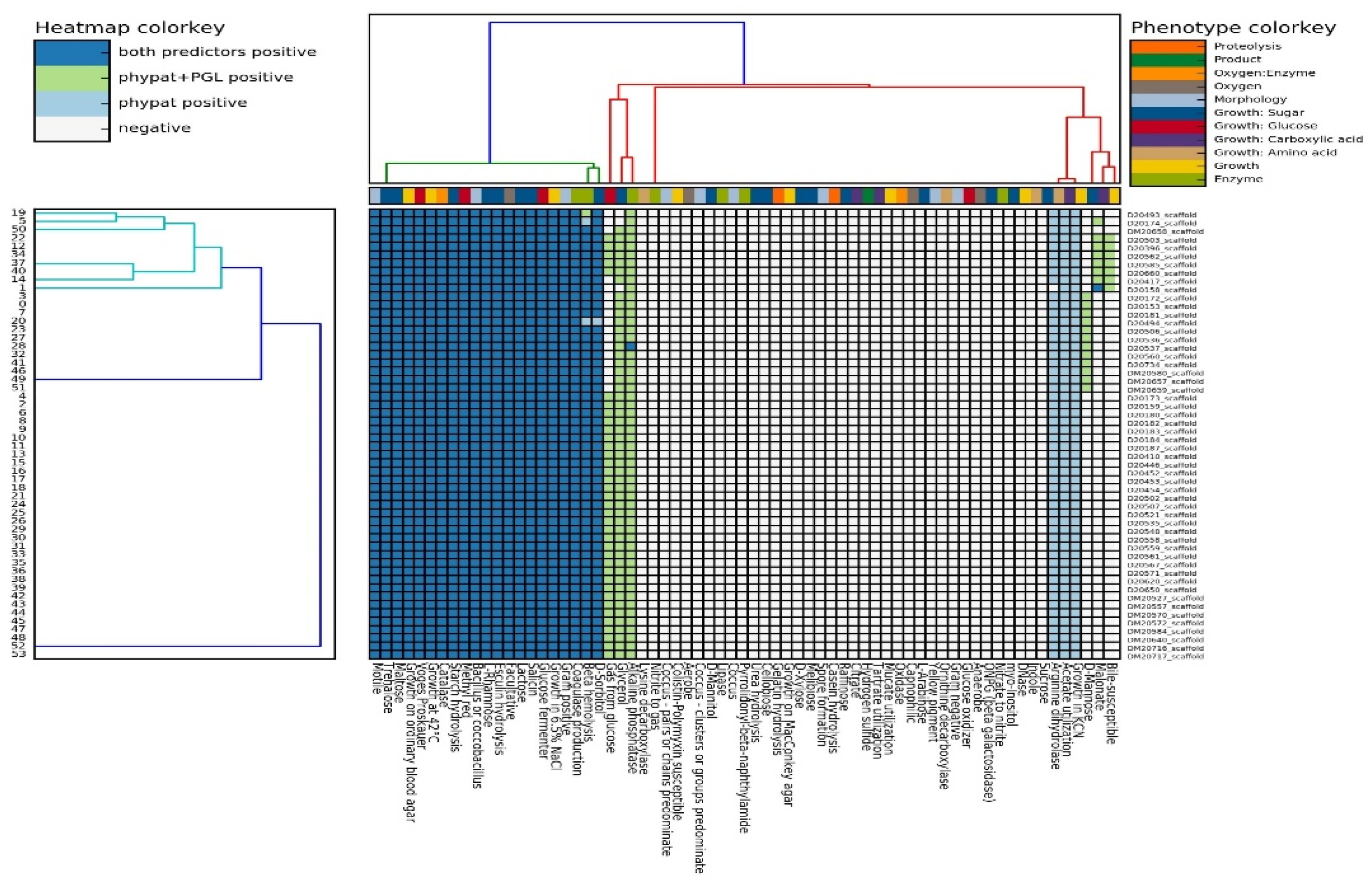
3.1. Phenotypic Characterization of L. monocytogenes Isolates
3.2. Antibiograms of L. monocytogenes Cheese and Surface Isolates
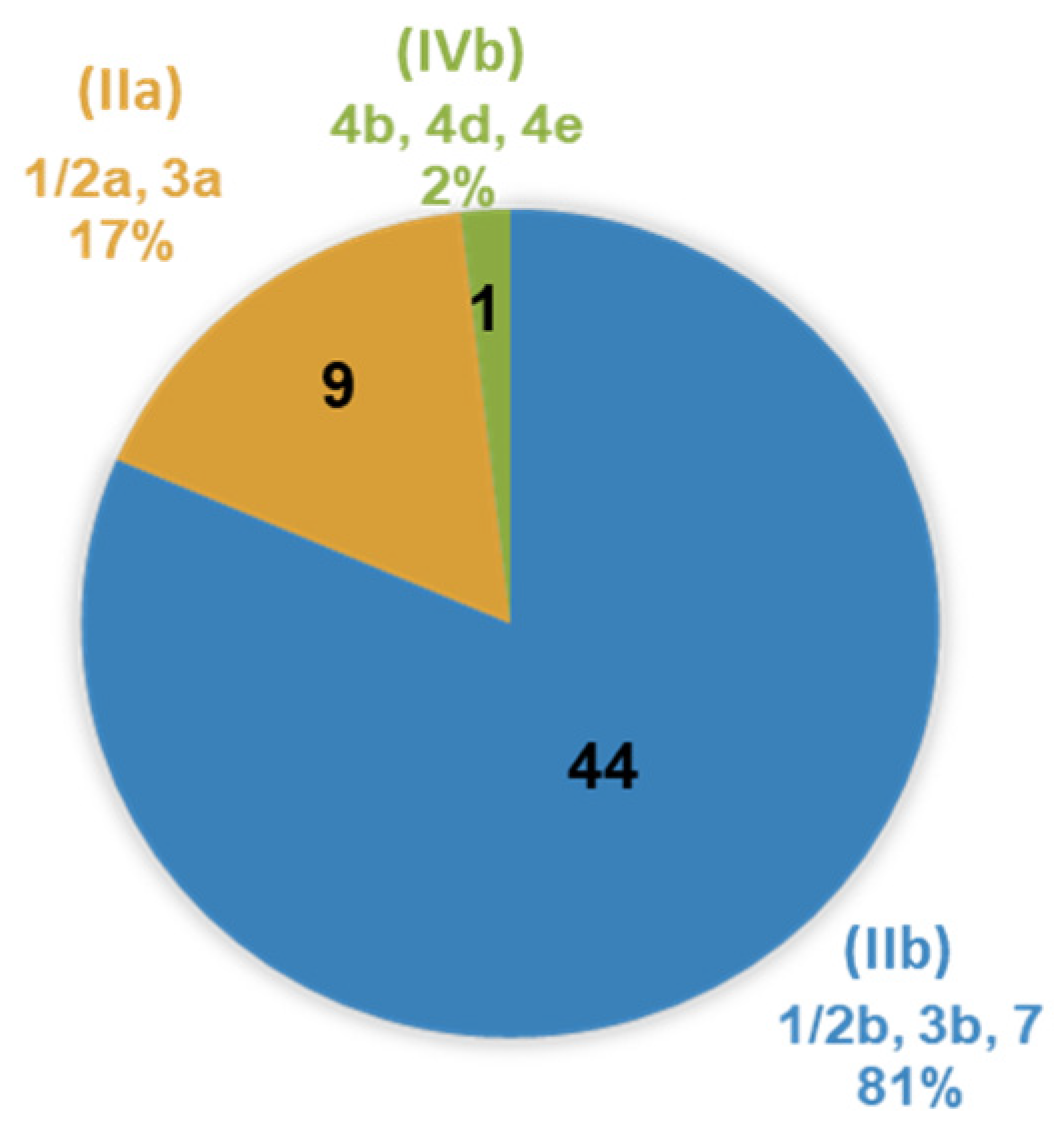
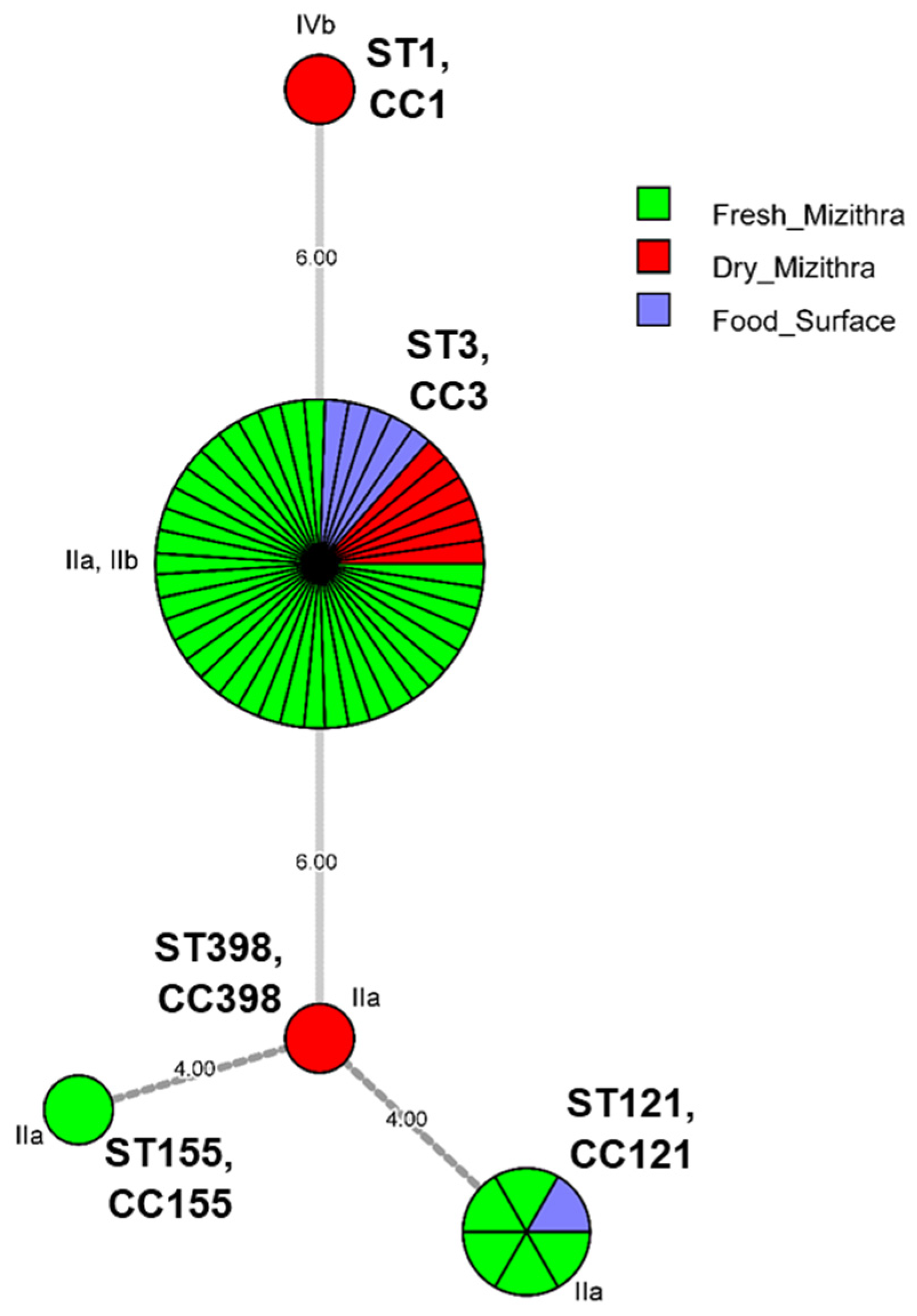
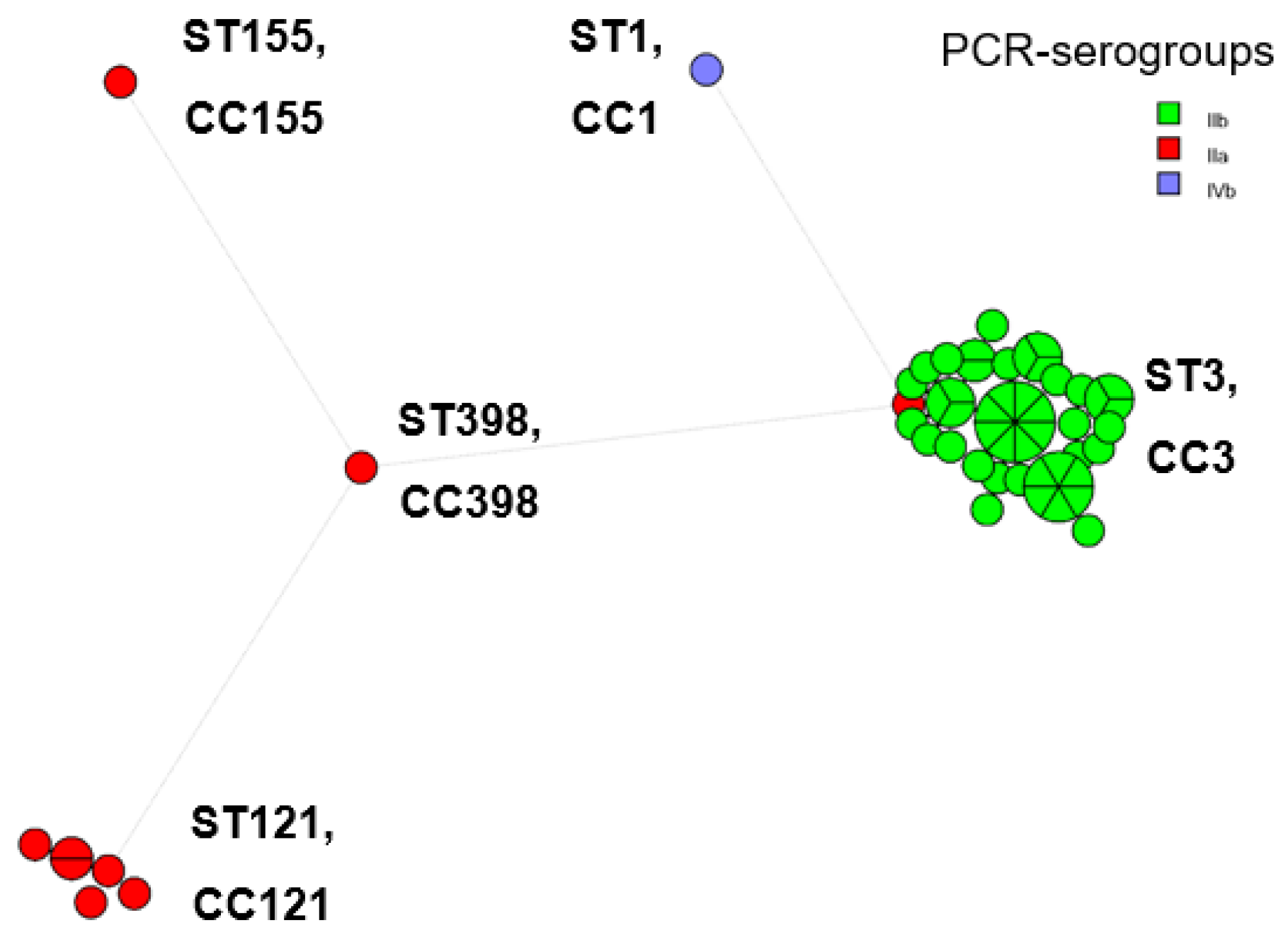
3.3. Genetic Diversity of L. monocytogenes Isolates
3.4. Analysis of Resistance, Virulence and Persistence of L. monocytogenes Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alegbeleye, O.O.; Sant’Ana, A.S. Survival and growth behaviour of Listeria monocytogenes in ready-to-eat vegetable salads. Food Control 2022, 138, 109023. [Google Scholar] [CrossRef]
- Alegbeleye, O.O.; Singleton, I.; Sant’Ana, A.S. Sources and contamination routes of microbial pathogens to fresh produce during field cultivation: A review. Food Microbiol. 2018, 73, 177–208. [Google Scholar] [CrossRef]
- Buchanan, R.L.; Gorris, L.G.M.; Hayman, M.M.; Jackson, T.C.; Whiting, R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 2017, 75, 1–13. [Google Scholar] [CrossRef]
- Huang, J.; Luo, Y.; Zhou, B.; Zheng, J.; Nou, X. Growth and survival of Salmonella enterica and Listeria monocytogenes on fresh-cut produce and their juice extracts: Impacts and interactions of food matrices and temperature abuse conditions. Food Control 2019, 100, 300–304. [Google Scholar] [CrossRef]
- Townsend, A.; Strawn, L.K.; Chapman, B.J.; Dunn, L.L. A systematic review of Listeria species and Listeria monocytogenes prevalence, persistence, and diversity throughout the fresh produce supply chain. Foods 2021, 10, 1427. [Google Scholar] [CrossRef] [PubMed]
- Acciari, V.A.; Ruolo, A.; Torresi, M.; Ricci, L.; Pompei, A.; Marfoglia, C.; Valente, F.M.; Gentorola, G.; Conte, A.; Salini, R.; et al. Genetic diversity of Listeria monocytogenes strains contaminating food and food producing environment as single based sample in Italy (retrospective study). Int. J. Food Microbiol. 2022, 366, 109562. [Google Scholar] [CrossRef]
- Manios, S.G.; Grivokostopoulos, N.C.; Bikouli, V.C.; Doultsos, D.A.; Zilelidou, E.A.; Gialitaki, M.A.; Skandamis, P.N. A 3-year hygiene and safety monitoring of a meat processing plant which uses materials of global origin. Int. J. Food Microbiol. 2015, 209, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Baltar, A.; Pérez-Boto, D.; Medina, M.; Montiel, R. Genomic diversity and characterization of Listeria monocytogenes from dry-cured ham processing plants. Food Microbiol. 2021, 99, 103779. [Google Scholar] [CrossRef] [PubMed]
- Zacharski, K.A.; Southern, M.; Ryan, A.; Adley, C.C. Evaluation of an environmental monitoring program for the microbial safety of air and surfaces in a dairy plant environment. J. Food Prot. 2018, 81, 1108–1116. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authortiy); ECDC (European Centre for Disease Prevention and Control). The European Union one health zoonoses report. EFSA J. 2021, 19, e06971. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States–Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, P.; Cilla, G.; López-Olaizola, M.; Vicente, D.; Marimón, J.M. Epidemiology and clinical features of listeriosis in Gipuzkoa, Spain, 2010-2020. Front. Microbiol. 2022, 13, 894334. [Google Scholar] [CrossRef] [PubMed]
- CDC (Centers for Disease Control and Prevention). Listeria (Listeriosis): Symptoms. Available online: https://www.cdc.gov/listeria/symptoms.html (accessed on 26 July 2022).
