Prevalence and Antibiotic Resistance Phenotypes of Pseudomonas spp. in Fresh Fish Fillets
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples Collection
- Five salmon (Salmo salar) fillets reared in Norway;
- Five plaice (Pleuronectes platessa) fillets caught using trawling in the Northeast Atlantic Ocean;
- Five northern cod (Gadus morhua) fillets caught using longlining in the Northeast Atlantic Ocean.
2.2. Samples Processing
2.3. Plate Counting and Statistical Analysis
2.4. Isolation of Pseudomonas spp. and Species Identification
2.5. Determination of Pseudomonas spp. Phenotypic Antibiotic Resistance
3. Results and Discussion
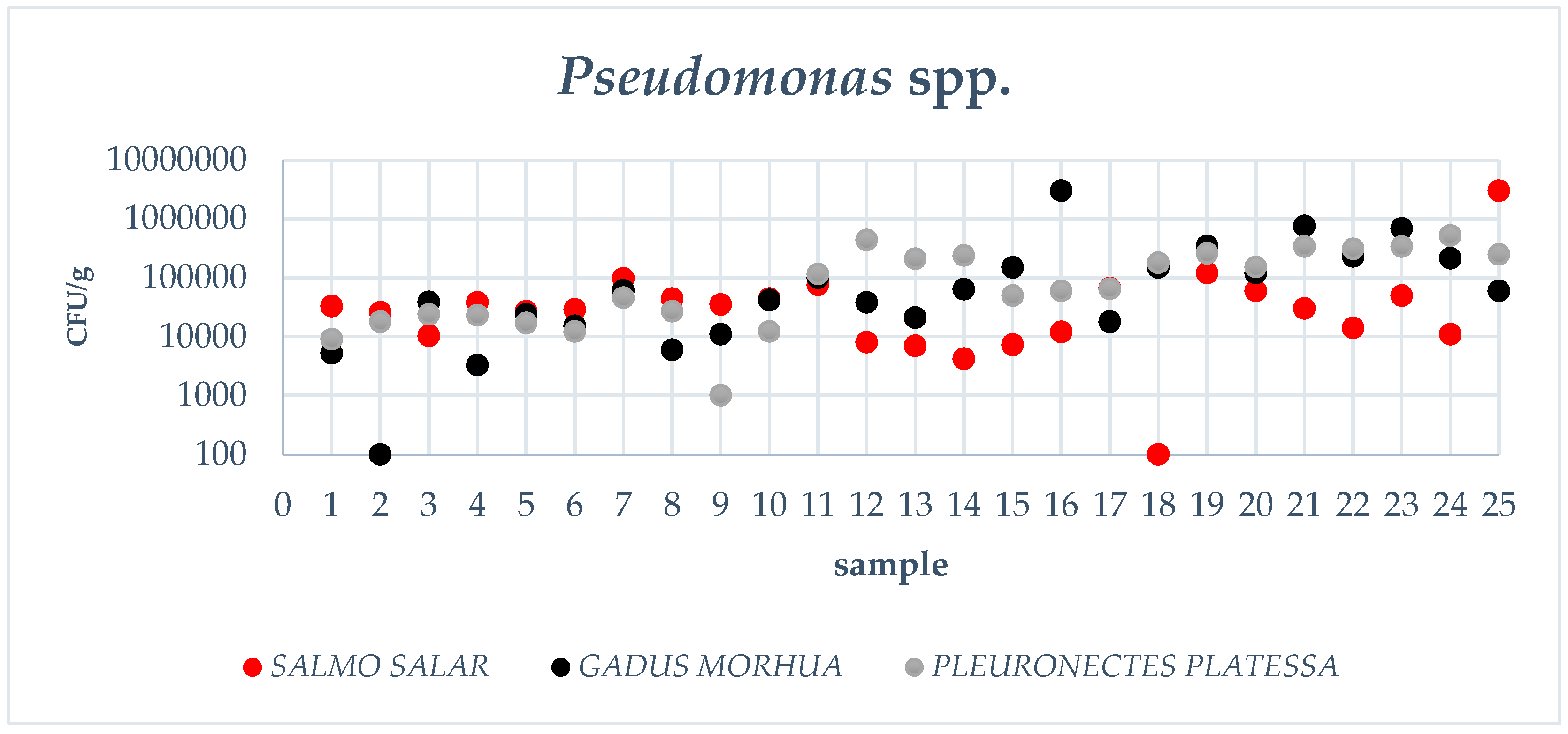
3.1. Pseudomonas spp. Count, Total Viable Count, Data Elaboration
3.2. Phenotypic Identification of Pseudomonas Species
3.3. Antimicrobial Resistance
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A



References
- Tryfinopoulou, P.; Tsakalidou, E.; Nychas, G.J.E. Characterization of Pseudomonas spp. associated with spoilage of gilt-head sea bream stored under various conditions. Appl. Environ. Microbiol. 2002, 68, 65–72. [Google Scholar] [CrossRef]
- Eissa, N.M.E.; Abou El-Ghiet, E.N.; Shaheen, A.A.; Abbass, A. Characterization of Pseudomonas species isolated from tilapia Oreochromis niloticus in Qaroun and Wadi-El-Rayan Lakes, Egypt. Glob. Vet. 2010, 5, 116–121. [Google Scholar] [CrossRef]
- Abd El Tawab, A.; Maarouf, A.; Ahmed, N. Detection of Virulence factors of Pseudomonas species isolated from fresh water fish by PCR. Benha Vet. Med. J. 2016, 30, 199–207. [Google Scholar] [CrossRef]
- Heir, E.; Moen, B.; Åsli, A.W.; Sunde, M.; Langsrud, S. Antibiotic resistance and phylogeny of Pseudomonas spp. Isolated over three decades from chicken meat in the norwegian food chain. Microorganisms 2021, 9, 207. [Google Scholar] [CrossRef]
- Gram, L.; Huss, H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef]
- De Jonghe, V.; Coorevits, A.; Van Hoorde, K.; Messens, W.; Van Landschoot, A.; De Vos, P.; Heyndrickx, M. Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 2011, 77, 460–470. [Google Scholar] [CrossRef]
- Doulgeraki, A.I.; Nychas, G.J.E. Monitoring the succession of the biota grown on a selective medium for pseudomonads during storage of minced beef with molecular-based methods. Food Microbiol. 2013, 34, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Mohareb, F.; Iriondo, M.; Doulgeraki, A.I.; Van Hoek, A.; Aarts, H.; Cauchi, M.; Nychas, G.J.E. Identification of meat spoilage gen (Eissa et al., 2010) e biomarkers in Pseudomonas putida using gene profiling. Food Control 2015, 57, 152–160. [Google Scholar] [CrossRef]
