Abstract
Staphylococcus aureus is one of the high-threat pathogens equipped with a repertoire of virulence factors making it responsible for many infections in humans, including foodborne diseases. The present study aims to characterize antibiotic resistance and virulence factors in foodborne S. aureus isolates, and to investigate their cytotoxic effects in human intestinal cells (HCT-116). Our results revealed methicillin resistance phenotypes (MRSA) along with the detection of mecA gene (20%) among tested foodborne S. aureus strains. Furthermore, 40% of tested isolates showed a strong ability for adhesion and biofilm formation. A high rate of exoenzymes production by tested bacteria was also registered. Additionally, treatment with S. aureus extracts leads to a significant decrease in HCT-116 cell viability, accompanied by a reduction in the mitochondrial membrane potential (MMP), as a result of reactive oxygen species (ROS) generation. Thereby, S. aureus food poisoning remains daunting and needs particular concern to prevent foodborne illness.
1. Introduction
Staphylococcus aureus is one of the major human pathogens characterized by a wide range of virulence factors along with a high ability to acquire resistance to various antibiotics leading to the constant emergence of new clones [1]. For instance, methicillin-resistant S. aureus (MRSA) is one of the most frequent phenotypes of antibiotic resistance due to the occurrence of the clinical use of methicillin [2]. Interestingly, MRSA is no longer only considered a nosocomial pathogen associated with healthcare settings, but has also become a major cause of community-associated infections and has created several reservoirs [3]. Various surveys clearly indicate the presence of MRSA in foods that pose an immediate risk to human health. For instance, 2% to 11.9% of meat samples tested in the Netherlands were contaminated with MRSA, 5% to 7.7% in USA and Canada, respectively, and 1.2% in Tunisia [4,5,6].
S. aureus uses various strategic ways to maintain an infection; one of them is biofilm formation [7]. This sessile form of subsistence allows this bacterium to avoid phagocyte attacks and protects them from antibiotics and disinfectants [8]. Successful colonization of biotic surfaces such as tissue cells or abiotic ones, such as medical devices of industrial materials, is provided via cell-wall-anchored and other S. aureus surface proteins, many of which belong to the microbial surface components recognizing adhesive matrix molecules (MSCRAMM) family [9]. Inside the infected organism, S. aureus uses a wide range of toxins, enzymes and surface proteins involved in an astounding series of mechanisms to avoid host immuno-system defenses and allows the invasion of cells and propagation through tissues [10].
S. aureus food poisoning is induced by released enterotoxins that cause intestinal activity disruption, which is characterized by numerous symptoms such as fever, nausea, diarrhea, vomiting, etc. [11,12]. Based on antigenic heterogeneity, more than 20 enterotoxins (SEA—SElV) have been discovered [13,14]. Among them, staphylococcal enterotoxin C, causing community-associated MRSA infections and staphylococcal enterotoxin B, is associated with food poisoning [15] and involved in reducing protective T-cell responses [16].
Human cells are permanently exposed to endogenous and exogenous factors such as radiations, chemicals and pathogens (e.g., bacteria), which are leading to oxidative stress through reactive oxygen species (ROS) generation [17]. This mechanism is a result of the imbalance between the intracellular accumulation of ROS and the ability of a biological system to detoxify these reactive products [18]. Inside organisms, produced ROS such as hydrogen peroxide, hydroxyl radical and superoxide radical [19,20,21] have different negative consequences such as cycle arrest, DNA and cell membrane damage and cell alterations in apoptosis [22]. On their side, S. aureus can directly and indirectly (via ROS) damage host DNA of which double-strand breaks are the most deleterious [17,18,19,20,21,22,23].
Here, we characterized S. aureus strains isolated from food products by the screening of methicillin resistance and the assessment of adhesion ability, hemolysin and exoenzymes production as well as the cytotoxic effect in intestinal epithelial HCT-116 cells.
2. Materials and Methods
2.1. Bacterial Strains
Ten S. aureus strains were acquired from the Microbiology Laboratory of the Sahline Hygiene Center in Monastir, Tunisia, and have been isolated from various foodborne samples (Cake, Cheese, Milk, Sausage, Meat and Chicken). The reference strain S. aureus ATCC 25923 was used as a control. Bacterial identification of tested strains was confirmed with biochemical methods, by subculturing in mannitol salt agar as the selective medium and using Gram staining, catalase, coagulase and DNase tests. The strains were maintained on Luria-Bertani (LB, Liofilchem, Milan, Italy) broth, and a set preserved as glycerol stocks at −80 °C. Prior to each assay, the strains were subcultured thrice and incubated at 37 °C for 24 h to ensure optimal growth.
2.2. Susceptibility to Antibiotics
Antimicrobial susceptibility testing was performed with the disk-diffusion method according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST). From an overnight culture of each S. aureus strains, a 0.5 McFarland standard bacteria solution was prepared were inoculated into a Mueller Hinton agar (MHA; Biolife, Milan, Italy) plate. The antimicrobial discs, purchased from Oxoid (Thermo Fisher Scientific, Basingstoke, UK), were penicillin G (P, 6 µg), cefoxitin (FOX, 30 µg), cefotaxim (CTX, 30 µg), kanamycin (K, 30 µg), tobramycin (TM, 10 µg), erythromycin (E, 15 µg), clindamycin (CMN, 2 µg), norfloxacine (NOR, 5 µg), ofloxacin (OFX, 5 µg), fusidic acid (FAD, 10 µg), rifampicin (RIF, 30 µg) and chloramphenicol (CHL, 30 µg). After incubation at 37 °C for 24 h, the diameters of the inhibition zones around the discs were determined.