- WHO (World Health Organization). Listeriosis. Available online: https://www.who.int/news-room/fact-sheets/detail/listeriosis (accessed on 26 July 2022).
- Adrião, A.; Vieira, M.; Fernandes, I.; Barbosa, M.; Sol, M.; Tenreiro, R.P.; Chambel, M.; Barata, B.; Zilhao, I.; Shama, G.; et al. Marked intra-strain variation in response of Listeria monocytogenes dairy isolates to acid or salt stress and the effect of acid or salt adaptation on adherence to abiotic surfaces. Int. J. Food Microbiol. 2008, 123, 142–150. [Google Scholar] [CrossRef]
- Beales, N. Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: A review. Compr. Rev. Food Sci. Food Saf. 2004, 3, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Chikindas, M.L. Listeria: A foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 2007, 113, 1–15. [Google Scholar] [CrossRef]
- Lakicevic, B.Z.; den Besten, H.M.W.; De Blase, D. Landscape of stress response and virulence genes among Listeria monocytogenes strains. Front. Microbiol. 2022, 12, 738470. [Google Scholar] [CrossRef]
- Myintzaw, P.; Pennone, V.; McAuliffe, O.; Begley, M.; Callanan, M. Correlation of organic acid tolerance and genotypic characteristics of Listeria monocytogenes food and clinical isolates. Food Microbiol. 2022, 104, 104004. [Google Scholar] [CrossRef]
- Osek, J.; Lachtara, B.; Wieczorek, K. Listeria monocytogenes—How this pathogen survives in food-production environments? Front. Microbiol. 2022, 13, 866462. [Google Scholar] [CrossRef] [PubMed]
- Skandamis, P.N.; Yoon, Y.; Stopforth, J.D.; Kendall, P.A.; Sofos, J.N. Heat and acid tolerance of Listeria monocytogenes after exposure to single and multiple sublethal stresses. Food Microbiol. 2008, 25, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Kokkoni, E.-A.; Andritsos, N.; Sakarikou, C.; Michailidou, S.; Argiriou, A.; Giaouris, E. Investigating transcriptomic induction of resistance and/or virulence in Listeria monocytogenes cells surviving sublethal antimicrobial exposure. Foods 2021, 10, 2382. [Google Scholar] [CrossRef]
- Pateiro, M.; Munekata, P.E.S.; Sant’Ana, A.S.; Domínguez, R.; Rodríguez-Lazaro, D.; Lorenzo, J.M. Application of essential oils as antimicrobial agents against spoilage and pathogenic microorganisms in meat products. Int. J. Food Microbiol. 2021, 337, 108966. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial biofilms and their implications in pathogenesis and food safety. Foods 2021, 10, 2117. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Chrysadakou, M.; Tzakoniati, A.; Bikouli, V.C.; Nychas, G.-J.; Skandamis, P.N. Variability of Listeria monocytogenes strains in biofilm formation on stainless steel and polystyrene materials and resistance to peracetic acid and quaternary ammonium compounds. Int. J. Food Microbiol. 2016, 237, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, B.; Cerf, O. Review—Persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 2011, 145, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, V.; Wiedmann, M.; Teixeira, P.; Stasiewicz, M.J. Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. J. Food Prot. 2014, 77, 150–170. [Google Scholar] [CrossRef]