- Caldera, L.; Franzetti, L.; Van Coillie, E.; De Vos, P.; Stragier, P.; De Block, J.; Heyndrickx, M. Identification, enzymatic spoilage characterization and proteolytic activity quantification of Pseudomonas spp. isolated from different foods. Food Microbiol. 2016, 54, 142–153. [Google Scholar] [CrossRef]
- Taliadourou, D.; Papadopoulos, V.; Domvridou, E.; Savvaidis, I.N.; Kontominas, M.G. Microbiological, chemical and sensory changes of whole and filleted Mediterranean aquacultured sea bass (Dicentrarchus labrax) stored in ice. J. Sci. Food Agric. 2003, 83, 1373–1379. [Google Scholar] [CrossRef]
- Carpentier, B.; Chassaing, D. Interactions in biofilms between Listeria monocytogenes and resident microorganisms from food industry premises. Int. J. Food Microbiol. 2004, 97, 111–122. [Google Scholar] [CrossRef]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 2012, 36, 893–916. [Google Scholar] [CrossRef]
- Giaouris, E.; Heir, E.; Desvaux, M.; Hébraud, M.; Møretrø, T.; Langsrud, S.; Doulgeraki, A.; Nychas, G.J.; Kacániová, M.; Czaczyk, K.; et al. Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 2015, 6, 841. [Google Scholar] [CrossRef]
- Sterniša, M.; Klančnik, A.; Smole Možina, S. Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 °C. J. Sci. Food Agric. 2019, 99, 4635–4641. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Fuhrer, T.; Chavarría, M.; Sánchez-Pascuala, A.; Sauer, U.; de Lorenzo, V. Reconfiguration of metabolic fluxes in Pseudomonas putida as a response to sub-lethal oxidative stress. ISME J. 2021, 15, 1751–1766. [Google Scholar] [CrossRef]
- Chen, C.; Malek, A.A.; Wargo, M.J.; Hogan, D.A.; Beattie, G.A. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 2010, 75, 29–45. [Google Scholar] [CrossRef]
- Sheng, L.; Wang, L. The microbial safety of fish and fish products: Recent advances in understanding its significance, contamination sources, and control strategies. Compr. Rev. Food Sci. Food Saf. 2021, 20, 738–786. [Google Scholar] [CrossRef] [PubMed]
- Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Official Journal of the European Union L338 22 December 2005. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32005R2073from=EN (accessed on 12 January 2023).