2.3. Detection of Methicillin Resistance Gene
The detection of the methicillin resistance gene (mecA gene), was achieved by PCR amplification of specific primers 5′GTAGAAATGACTGAACGTCCGATAA-3′ (Forward) and 5′CCAATTCCACATTGTTTCGGTCTAA-3′ (Reverse). The standard thermal lyses technique was used for genomic DNA extraction from foodborne S. aureus isolates. Then, the PCR protocol was carried out as previously described [24]. Amplified PCR products were analyzed on a 2% (w/v) agarose gel stained with ethidium bromide (0.5 g mL−1) and photographed using a Gel Doc XR instrument (Bio-rad, New Castle, DE, USA). Reference strain S. aureus ATCC 43300 was used as a positive control (MRSA).
2.4. Adhesive Abilities of Tested Strains
Exopolysaccharides (Slime) production by tested isolates (n = 10) was assessed on Congo red agar (CRA) medium as previously described [25]. Black colonies with a rough surface on CRA plate revealed slime-producing ability of tested strains, whereas non-producing strains gave red colonies with a smooth surface.
The capacity of tested strains to form biofilms was assessed using the crystal violet assay on polystyrene-96 well plates, as previously described [26]. Reference strain S. aureus ATCC 25923 was used as a positive control. Biofilm formation was classified.as highly positive (OD570 ≥1), low-grade positive (0.1 ≤ OD570 < 1), or negative (OD570 < 0.1).
2.5. Hydrophobicity of Bacterial Cell Surfaces
The microbial adhesion to solvent (MATS) test was used to assess the cell surface hydrophobicity of S. aureus strains [27]. It entailed determining the cells’ affinity for apolar solvents (hexadecane). Firstly, bacterial cells were harvested by centrifugation at 7000× g for 5 min and resuspended in 0.01 M potassium phosphate buffer with Abs600 nm = 0.4 (108 CFU mL−1 cell density) (Abs1) (pH 6.5). Then, bacterial suspension was vortexed for 90 s with hexadecane in a 1:6 (0.4/2.4 v/v) ratio to generate an emulsion and was left for 20 min to allow two separate phases. The aqueous phase absorbance (Abs2) was measured and the percentage of adhesion was calculated as: % adhesion = (1-Abs2/Abs1) × 100. According to Abasolo-Pacheco et al. [28], a percentage greater than 70% indicates that the tested microorganism is hydrophobic, values from 30 to 70% indicate it is weakly hydrophobic, and values less than 30% indicate it is hydrophilic.
2.6. Secretion of Exo-Enzymes Hemolysins
The capacity of foodborne S. aureus strains to generate hydrolytic enzymes was assessed after inoculating bacterial cultures on TSA medium (Biorad) supplemented with: 1% (w/v) skim milk for caséinase, 1% (w/v) gelatin for gelatinase, 5% (w/v) starch for amylase, Tween 80 for lipase and 5% (v/v) egg yolk for lecithinase [29]. The appearance of a distinct halo surrounding the colonies after 24 h of incubation at 37 °C confirms the presence of the target exo-enzyme. Regarding the hemolytic activity, it was assessed on bacteriological agar with 5% sheep’s blood [30].
2.7. Cytotoxicity on HCT116 Cells
S. aureus isolates were inoculated in tryptic soy broth (TSB, BioRad, Marnes-la-Coquette, France) and incubated in sterile 15 mL test tubes at 37 °C for 18–24 h. After centrifuging the bacterial cultures at 3000 rpm for 15 min, the supernatant was filtered through a 0.22 µm pore size filter membrane (Millipore, Merck KGaA, Darmstadt, Germany).
Human colon cancer cells (HCT116) were grown in a cell culture medium (RPMI; Sigma, Burlington, MA, USA) enriched with 10% fetal calf serum (FCS), 1% L-glutamine (200 mM), 1% penicillin (100 IU/mL) and streptomycin (100 IU/mL) at 37 °C with 5% CO2. On alternating days, the medium was changed. Then, on 96-well tissue culture plates, confluent monolayers of HCT116 cells were washed with PBS and 50 µL aliquots of RPMI were added to each well. Then, 50 µL of bacterial filtrates from each strain were introduced to HCT116 cell monolayers that had been previously washed in PBS and cultured for 24 h at 37 °C in 5% CO2. As a control, wells containing solely RPMI were used.
The MTT test was used to evaluate cell viability, as previously reported [31]. After washing with PBS, cells were treated for 1 h at 37 °C with 5-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma). After removing the supernatants, the cells were treated with 100 µL of DMSO to dissolve the formazan crystals generated in the live metabolically active cells. The absorbance of each well of each tested isolate was measured using a microplate reader at 540 nm (Bioteck, Elx 800). The cytotoxicity percentage was computed as follows: % Cytotoxicity = (1-A540 of infected culture/A540 of control) × 100 [32].
2.8. Oxidative Status Evaluation
HCT-116 cells were grown at 105 cells/well on 24-well culture plates (Polylabo, Strasbourg, France) for 24 h of incubation. Following that, the cells were treated for 24 h at 37 °C with various bacterial supernatants (50 µL/well). As a positive control, H2O2 (20 M) was employed. Following incubation, the cells were treated with 20 M DCFH-DA. Intracellular generation of reactive oxygen species (ROS) was measured after 30 min of incubation at 37 °C using a fluorimeter (Biotek FL 800) with an excitation wavelength of 485 nm and an emission wavelength of 522 nm. The non-fluorescent (DCFH-DA) product is converted to the highly fluorescent 2,7-dichlorofluorescein product (DCF) (lmax = 522 nm) in many processes. The fluorescent probe is degraded by intracellular esterases after diffusing across the cell membrane, converting nonfluorescent dichlorofluorescein (DCFH) to fluorescent DCF, which is contained inside the cells and oxidized by peroxides in the presence of ROS [33]. The amount of ROS produced intracellularly is proportional to the intensity of DCF fluorescence.