- Toro, M.; Williams-Vergara, J.; Solar, C.; Quesille-Villalobos, A.M.; Kwon, H.J.; Navarrete, P.; Meng, J.; Chen, Y.; Reyes-Jara, A. Evaluation of the persistence and characterization of Listeria monocytogenes in foodservice operations. Foods 2022, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Kousta, M.; Mataragas, M.; Skandamis, P.; Drosinos, E.H. Prevalence and sources of cheese contamination with pathogens at farm and processing levels. Food Control 2010, 21, 805–815. [Google Scholar] [CrossRef]
- Andritsos, N.D.; Kallitsis, T.; Roukas, D. Growth potential of Listeria monocytogenes in ready-to-eat Feta cheese-based sauce stored at 4 °C. J. Food Saf. 2019, 39, e12599. [Google Scholar] [CrossRef]
- Angelidis, A.S.; Boutsiouki, P.; Papageorgiou, D.K. Loss of viability of Listeria monocytogenes in contaminated processed cheese during storage at 4, 12 and 22 °C. Food Microbiol. 2010, 27, 809–818. [Google Scholar] [CrossRef]
- Giannou, E.; Kakouri, A.; Matijašic, B.B.; Rogelj, I.; Samelis, J. Fate of Listeria monocytogenes on fully ripened Greek Graviera cheese stored at 4, 12, or 25 °C in air or vacuum packages: In situ PCR detection of a cocktail of bacteriocins potentially contributing to pathogen inhibition. J. Food. Prot. 2009, 72, 531–538. [Google Scholar] [CrossRef]
- Jesus, A.L.T.; Fernandes, M.S.; Kamimura, B.A.; Prado-Silva, L.; Silva, R.; Esmerino, E.A.; Cruz, A.G.; Sant’Ana, A.S. Growth potential of Listeria monocytogenes in probiotic cottage cheese formulations with reduced sodium content. Food Res. Int. 2016, 81, 180–187. [Google Scholar] [CrossRef]
- Lahou, E.; Uyttendaele, M. Growth potential of Listeria monocytogenes in soft, semi-soft and semi-hard artisanal cheeses after post-processing contamination in deli retail establishments. Food Control 2017, 76, 13–23. [Google Scholar] [CrossRef]
- Mataragas, M.; Stergiou, V.; Nychas, G.-J.E. Modeling survival of Listeria monocytogenes in the traditional Greek soft cheese Katiki. J. Food Prot. 2008, 71, 1835–1845. [Google Scholar] [CrossRef] [PubMed]
- Rogga, K.J.; Samelis, J.; Kakouri, A.; Katsiari, M.C.; Savvaidis, I.N.; Kontominas, M.G. Survival of Listeria monocytogenes in Galotyri, a traditional Greek soft acid-curd cheese, stored aerobically at 4 °C and 12 °C. Int. Dairy J. 2005, 15, 59–67. [Google Scholar] [CrossRef]
- Samelis, J.; Giannou, E.; Lianou, A. Assuring growth inhibition of listerial contamination during processing and storage of traditional Greek Graviera cheese: Compliance with the new European Union regulatory criteria for Listeria monocytogenes. J. Food Prot. 2009, 72, 2264–2271. [Google Scholar] [CrossRef]
- Papageorgiou, D.K.; Bori, M.; Mantis, A. Growth of Listeria monocytogenes in the whey cheeses Myzithra, Anthotyros, and Manouri during storage at 5, 12, and 22 °C. J. Food Prot. 1996, 59, 1193–1199. [Google Scholar] [CrossRef]
- Sameli, N.; Samelis, J. Growth and biocontrol of Listeria monocytogenes in Greek Anthotyros whey cheese without or with a crude enterocin A-B-P extract: Interactive effects of the native spoilage microbiota during vacuum-packed storage at 4 °C. Foods 2022, 11, 334. [Google Scholar] [CrossRef]
- Vasileiadi, N.; Lappa, A.; Koukouvinos, C.; Tsironi, T.; Mandilara, G. Challenge test for assessing the growth potential of Listeria monocytogenes in Greek soft cheese (Anthotyros). Appl. Sci. 2022, 12, 12349. [Google Scholar] [CrossRef]