- FAO. The State of World Fisheries and Aquaculture 2022. In Towards Blue Transformation; FAO: Rome, Italy, 2022. [Google Scholar] [CrossRef]
- The European Market Observatory for Fisheries and Aquaculture. Available online: https://www.eumofa.eu/it/data (accessed on 12 January 2023).
- Schar, D.; Klein, E.Y.; Laxminarayan, R.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in aquaculture. Sci. Rep. 2020, 10, 918. [Google Scholar] [CrossRef] [PubMed]
- Scott, A.; McEwen, P.J.C. Antimicrobial resistance: A One Health perspective. Trans. R. Soc. Trop. Med. Hyg. 2018, 111, 255–260. [Google Scholar] [CrossRef]
- Marshall, B.M.; Levy, S.B. Food animals and antimicrobials: Impacts on human health. Clin. Microbiol. Rev. 2011, 24, 718–733. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, S.; Hoffer, C.; Youf, R.; Dauvergne, E.; Webb, H.E.; Brauge, T.; Tran, M.-L.; Midelet, G.; Granier, S.A.; Haenni, M.; et al. High Throughput Screening of Antimicrobial Resistance Genes in Gram-Negative Seafood Bacteria. Microorganisms 2022, 10, 1225. [Google Scholar] [CrossRef]
- Levy, S.B.; Bonnie, M. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Kačániová, M.; Terentjeva, M.; Vukovic, N.; Puchalski, C.; Roychoudhury, S.; Kunová, S.; Klūga, A.; Tokár, M.; Kluz, M.; Ivanišová, E. The antioxidant and antimicrobial activity of essential oils against Pseudomonas spp. isolated from fish. Saudi Pharm. J. 2017, 65, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef]
- Luczkiewicz, A.; Kotlarska, E.; Artichowicz, W.; Tarasewicz, K.; Fudala-Ksiazek, S. Antimicrobial resistance of Pseudomonas spp. isolated from wastewater and wastewater-impacted marine coastal zone. Environ. Sci. Pollut. Res. 2015, 22, 19823–19834. [Google Scholar] [CrossRef]
- King, D.T.; Sobhanifar, S.; Strynadka, N.C.J. Handbook of Antimicrobial Resistance. In Handbook of Antimicrobial Resistance; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–22. [Google Scholar] [CrossRef]
- Elbehiry, A.; Marzouk, E.; Aldubaib, M.; Moussa, I.; Abalkhail, A.; Ibrahem, M.; Hamada, M.; Sindi, W.; Alzaben, F.; Almuzaini, A.M.; et al. Pseudomonas species prevalence, protein analysis, and antibiotic resistance: An evolving public health challenge. AMB Express 2022, 12, 53. [Google Scholar] [CrossRef]
- Kačániová, M.; Klūga, A.; Kántor, A.; Medo, J.; Žiarovská, J.; Puchalski, C.; Terentjeva, M. Comparison of MALDI-TOF MS Biotyper and 16S rDNA sequencing for the identification of Pseudomonas species isolated from fish. Microb. Pathog. 2019, 132, 313–318. [Google Scholar] [CrossRef]
- Fazeli, N.; Momtaz, H. Virulence gene profiles of multidrug-resistant Pseudomonas aeruginosa isolated from Iranian hospital infections. Iran. Red Crescent Med. J. 2014, 16, e15722. [Google Scholar] [CrossRef]
- Shabana, B.M.; Elkenany, R.M.; Younis, G. Sequencing and multiple antimicrobial resistance of Pseudomonas fluorescens isolated from Nile tilapia fish in Egypt. Braz. J. Biol. 2022, 84, 1–9. [Google Scholar] [CrossRef]
- EUCAST Disk Diffusion Test Methodology. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology (accessed on 12 January 2023).