2.9. Mitochondrial Membrane Potential (MMP) Assay
The absorption of the cationic fluorescent dye rhodamine-123 was used to calculate MMP [20]. In a typical experiment, seeded cells in 96-well culture plates were treated with various S. aureus supernatants for 24 h, after which the cells were thoroughly washed with PBS and 100 µL of rhodamine-123 (1 mM) in PBS was reintroduced on the plates. For 15 min, cells were reintroduced to the incubator (37 °C, 5% CO2). The supernatant PBS (which included unabsorbed rhodamine-123) was then removed and replaced with fresh PBS. Fluorimetric detection was used to determine rhodamine-123 uptake. The results were reported as a percentage of rhodamine fluorescence uptake relative to negative control cell fluorescence.
2.10. Statistical Analyses
All experiments were performed in triplicate and results were presented as mean ± SD. Statistical differences between the control and tests were determined and compared using analysis of variance (ANOVA). Differences were considered significant at p < 0.05.
3. Results
3.1. Susceptibility Profiles of S. aureus Strains and PCR- Detection of mecA Gene
Results of the susceptibility to antibiotics of foodborne S. aureus strains are summarized in Table 1. Comparing with the standard limits (CASFM), we noted that tested strains showed strong resistant to penicillin G (100%), 20% of them are resistance to cefoxitin, cefotaxime and kanamycine, 10% to tobramycine, erythromycine and clindamycine. No resistance was registered against the rest of the drugs.

Table 1.
Susceptibility profiles of foodborne S. aureus strains and PCR-detection of mecA gene.
Regarding the prevalence of methicillin resistance phenotype among tested S. aureus isolates, our results revealed that two strains (896 and 853) out of ten (20%) were found in MRSA since they are resistant to cefoxitin (FOX). Molecular confirmation of this result was carried out by PCR test revealing the detection of the mecA gene in the MRSA 896 and 853 strains (Figure 1; Table 1).
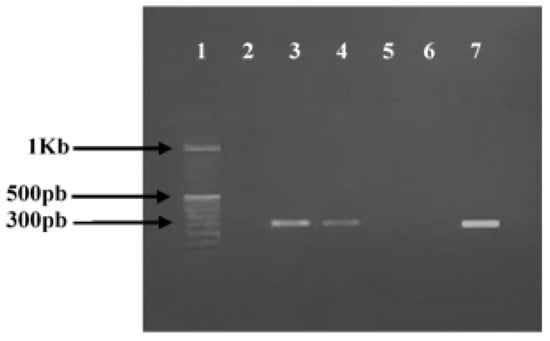
Figure 1.
Agarose gel electrophoresis of polymerase chain reaction (PCR) amplification of mecA gene. Lanes 1, 100 bp DNA molecular size marker; lane 2, negative control; lanes 3–7, PCR amplicons obtained with DNA amplification of S. aureus strains, respectively: ATCC 43300 (Positive control); 896; 976″; 977″; 853.
3.2. Adhesive Properties of Tested Strains
In this part of our study, we have determined the MATS of 10 S. aureus strains. Our data (Table 2) indicate that the affinity to hexadecane was low suggesting a hydrophilic character for all the studied strains (hydrophobicity ˂ 30%) excepting S. aureus 976′ that was weakly hydrophobic (30% ≤ hydrophobicity ˂ 70%).

Table 2.
Cell surface hydrophobicity, slime production and biofilm formation ability of studied strains.
Regarding the ability of studied foodborne pathogens to produce exopolysaccharides (EPS) on the CRA plates, our results showed that three out of ten (30%) were slime producers developing positive and variable phenotypes (Figure 2). The other strains showed a negative phenotype (70%) (Table 2).
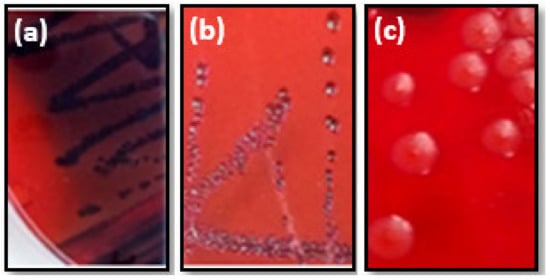
Figure 2.
Various morphotypes of S. aureus strains cultivated on CRA plates. Black colonies (a) or red colonies with black center (b) indicated positive morphotype. While red colonies (c) bacteria revealed negative morphotype.
The result of OD570 presented in Table 2, showed that fore strains (40%) isolated from various types of foods were highly biofilm positive (OD570 ≥ 1). The rest of the strains showed a low-grade biofilm formation (0.1 ≤ OD570 < 1).
3.3. Hemolysin and Hydrolytic Enzymes Production
The resulting enzymes secretion showed that all S. aureus tested strains were positive for Lecithinase, Caseinase and Amylase (100%). Lipase activity was detected in eight strains (80%) and we noted that among the ten S. aureus strains, four were Gelatinase producers (40%). Regarding hemolysin activity, seven out of ten tested strains were beta-hemolytic (70%) (Table 3).

Table 3.
Exoenzymes production and hemolysis of studied strains.