- Angelidis, A.S.; Georgiadou, S.S.; Zafeiropoulou, V.; Velonakis, E.N.; Papageorgiou, D.K.; Vatopoulos, A. A survey of soft cheeses in Greek retail outlets highlights a low prevalence of Listeria spp. Dairy Sci. Technol. 2012, 92, 189–201. [Google Scholar] [CrossRef]
- Martinez-Rios, V.; Dalgaard, P. Prevalence of Listeria monocytogenes in European cheeses: A systematic review and meta-analysis. Food Control 2018, 84, 205–214. [Google Scholar] [CrossRef]
- Datta, A.R.; Burall, L.S. Serotype to genotype: The changing landscape of listeriosis outbreak investigations. Food Microbiol. 2018, 75, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Kathariou, S. Listeria monocytogenes virulence and pathogenicity, a food safety perspective. J. Food Prot. 2002, 65, 1811–1829. [Google Scholar] [CrossRef] [PubMed]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Liu, D. Epidemiology. Handbook of Listeria monocytogenes; Liu, D., Ed.; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2006; pp. 27–60. [Google Scholar]
- Andritsos, N.D.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serogroup 1/2 strains have a competitive growth advantage over serotype 4b during refrigerated storage of an artificially contaminated ready-to-eat pork meat product. Appl. Sci. 2021, 11, 6096. [Google Scholar] [CrossRef]
- Zilelidou, E.; Karmiri, C.-V.; Zoumpopoulou, G.; Mavrogonatou, E.; Kletsas, D.; Tsakalidou, E.; Papadimitriou, K.; Drosinos, E.; Skandamis, P. Listeria monocytogenes strains underrepresented during selective enrichment with an ISO method might dominate during passage through simulated gastric fluid and in vitro infection of Caco-2 cells. Appl. Environ. Microbiol. 2016, 82, 6846–6858. [Google Scholar] [CrossRef]
- Muhterem-Uyar, M.; Dalmasso, M.; Bolocan, A.S.; Hernandez, M.; Kapetanakou, A.E.; Kuchta, T.; Manios, S.G.; Melero, B.; Minarovičová, J.; Nicolau, A.I.; et al. Environmental sampling for Listeria monocytogenes control in food processing facilities reveals three contamination scenarios. Food Control 2015, 51, 94–107. [Google Scholar] [CrossRef]
- Hadjilouka, A.; Andritsos, N.D.; Paramithiotis, S.; Mataragas, M.; Drosinos, E.H. Listeria monocytogenes serotype prevalence and biodiversity in diverse food products. J. Food Prot. 2014, 77, 2115–2120. [Google Scholar] [CrossRef]
- Henri, C.; Félix, B.; Guiller, L.; Leekitcharoenphon, P.; Michelon, D.; Mariet, J.-F.; Aarestrup, F.M.; Mistou, M.-Y.; Hendriksen, R.S.; Roussel, S. Population genetic structure of Listeria monocytogenes strains as determined by pulsed-field gel electrophoresis and multilocus sequence typing. Appl. Environ. Microbiol. 2016, 82, 5720–5728. [Google Scholar] [CrossRef]
- Stessl, B.; Fricker, M.; Fox, E.; Karpiskova, R.; Demnerova, K.; Jordan, K.; Ehling-Schulz, M.; Wagner, M. Collaborative survey of the colonization of different types of cheese-processing facilities with Listeria monocytogenes. Foodborne Pathog. Dis. 2014, 11, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Dessai, U.; McGarry, S.; Gerner-Smidt, P. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef]
- Ribot, E.M.; Freeman, M.; Hise, K.B.; Gerner-Smidt, P. Pulse-Net: Entering the age of next-generation sequencing. Foodborne Pathog. Dis. 2019, 16, 451–456. [Google Scholar] [CrossRef]