- Simonsen, G.S.; Blix, H.S.; Grave, K.; Urdahl, A.M. NORM/NORM-VET 2020: Usage of Antimicrobial Agents and Occurrence of Antimicrobial Resistance in Norway; Norwegian Institute of Public Health: Tromsø/Oslo, Norway, 2021. [Google Scholar]
- WHO. Critically Important Antimicrobials for Human Medicine (6th revision). Available online: https://www.who.int/publications/i/item/9789241515528 (accessed on 12 January 2023).
- Silbande, A.; Adenet, S.; Smith-Ravin, J.; Joffraud, J.J.; Rochefort, K.; Leroi, F. Quality assessment of ice-stored tropical yellowfin tuna (Thunnus albacares) and influence of vacuum and modified atmosphere packaging. Food Microbiol. 2016, 60, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Speck, M.L.; Ray, B. Effects of Freezing and Storage on Microorganisms in Frozen Foods: A Review. J. Food Prot. 1997, 40, 333–336. [Google Scholar] [CrossRef]
- Baña, Z.; Abad, N.; Uranga, A.; Azúa, I.; Artolozaga, I.; Unanue, M.; Iriberri, J.; Arrieta, J.M.; Ayo, B. Recurrent seasonal changes in bacterial growth efficiency, metabolism and community composition in coastal waters. Environ Microbiol. 2020, 22, 369–380. [Google Scholar] [CrossRef]
- Thomassen, G.M.B.; Reiche, T.; Tennfjord, C.E.; Mehli, L. Antibiotic Resistance Properties among Pseudomonas spp. Associated with Salmon Processing Environments. Microorganisms 2022, 10, 1420. [Google Scholar] [CrossRef] [PubMed]
- Livermore, D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin. Infect. Dis. 2002, 34, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Du, W.X.; Kim, J.; Cornell, J.A.; Huang, T.S.; Marshall, M.R.; Wei, C.I. Microbiological 2002, sensory, and electronic nose evaluation of yellowfin tuna under various storage conditions. J. Food Prot. 2001, 64, 2027–2036. [Google Scholar] [CrossRef] [PubMed]