3.4. Toxicity in HCT116 Cells
HCT-116 cells were treated separately with supernatant from tested S. aureus strains for 24 h and cell viability was determined by MTT assay (Figure 3). Our results showed that four isolates (976′; 976″; 977′; 977″) from the total tested strains induced more than 50% of cell toxicity in HCT116 cells (p < 0.05). The cytotoxicity potential of these strains, isolated from meat and chicken, is classified as moderate (between 50–85%).
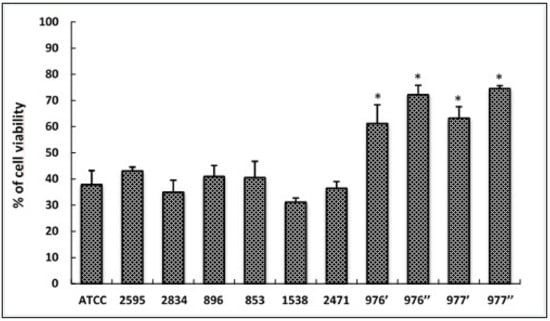
Figure 3.
Cytotoxic effects of foodborne S. aureus strains on HCT116 cells treated with different bacterial extracts for 24 h. Cell viability was determined using the MTT assay and expressed as percentages of viability. ATCC referred to the reference strain S. aureus ATCC 25923. Data are expressed as the mean ± SD. of three independent experiments. *: Values are significantly different (p < 0.05) from other strains.
3.5. ROS Generation in Infected Cells
To check the oxidative stress status in HCT-116 cells in response to treatment with different bacterial extracts, we measured the production of fluorescent DCF (the result of DCFH oxidation by a variety of peroxides). As shown in Figure 4, our results revealed that tested extracts, from different S. aureus strains, induced the significant increase in ROS generation (p < 0.05) when compared to the control (untreated cells). Additionally, it was deduced that bacterial extracts from 976′, 976″ and 977″ strains strongly induced oxidative stress in HCT-116 cells, triggering a high production of ROS close to that of hydrogen peroxide (H2O2; positive control).
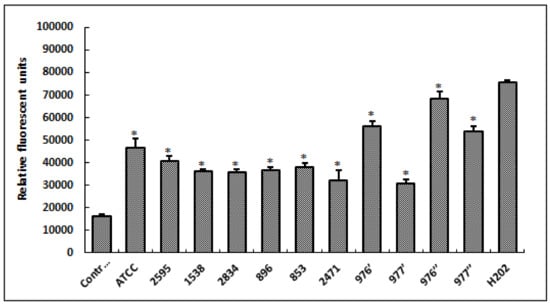
Figure 4.
Levels of relative fluorescent DCF production, after 24 h of cell exposure to different S. aureus extracts. Fluorescent DCF is the result of DCFH oxidation by a variety of peroxides. H2O2, at 20 μM, was used as a positive control. ATCC referred to the reference strain S. aureus ATCC 25923. Data are expressed as the mean ± SD of three independent experiments. Values are significantly different (* p < 0.05) from control (untreated cells) and H2O2.
3.6. Loss of Mitochondrial Transmembrane Potential
After cell exposure to different bacterial extracts, from different S. aureus strains, a significant decrease in MMP (p < 0.05) was observed compared to the control (untreated cells), indicating that mitochondria were depolarized (Figure 5).
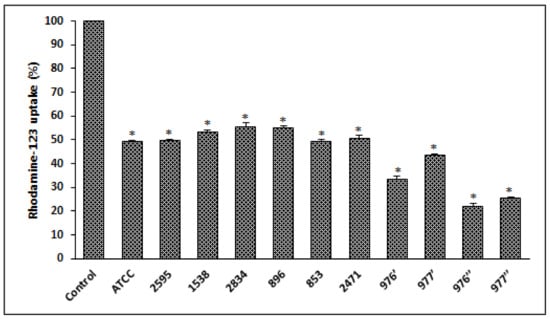
Figure 5.
S. aureus extracts induces a loss of mitochondrial transmembrane potential on HCT116 cells treated for 24 h and evaluated with rhodamine-123. ATCC referred to the reference strain S. aureus ATCC 25923. Data are expressed as the mean ± SD of three independent experiments. Values are significantly different (* p < 0.05) from control (untreated cells).
4. Discussion
S. aureus is one of the main causes of hospital and community infections [34]. The virulence of this opportunistic bacterium is a multifactorial process requiring the involvement of a variety of cellular components regulated in a perfectly coordinated manner [35]. Additionally, the unreasonable administration of antibiotics is at the origin of the incidence of MRSA infections, which have increased in recent years [36]. In the present study, we detected by simplex PCR the presence of mecA gene in 20% of foodborne S. aureus isolates, exhibiting phenotypic resistance to cefoxitin. Our finding is in agreement with previous stud dealing with the characterization of different S. aureus strains collected from different Tunisian food origins and showing that 1.2% of the studied strains were MRSA [6]. However, other reports showed that the percentage of MRSA prevalence can reach 38% [37]. It is well known that a wide range of staphylococcal species harbor the mecA gene encoding a second penicillin-binding protein (PBP2a), involved in methicillin resistance through the target modification mechanism [38]. Responsible for methicillin resistance, this gene is carried by a unique class of mobile genetic elements: the staphylococcal cassette chromosome (SCCmec) [39]. The mediators of methicillin resistance have been described with an ability to genes transfer from one species to another [40]. Particularly, the mecA gene has been shown to be transferred from coagulase-negative Staphylococcus species (SCoN) to S. aureus species in vivo, accounting for the emergence of more efficient MRSA clones with high adherence and invasion capabilities [41,42].