- Alvarez-Molina, A.; Cobo-Díaz, J.F.; López, M.; Prieto, M.; de Toro, M.; Alvarez-Ordóñez, A. Unraveling the emergence and population diversity of Listeria monocytogenes in a newly built meat facility through whole genome sequencing. Int. J. Food Microbiol. 2021, 340, 109043. [Google Scholar] [CrossRef]
- Palaiodimou, L.; Fanning, S.; Fox, E.M. Genomic insights into persistence of Listeria monocytogenes in the food processing environment. J. Appl. Microbiol. 2021, 131, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Unrath, N.; McCabe, E.; Macori, G.; Fanning, S. Application of whole genome sequencing to aid in deciphering the persistence potential of Listeria monocytogenes in food production environments. Microorganisms 2021, 9, 1856. [Google Scholar] [CrossRef]
- ISO 18593:2004; Microbiology of Food and Animal Feeding Stuffs–Horizontal Methods for Sampling Techniques from Surfaces Using Contact Plates and Swabs. ISO (International Organization for Standardization): Geneva, Switzerland, 2004.
- ISO 18593:2018; Microbiology of the Food Chain–Horizontal Methods for Surface Sampling. ISO (International Organization for Standardization): Geneva, Switzerland, 2018.
- ISO 11290-1:1996/Amd. 1:2004; Microbiology of Food and Animal Feeding Stuffs–Horizontal Method for the Detection and Enumeration of Listeria monocytogenes—Part 1: Detection Method. Amendment 1: Modification of the Isolation Media and the Haemolysis Test, and Inclusion of Precision Data. ISO (International Organization for Standardization): Geneva, Switzerland, 2004.
- ISO 11290-1:2017; Microbiology of the Food Chain–Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. ISO (International Organization for Standardization): Geneva, Switzerland, 2017.
- EUCAST (European Committee on Antimicrobial Susceptibility Testing). Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. 2022. Available online: https://www.eucast.org (accessed on 26 July 2022).
- Marroki, A.; Bousmaha-Marroki, L. Antibiotic resistance diagnostic methods for pathogenic bacteria. In Encyclopedia of Infection and Immunity, 1st ed.; Editor Rezaei, N., Ed.; Elsevier Science Publishing Co. Inc.: New York, NY, USA, 2022; Volume 4, pp. 320–341. [Google Scholar]
- Syrokou, M.K.; Paramithiotis, S.; Skandamis, P.N.; Drosinos, E.H.; Bosnea, L.; Mataragas, M. High-quality draft genome sequence data of six Lactiplantibacillus plantarum subsp. argentoratensis strains isolated from various Greek wheat sourdoughs. Data Brief 2021, 37, 107172. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Salzberg, S.L. SkewIT: The Skew Index Test for large-scale GC Skew analysis of bacterial genomes. PLoS Comput. Biol. 2020, 16, e1008439. [Google Scholar] [CrossRef]
- Weimann, A.; Mooren, K.; Frank, J.; Pope, P.B.; Bremges, A.; McHardy, A.C. From genomes to phenotypes: Traitar, the microbial trait analyzer. mSystems 2016, 1, e00101-16. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, Y.; Pouillot, R.; Dennis, S.; Xian, Z.; Luchansky, J.B.; Porto-Fett, A.C.S.; Lindsay, J.A.; Hammack, T.S.; Allard, M.; et al. Genetic diversity and profiles of genes associated with virulence and stress resistance among isolates from the 2010-2013 interagency Listeria monocytogenes market basket survey. PLoS ONE 2020, 15, e0231393. [Google Scholar] [CrossRef] [PubMed]
- Mafuna, T.; Matle, I.; Magwedere, K.; Pierneef, R.E.; Reva, O.N. Whole genome-based characterization of Listeria monocytogenes isolates recovered from the food chain in South Africa. Front. Microbiol. 2021, 12, 669287. [Google Scholar] [CrossRef] [PubMed]