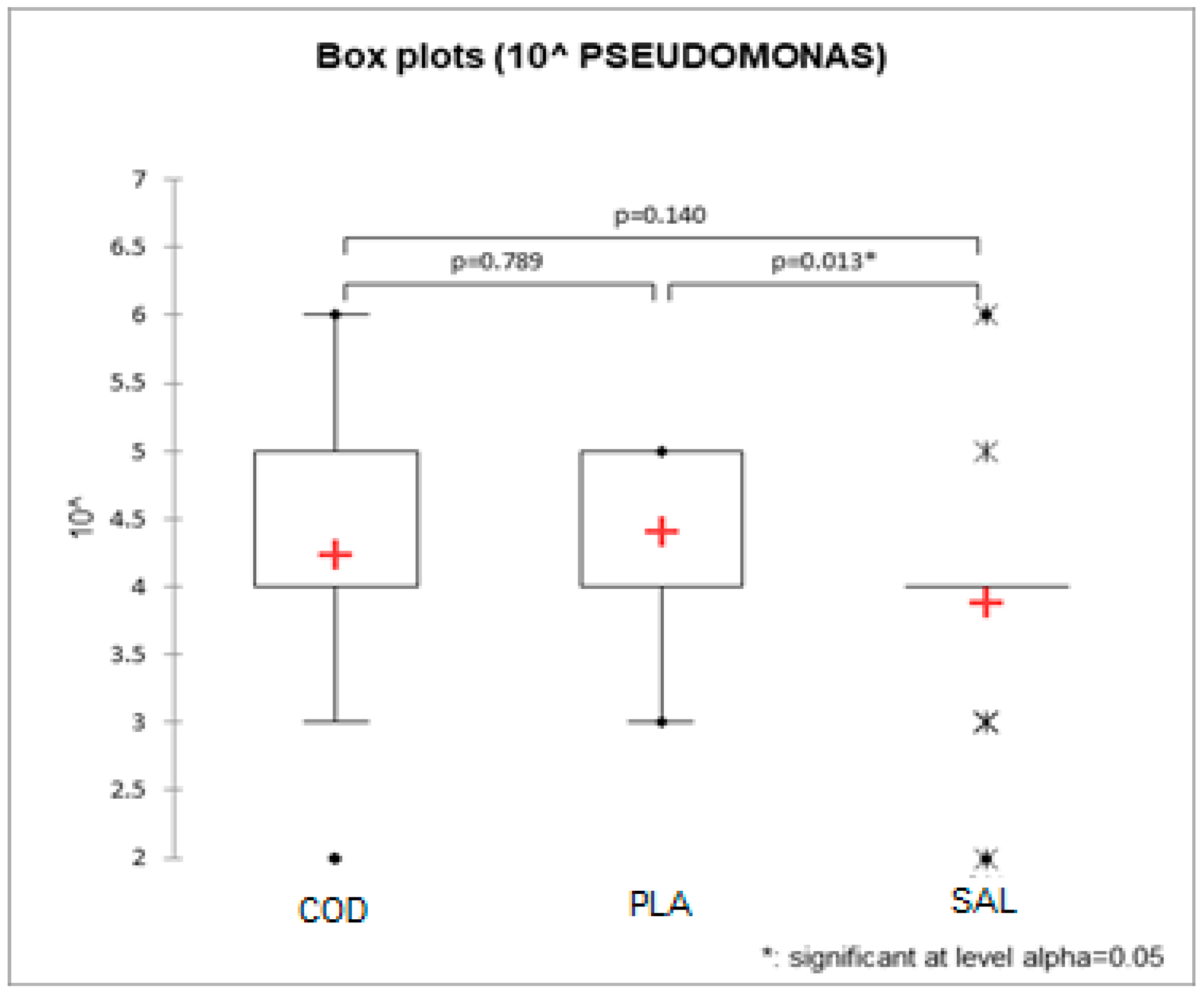
| Fish Fillets | TVC Mean Value (cfu/g) | Pseudomonas spp. Mean Value (cfu/g) |
|---|---|---|
| Gadus morhua | 2.57 × 105 (1.83–3.61 × 105) a | 4.67 × 104 (2.68–8.16 × 104) a,b |
| Pleuronectes platessa | 2.56 × 105 (1.82–3.60 × 105) a | 6.71 × 104 (3.85–1.17 × 104) a |
| Salmo salar | 1.07 × 105 (7.65–1.51 × 105) b | 2.33 × 104 (1.34–4.06 × 104) b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ben Mhenni, N.; Alberghini, G.; Giaccone, V.; Truant, A.; Catellani, P. Prevalence and Antibiotic Resistance Phenotypes of Pseudomonas spp. in Fresh Fish Fillets. Foods 2023, 12, 950. https://doi.org/10.3390/foods12050950
Ben Mhenni N, Alberghini G, Giaccone V, Truant A, Catellani P. Prevalence and Antibiotic Resistance Phenotypes of Pseudomonas spp. in Fresh Fish Fillets. Foods. 2023; 12(5):950. https://doi.org/10.3390/foods12050950
Chicago/Turabian StyleBen Mhenni, Nesrine, Giulia Alberghini, Valerio Giaccone, Alessandro Truant, and Paolo Catellani. 2023. "Prevalence and Antibiotic Resistance Phenotypes of Pseudomonas spp. in Fresh Fish Fillets" Foods 12, no. 5: 950. https://doi.org/10.3390/foods12050950
APA StyleBen Mhenni, N., Alberghini, G., Giaccone, V., Truant, A., & Catellani, P. (2023). Prevalence and Antibiotic Resistance Phenotypes of Pseudomonas spp. in Fresh Fish Fillets. Foods, 12(5), 950. https://doi.org/10.3390/foods12050950