Regardless, the potential implications for MRSA reservoirs in food products demand careful monitoring of the epidemiology of this strain to design appropriate control measures before a catastrophe occurs [3]. MRSA epidemiology has increased considerably with the appearance of new strains resistant to antibiotic treatments. The majority of MRSA infections are related to biofilm formation, which is considered one of the most important virulence factors [43,44]. It is well known that most infections caused by S. aureus are associated with biofilm formation due to the particular characteristic of this microbial architecture allowing a high resistance to antibiotics and disinfectants, compared to planktonic forms [45]. Our results showed that 40% of isolates from various food samples were highly biofilm positive. A recent investigation carried out in Bangladesh showed that 21% of S. aureus isolates from different food sources are biofilm producers [46]. While other reports revealed that 72% of foodborne S. aureus isolates in China produced biofilms [47]. One of the major characters for bacterial adhesion and biofilm formation is cell surface hydrophobicity [48,49]. Here, we evaluated the microbial adhesion to solvent and the results revealed a hydrophilic character for the majority of studied strains, which is in agreement with another founding reporting that the S. aureus surface was hydrophilic [50,51].
The production of hydrolytic enzymes by S. aureus is an essential factor playing a critical role in virulence. In fact, they contribute to the invasion process, host tissue damage and even the propagation to other organs, regardless of its primary ecological niche [52]. Protease enzymes can change the quality of food rich in proteins resulting in a decreased shelf life of foods and their product [53]. Lipases produced from S. aureus could increase the rate of food deterioration through their action on lipids causing accumulation of intermediate and products that change the flavor of foods [54]. Our results showed that S. aureus tested strains were 100%, positive for lecithinase, caseinase and amylase and 80% for lipase. Similarly, lipase and gelatinase were identified in 82% and 88% of S. aureus strains isolated from Food handlers, respectively [55]. It was reported that 78%, 81% and 51% of S. aureus clinical isolates produced gelatinase, protease and lipase, respectively [56].
Cytotoxicity of the foodborne S. aureus strains was investigated using the MTT assay. After 24 HCT116 cell infections, 40% of S. aureus strains showed moderate cytotoxicity while the rest of the isolates (60%) showed a low cytotoxic effect on treated cells. Other findings showed that 76.2% of oral S. aureus strains revealed moderate cytotoxic effects on epithelial cells [57]. Numerous exoenzymes involved in the host component degradation and various toxins damaging the host tissue are secreted by S. aureus, along with a wide array of cell surface proteins implicated in the virulence and pathogenicity of this bacterium [58]. Through this study, a significant positive correlation (R = 0.95; p ˂ 0.001) was noted between bacterial exoenzymes activities and the cytotoxicity levels of foodborne S. aureus isolates. In fact, the qualitative assay of the soluble exo-enzymes secretion showed that the highest cytotoxic strains produced 100% of all sought enzymes.
Reactive oxygen species (ROS) are critical intermediates in oxidative metabolism in biological systems. Nonetheless, when oxidative stress occurs, ROS are produced in excess, which can harm cells by oxidizing lipids, altering DNA and destroying proteins [59]. The findings of intracellular ROS measurements clearly reveal that cell exposure to bacterial extract for 24 h significantly increases the quantity of these indicators. Under our experimental circumstances, the loss of cell viability after treatment with foodborne S. aureus extracts might be attributed to oxidative stress in the form of ROS production. In reality, oxidative stress is a phrase often used to describe the imbalance between ROS concentrations and cell anti-oxidative defense systems. The excessive generation of ROS changes. Excessive ROS generation affects cellular defenses, produces several harms and plays a significant role in carcinogenesis [60,61]. In terms of mitochondrial membrane potential (MMP) measurements in treated HCT116 cells, rhodamine-123 was utilized, because dynamic assessments of MMP, both in vitro and in situ, are proportional to the uptaken fluorophore [62], and MMP change is invariably related with cell death [63]. After 24 h of treatment, HCT116 cell death caused by bacterial extracts from distinct S. aureus strains was related to a substantial drop in MPP. It is well known that mitochondria produce the majority of ROS in cells, resulting in cell death [59]. Furthermore, it has been established that oxidative stress and MMP decline always occur concurrently [64], which is consistent with our findings.
5. Conclusions
In the present study, we detected methicillin resistance phenotype along with the presence of the mecA gene among tested foodborne S. aureus isolates. A high ability of biofilm formation and exoenzymes production was also registered. Additionally, tested strains exhibited cytotoxic effects on treated HCT-116 cells, predominately by ROS generation, which, in turn, induces mitochondrial dysfunction and leads to cell death. Hence, food contamination with S. aureus pathogen represented a real risk of acute infection, which can be avoided by hygienic quality instauration.