- Pyz-Łukasik, R.; Paszkiewicz, W.; Kiełbus, M.; Ziomek, M.; Gondek, M.; Domaradzki, P.; Michalak, K.; Pietras-Ożga, D. Genetic diversity and potential virulence of L. monocytogenes isolates originating from Polish artisanal cheeses. Foods 2022, 11, 2805. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, G.; Yang, J.; Zhao, L.; Jiang, Y.; Guo, D.; Wang, X.; Zhi, S.; Xu, X.; Dong, Q.; et al. Prevalence, antibiotic resistance, and molecular epidemiology of Listeria monocytogenes isolated from imported foods in China during 2018 to 2020. Int. J. Food Microbiol. 2022, 382, 109916. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ji, Q.; Li, S.; Liu, M. Prevalence and genetic diversity of Listeria monocytogenes isolated from retail pork in Wuhan, China. Front. Microbiol. 2021, 12, 620482. [Google Scholar] [CrossRef]
- Burnett, E.; Kucerova, Z.; Freeman, M.; Kathariou, S.; Chen, J.; Smikle, M. Whole-genome sequencing reveals multiple subpopulations of dominant and persistent lineage I isolates of Listeria monocytogenes in two meat processing facilities during 2011–2015. Microorganisms 2022, 10, 1070. [Google Scholar] [CrossRef]
- Figueroa-López, A.M.; Maldonado-Mendoza, I.E.; López-Cervantes, J.; Verdugo-Fuentes, A.A.; Ruiz-Vega, D.A.; Cantú-Soto, E. Prevalence and characterization of Listeria monocytogenes isolated from pork meat and on inert surfaces. Braz. J. Microbiol. 2019, 50, 817–824. [Google Scholar] [CrossRef]
- Kayode, A.J.; Okoh, A.I. Assessment of multidrug-resistant Listeria monocytogenes in milk and milk product and One Health perspective. PLoS ONE 2022, 17, e0270993. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, Z.; Wang, H.; Wang, P.; Lan, R.; Deng, J.; Miao, Y.; Wang, Y.; Wang, Y.; Xu, J.; et al. A 12-month longitudinal study of Listeria monocytogenes contamination and persistence in pork retail markets in China. Food Control 2017, 76, 66–73. [Google Scholar] [CrossRef]
- Gulel, G.T.; Gucukoglu, A.; Cadirci, O.; Saka, E.; Alisarli, M. Serotyping and antibiotic resistance of Listeria monocytogenes isolated from raw water buffalo milk and milk products. J. Food Sci. 2020, 85, 2889–2895. [Google Scholar] [CrossRef]
- Kevenk, T.H.; Gulel, G.T. Prevalence, antimicrobial resistance and serotype distribution of Listeria monocytogenes isolated from raw milk and dairy products. J. Food Saf. 2016, 36, 11–18. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Al-Holy, M.A.; Shahbaz, H.M.; Al-Nabulsi, A.A.; Ghoush, M.H.A.; Osaili, T.M.; Ayyash, M.M.; Holley, R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: A comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1277–1292. [Google Scholar] [CrossRef]
- Rodríguez-Melcón, C.; Alonso-Calleja, C.; García-Fernández, C.; Carballo, J.; Capita, R. Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC) for twelve antimicrobials (biocides and antibiotics) in wight strains of Listeria monocytogenes. Biology 2022, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Bland, R.; Waite-Cusic, J.; Weisberg, A.J.; Riutta, E.R.; Chang, J.H.; Kovacevic, J. Adaptation to a commercial quaternary ammonium compound sanitizer leads to cross-resistance to select antibiotics in Listeria monocytogenes isolated from fresh produce environments. Front. Microbiol. 2022, 12, 782920. [Google Scholar] [CrossRef]