Author Contributions
Conceptualization, A.M. and S.A.-E.; methodology, A.M., H.H., M.B.A., Z.M.A.-H. and D.A.A.-Q.; validation, A.M. and S.A.-E.; writing—original draft preparation, A.M., H.H. and M.B.A.; writing—review and editing, A.M., H.H., M.B.A., Z.M.A.-H. and D.A.A.-Q.; supervision, A.M., H.H. and S.A.-E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
This work was supported by the Ministry of Higher Education and Scientific Research (MHESR) of Tunisia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lakhundi, S.; Zhang, K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Diekema, D.J.; Pfaller, M.A.; Schmitz, F.J.; Smayevsky, J.; Bell, J.; Jones, R.N.; Beach, M.; SENTRY Partcipants Group. Survey of infections due to Staphylococcus species: Frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 2001, 32 (Suppl. S2), S114–S132. [Google Scholar]
- Romero, L.C.; de Souza da Cunha, M.L.R. Insights into the epidemiology of community-associated methicillin-resistant Staphylococcus aureus in special populations and at the community-healthcare interface. Braz. J. Infect. Dis. 2021, 25, 101636. [Google Scholar] [CrossRef]
- Pu, S.; Han, F.; Ge, B. Isolation and Characterization of Methicillin-Resistant Staphylococcus aureus Strains from Louisiana Retail Meats. Appl. Environ. Microbiol. 2009, 75, 265–267. [Google Scholar] [CrossRef]
- Kluytmans, J.A. Methicillin-resistant Staphylococcus aureus in food products: Cause for concern or case for complacency? Clin. Microbiol. Infect. 2010, 16, 11–15. [Google Scholar] [CrossRef]
- Chairat, S.; Gharsa, H.; Lozano, C.; Gómez-Sanz, E.; Gómez, P.; Zarazaga, M.; Boudabous, A.; Torres, C.; Ben Slama, K. Characterization of Staphylococcus aureus from Raw Meat Samples in Tunisia: Detection of Clonal Lineage ST398 from the African Continent. Foodborne Pathog. Dis. 2015, 12, 686–692. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcal Biofilms. Microbiol. Spectr. 2018, 6, 4. [Google Scholar] [CrossRef]
- Thurlow, L.R.; Hanke, M.L.; Fritz, T.; Angle, A.; Aldrich, A.; Williams, S.H.; Engebretsen, I.L.; Bayles, K.W.; Horswill, A.R.; Kielian, T. Staphylococcus aureus Biofilms Prevent Macrophage Phagocytosis and Attenuate Inflammation In Vivo. J. Immunol. 2011, 186, 6585–6596. [Google Scholar] [CrossRef]
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Hennekinne, J.A.; De Buyser, M.L.; Dragacci, S. Staphylococcus aureus and its food poisoning toxins: Characterization and outbreak investigation. FEMS Microbiol. Rev. 2012, 36, 815–836. [Google Scholar] [CrossRef]
- Hu, D.-L.; Li, S.; Fang, R.; Ono, H.K. Update on molecular diversity and multipathogenicity of staphylococcal superantigen toxins. Anim. Dis. 2021, 1, 7. [Google Scholar] [CrossRef]
- Kong, C.; Neoh, H.-M.; Nathan, S. Targeting Staphylococcus aureus Toxins: A Potential form of Anti-Virulence Therapy. Toxins 2016, 8, 72. [Google Scholar] [CrossRef]
- Fisher, E.L.; Otto, M.; Cheung, G.Y.C. Basis of Virulence in Enterotoxin-Mediated Staphylococcal Food Poisoning. Front. Microbiol. 2018, 9, 436. [Google Scholar] [CrossRef]
- Ahmad-Mansour, N.; Loubet, P.; Pouget, C.; Dunyach-Remy, C.; Sotto, A.; Lavigne, J.-P.; Molle, V. Staphylococcus aureus Toxins: An Update on Their Pathogenic Properties and Potential Treatments. Toxins 2021, 13, 677. [Google Scholar] [CrossRef]
- Llewelyn, M.; Cohen, J. Superantigens: Microbial agents that corrupt immunity. Lancet Infect. Dis. 2002, 2, 156–162. [Google Scholar] [CrossRef]
- Deplanche, M.; Mouhali, N.; Nguyen, M.-T.; Cauty, C.; Ezan, F.; Diot, A.; Raulin, L.; Dutertre, S.; Langouet, S.; Legembre, P.; et al. Staphylococcus aureus induces DNA damage in host cell. Sci. Rep. 2019, 9, 7694. [Google Scholar] [CrossRef]
- Pizzino, G.; Irrera, N.; Cucinotta, M.; Pallio, G.; Mannino, F.; Arcoraci, V.; Squadrito, F.; Altavilla, D.; Bitto, A. Oxidative Stress: Harms and Benefits for Human Health. Oxid. Med. Cell. Longev. 2017, 2017, 8416763. [Google Scholar] [CrossRef]
- Cathcart, R.; Schwiers, E.; Ames, B.N. Detection of picomole levels of hydroperoxides using a fluorescent dichlorofluorescein assay. Anal. Biochem. 1983, 134, 111–116. [Google Scholar] [CrossRef]
- Debbasch, C.; Brignole, F.; Pisella, P.J.; Warnet, J.M.; Rat, P.; Baudouin, C. Quaternary ammoniums and other preservatives’ contribution in oxidative stress and apoptosis on Chang conjunctival cells. Investig. Opthalmology Vis. Sci. 2001, 42, 642–652. [Google Scholar]
- Gomes, A.; Fernandes, E.; Lima, J.L.F.C. Fluorescence probes used for detection of reactive oxygen species. J. Biochem. Biophys. Methods 2005, 65, 45–80. [Google Scholar] [CrossRef] [PubMed]
- Dayem, A.A.; Hossain, M.K.; Lee, S.B.; Kim, K.; Saha, S.K.; Yang, G.-M.; Choi, H.Y.; Cho, S.-G. The Role of Reactive Oxygen Species (ROS) in the Biological Activities of Metallic Nanoparticles. Int. J. Mol. Sci. 2017, 18, 120. [Google Scholar] [CrossRef]
- Kaina, B. DNA damage-triggered apoptosis: Critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem. Pharmacol. 2003, 66, 1547–1554. [Google Scholar] [CrossRef]