- Guérin, A.; Bridier, A.; Le Grandois, P.; Sévellec, Y.; Palma, F.; Félix, B.; LISTADAPT Study Group; Roussel, S.; Soumet, C. Exposure to quaternary ammonium compounds selects resistance to ciprofloxacin in Listeria monocytogenes. Pathogens 2021, 10, 220. [Google Scholar] [CrossRef] [PubMed]
- Kode, D.; Nannapaneni, R.; Bansal, M.; Chang, S.; Cheng, W.-H.; Sharma, C.S.; Kiess, A. Low-level tolerance to fluoroquinolone antibiotic ciprofloxacin in QAC-adapted subpopulations of Listeria monocytogenes. Microorganisms 2021, 9, 1052. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, Y.; Zhang, P.; Niu, Y.; Chen, Q.; Ma, X. Genomic characterization of clinical Listeria monocytogenes isolates in Beijing, China. Front. Microbiol. 2021, 12, 751003. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic characterization of Listeria monocytogenes isolated from ready-to-eat meat and meat processing environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef]
- Maury, M.M.; Bracq-Dieye, H.; Huang, L.; Vales, G.; Lavina, M.; Thouvenot, P.; Disson, O.; Leclercq, A.; Brisse, S.; Lecuit, M. Hypervirulent Listeria monocytogenes clones’ adaptation to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019, 10, 2488. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Collin, H. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Morón-García, A.; Palacios-Gorba, C.; Dessaux, C.; García-del Portillo, F.; Pucciarelli, M.G.; Ortega, A.D. Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 2021, 12, 2509–2545. [Google Scholar] [CrossRef]
- Van Stelten, A.; Roberts, A.R.; Manuel, C.S.; Nightingale, K.K. Listeria monocytogenes isolates carrying virulence-attenuated mutations in internalin A are commonly isolated from ready-to-eat food processing plant and retail environments. J. Food Prot. 2016, 79, 1733–1740. [Google Scholar] [CrossRef]
- Varsaki, A.; Ortiz, S.; Santorum, P.; López, P.; López-Alonso, V.; Hernández, M.; Abad, D.; Rodríguez-Grande, J.; Ocampo-Sosa, A.A.; Marínez-Suárez, J.V. Prevalence and population diversity of Listeria monocytogenes isolated from dairy cattle farms in the Cantabria region of Spain. Animals 2022, 12, 2477. [Google Scholar] [CrossRef] [PubMed]
- Schiavano, G.F.; Ateba, C.N.; Petruzzelli, A.; Mele, V.; Amagliani, G.; Guidi, F.; De Santi, M.; Pomilio, F.; Blasi, G.; Gattuso, A.; et al. Whole-genome sequencing characterization of virulence profiles of Listeria monocytogenes food and human isolates and in vitro adhesion/invasion assessment. Microorganisms 2022, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Begley, M.; Hill, C.; Gahan, C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010, 109, 984–995. [Google Scholar] [CrossRef]
- Schmitz-Esser, S.; Müller, A.; Stessl, B.; Wagner, M. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 2015, 6, 380. [Google Scholar] [CrossRef] [PubMed]
- Guidi, F.; Orsini, M.; Chiaverini, A.; Torresi, M.; Centorame, P.; Acciari, V.A.; Salini, R.; Palombo, B.; Brandi, G.; Amagliani, G.; et al. Hypo- and hyper-virulent Listeria monocytogenes clones persisting in two different food processing plants of central Italy. Microorganisms 2021, 9, 376. [Google Scholar] [CrossRef]
- Hurley, D.; Luque-Sastre, L.; Parker, C.T.; Huynh, S.; Eshwar, A.K.; Nguyen, S.V.; Andrews, N.; Moura, A.; Fox, E.M.; Jordan, K.; et al. Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 2019, 4, e00252-19. [Google Scholar] [CrossRef] [PubMed]