- Geha, D.J.; Uhl, J.R.; Gustaferro, C.A.; Persing, D.H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J. Clin. Microbiol. 1994, 32, 1768–1772. [Google Scholar] [CrossRef] [PubMed]
- Touati, A.; Achour, W.; Abbassi, M.S.; Ben Hassen, A. Détection des gènes ica et de la production de slime parmi des souches de Staphylococcus epidermidis isolées d’infections liées aux cathéters chez des patients neutropéniques [Detection of ica genes and slime production in a collection of Staphylococcus epidermidis strains from catheter-related infections in neutropenic patients]. Pathol. Biol. 2007, 55, 277–282. [Google Scholar] [PubMed]
- Mack, D.; Bartscht, K.; Fischer, C.; Rohde, H.; de Grahl, C.; Dobinsky, S.; Horstkotte, M.A.; Kiel, K.; Knobloch, J.K.-M. Genetic and biochemical analysis of Staphylococcus epidermidis biofilm accumulation. Methods Enzym. 2001, 336, 215–239. [Google Scholar] [CrossRef]
- Bellon-Fontaine, M.-N.; Rault, J.; van Oss, C. Microbial adhesion to solvents: A novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surfaces B Biointerfaces 1996, 7, 47–53. [Google Scholar] [CrossRef]
- Abasolo-Pacheco, F.; Saucedo, P.E.; Mazón-Suástegui, J.M.; Tovar-Ramírez, D.; Araya, R.; Ramírez-Orozco, J.M.; Campa-Córdova, A.I. Isolation and use of beneficial microbiota from the digestive tract of lions-paw scallop Nodipecten subnodosus and winged pearl oyster Pteria sterna in oyster aquaculture. Aqua. Res. 2016, 47, 3042–3051. [Google Scholar] [CrossRef]
- Ben Kahla-Nakbi, A.; Chaieb, K.; Bakhrouf, A. Investigation of several virulence properties among Vibrio alginolyticus strains isolated from diseased cultured fish in Tunisia. Dis. Aquat. Org. 2009, 86, 21–28. [Google Scholar] [CrossRef]
- Stulik, L.; Malafa, S.; Hudcova, J.; Rouha, H.; Henics, B.Z.; Craven, D.E.; Sonnevend, A.M.; Nagy, E. α-Hemolysin Activity of Methicillin-Susceptible Staphylococcus aureus Predicts Ventilator-associated Pneumonia. Am. J. Respir. Crit. Care Med. 2014, 190, 1139–1148. [Google Scholar] [CrossRef]
- Saliba, A.M.; Filloux, A.; Ball, G.; Silva, A.S.V.; Assis, M.-C.; Plotkowski, M.-C. Type III secretion-mediated killing of endothelial cells by Pseudomonas aeruginosa. Microb. Pathog. 2002, 33, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Kishi, K.; Hirai, K.; Hiramatsu, K.; Yamasaki, T.; Nasu, M. Macrolides and clindamycin suppress the release of Shiga-like toxins from Escherichia coli O157:H7 in vitro. Int. J. Antimicrob. Agents 2000, 15, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wong, Y.-S. Selenocystine induces caspase-independent apoptosis in MCF-7 human breast carcinoma cells with involvement of p53 phosphorylation and reactive oxygen species generation. Int. J. Biochem. Cell Biol. 2009, 41, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Gould, I. Costs of hospital-acquired methicillin-resistant Staphylococcus aureus (MRSA) and its control. Int. J. Antimicrob. Agents 2006, 28, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Dinges, M.M.; Orwin, P.M.; Schlievert, P.M. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 2000, 13, 16–34. [Google Scholar] [CrossRef]
- Borg, M.A.; Camilleri, L. What Is Driving the Epidemiology of Methicillin-Resistant Staphylococcus aureus Infections in Europe? Microb. Drug Resist. 2021, 27, 889–894. [Google Scholar] [CrossRef]
- Pereira, V.; Lopes, C.; Castro, A.; Silva, J.; Gibbs, P.; Teixeira, P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol. 2009, 26, 278–282. [Google Scholar] [CrossRef]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The Basis for Resistance to β-Lactam Antibiotics by Penicillin-binding Protein 2a of Methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef]
- Itou, T.; Katayama, Y.; Hiramatsu, K. A new mobile genetic element, staphylococcal cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000, 55, 1549–1555. [Google Scholar]
- Schwendener, S.; Cotting, K.; Perreten, V. Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci. Rep. 2017, 7, srep43797. [Google Scholar] [CrossRef]
- Harrison, E.M.; Paterson, G.K.; Holden, M.T.; Larsen, J.; Stegger, M.; Larsen, A.R.; Petersen, A.; Skov, R.L.; Christensen, J.M.; Bak Zeuthen, A.; et al. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol. Med. 2013, 5, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Szczuka, E.; Krzymińska, S.; Bogucka, N.; Kaznowski, A. Multifactorial mechanisms of the pathogenesis of methicillin-resistant Staphylococcus hominis isolated from bloodstream infections. Antonie Leeuwenhoek 2017, 111, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Rachid, S.; Ohlsen, K.; Wallner, U.; Hacker, J.; Hecker, M.; Ziebuhr, W. Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 2000, 182, 6824–6826. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.; Almeida, L.; Gaio, V.; Cerca, N.; Manageiro, V.; Caniça, M.; Capelo, J.L.; Igrejas, G.; Poeta, P. Biofilm Formation of Multidrug-Resistant MRSA Strains Isolated from Different Types of Human Infections. Pathogens 2021, 10, 970. [Google Scholar] [CrossRef] [PubMed]