- Vu, H.T.K.; Stasiewicz, M.J.; Benjakul, S.; Vongkamjan, K. Genomic analysis of prophages recovered from Listeria monocytogenes lysogens found in seafood and seafood-related environment. Microorganisms 2021, 9, 1354. [Google Scholar] [CrossRef]
- Verghese, B.; Lok, M.; Wen, J.; Alessandria, V.; Chen, Y.; Kathariou, S.; Knabel, S. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Appl. Environ. Microbiol. 2011, 77, 3279–3292. [Google Scholar] [CrossRef]
- Harrand, A.S.; Jagadeesan, B.; Baert, L.; Wiedmann, M.; Orsi, R.H. Evolution of Listeria monocytogenes in a food processing plant involves limited single-nucleotide substitutions but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 2020, 86, e02493-19. [Google Scholar] [CrossRef]
- Brown, P.; Chen, Y.; Siletzky, R.; Parsons, C.; Jaykus, L.-A.; Eifert, J.; Ryser, E.; Logue, C.M.; Stam, C.; Brown, E.; et al. Harnessing whole genome sequence data for facility-specific signatures for Listeria monocytogenes: A case study with turkey processing plants in the United States. Front. Sustain. Food Syst. 2021, 5, 742353. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Chen, C.-C.; Yang, C.-H.; Hsieh, H.-Y.; He, J.-X.; Lin, H.-H.; Lee, C.-C. LmTraceMap: A Listeria monocytogenes fast-tracing platform for global surveillance. PLoS ONE 2022, 17, e0267972. [Google Scholar] [CrossRef]
- Lachtara, B.; Wieczorek, K.; Osek, J. Genetic diversity and relationships of Listeria monocytogenes serogroup IIa isolated in Poland. Microorganisms 2022, 10, 532. [Google Scholar] [CrossRef] [PubMed]
- Bolocan, A.S.; Oniciuc, E.A.; Alvarez-Ordóñez, A.; Wagner, M.; Rychli, K.; Jordan, K.; Nicolau, A.I. Putative cross-contamination routes of Listeria monocytogenes in a meat processing facility in Romania. J. Food Prot. 2015, 78, 1664–1674. [Google Scholar] [CrossRef] [PubMed]
- Stessl, B.; Ruppitsch, W.; Wagner, M. Listeria monocytogenes post-outbreak management—When could a food production be considered under control again? Int. J. Food Microbiol. 2022, 379, 109844. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, G.; Orsi, R.H.; Estrada, E.; Strawn, L.; Wiedmann, M. Whole-genome sequencing-based characterization of Listeria isolates from produce packinghouses and fresh-cut facilities suggests both persistence and reintroduction of fully virulent L. monocytogenes. Appl. Environ. Microbiol. 2022, 88, e0117722. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andritsos, N.D.; Mataragas, M. Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods 2023, 12, 1200. https://doi.org/10.3390/foods12061200
Andritsos ND, Mataragas M. Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods. 2023; 12(6):1200. https://doi.org/10.3390/foods12061200
Chicago/Turabian StyleAndritsos, Nikolaos D., and Marios Mataragas. 2023. "Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility" Foods 12, no. 6: 1200. https://doi.org/10.3390/foods12061200
APA StyleAndritsos, N. D., & Mataragas, M. (2023). Characterization and Antibiotic Resistance of Listeria monocytogenes Strains Isolated from Greek Myzithra Soft Whey Cheese and Related Food Processing Surfaces over Two-and-a-Half Years of Safety Monitoring in a Cheese Processing Facility. Foods, 12(6), 1200. https://doi.org/10.3390/foods12061200