- Sandasi, M.; Leonard, C.; Viljoen, A. The in vitro antibiofilm activity of selected culinary herbs and medicinal plants against Listeria monocytogenes. Lett. Appl. Microbiol. 2010, 50, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Ballah, F.M.; Islam, M.S.; Rana, M.L.; Ferdous, F.B.; Ahmed, R.; Pramanik, P.K.; Karmoker, J.; Ievy, S.; Sobur, M.A.; Siddique, M.P.; et al. Phenotypic and Genotypic Detection of Biofilm-Forming Staphylococcus aureus from Different Food Sources in Bangladesh. Biology 2022, 11, 949. [Google Scholar] [CrossRef]
- Chen, Q.; Xie, S.; Lou, X.; Cheng, S.; Liu, X.; Zheng, W.; Zheng, Z.; Wang, H. Biofilm formation and prevalence of adhesion genes among Staphylococcus aureus isolates from different food sources. MicrobiologyOpen 2020, 9, e00946. [Google Scholar] [CrossRef]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef]
- Mirani, Z.A.; Fatima, A.; Urooj, S.; Aziz, M.; Khan, M.; Abbas, T. Relationship of cell surface hydrophobicity with biofilm formation and growth rate: A study on Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli. Iran. J. Basic Med. Sci. 2018, 21, 760–769. [Google Scholar] [CrossRef]
- Hamadi, F.; Latrache, H.; Mabrrouki, M.; Elghmari, A.; Outzourhit, A.; Ellouali, M.; Chtaini, A. Effect of pH on distribution and adhesion of Staphylococcus aureus to glass. J. Adhes. Sci. Technol. 2005, 19, 73–85. [Google Scholar] [CrossRef]
- Kouidhi, B.; Zmantar, T.; Hentati, H.; Bakhrouf, A. Cell surface hydrophobicity, biofilm formation, adhesives properties and molecular detection of adhesins genes in Staphylococcus aureus associated to dental caries. Microb. Pathog. 2010, 49, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, H.P.; Prasad, U.V.; Yeswanth, S.; Swarupa, V.; Prasad, O.H.; Narasu, M.L.; Sarma, P.V.G.K. Molecular characterization of α-amylase from Staphylococcus aureus. Bioinformation 2013, 9, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Shaw, L.; Golonka, E.; Potempa, J.; Foster, S.J. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 2004, 150 Pt 1, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.-E.; Dijkstra, B.W.; Reetz, M.T. Bacterial Biocatalysts: Molecular Biology, Three-Dimensional Structures, and Biotechnological Applications of Lipases. Annu. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef]
- Castro, A.; Santos, C.; Meireles, H.; Silva, J.; Teixeira, P. Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. J. Infect. Public Health 2016, 9, 153–160. [Google Scholar] [CrossRef]
- Amit, K.; Parimal, D.; Chandradipa, G. Biochemical and Molecular Analysis of Staphylococcus aureus Clinical Isolates from Hospitalized Patients. Can. J. Infect. Dis. Med. Microbiol. 2016, 2016, 9041636. [Google Scholar]
- Merghni, A.; Ben Nejma, M.; Helali, I.; Hentati, H.; Bongiovanni, A.; Lafont, F.; Aouni, M.; Mastouri, M. Assessment of adhesion, invasion and cytotoxicity potential of oral Staphylococcus aureus strains. Microb. Pathog. 2015, 86, 1–9. [Google Scholar] [CrossRef]
- Saïd-Salim, B.; Dunman, P.M.; McAleese, F.M.; Macapagal, D.; Murphy, E.; McNamara, P.J.; Arvidson, S.; Foster, T.J.; Projan, S.J.; Kreiswirth, B.N. Global Regulation of Staphylococcus aureus Genes by Rot. J. Bacteriol. 2003, 185, 610–619. [Google Scholar] [CrossRef]
- Hamdi, H.; Ben Salem, I.; Ben Othmène, Y.; Annabi, E.; Abid-Essefi, S. The involvement of ROS generation on Epoxiconazole-induced toxicity in HCT116 cells. Pestic. Biochem. Physiol. 2018, 148, 62–67. [Google Scholar] [CrossRef]
- Yu, J.-Y.; Zheng, Z.-H.; Son, Y.O.; Shi, X.; Jang, Y.-O.; Lee, J.-C. Mycotoxin zearalenone induces AIF- and ROS-mediated cell death through p53- and MAPK-dependent signaling pathways in RAW264.7 macrophages. Toxicol. In Vitro 2011, 25, 1654–1663. [Google Scholar] [CrossRef]
- Schwarzbacherová, V.; Wnuk, M.; Lewinska, A.; Potocki, L.; Zebrowski, J.; Koziorowski, M.; Holečková, B.; Šiviková, K.; Dianovský, J. Evaluation of cytotoxic and genotoxic activity of fungicide formulation Tango Super in bovine lymphocytes. Environ. Pollut. 2017, 220 Pt A, 255–263. [Google Scholar] [CrossRef]
- Cao, J.; Liu, Y.; Jia, L.; Zhou, H.M.; Kong, Y.; Yang, G.; Jiang, L.P.; Li, Q.J.; Zhong, L.F. Curcumin induces apoptosis through mitochondrial hyperpolarization and mtDNA damage in human hepatoma G2 cells. Free Radic. Biol. Med. 2007, 43, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xiong, W.; Wan, J.; Sun, X.; Xu, H.; Yang, X. The decrease of PAMAM dendrimer-induced cytotoxicity by PEGylation via attenuation of oxidative stress. Nanotechnology 2009, 20, 105103. [Google Scholar] [CrossRef] [PubMed]
- Amengual, J.; Lobo, G.P.; Golczak, M.; Li, H.N.; Klimova, T.; Hoppel, C.L.; Wyss, A.; Palczewski, K.; von Lintig, J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011, 25, 948–959. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).