Legume Fingerprinting through Lipid Composition: Utilizing GC/MS with Multivariate Statistics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Composition
2.2. Lipid Extraction
2.3. GC/MS Analysis
2.4. Data Acquisition
2.5. Data Analysis
3. Results
3.1. Data Acquisition
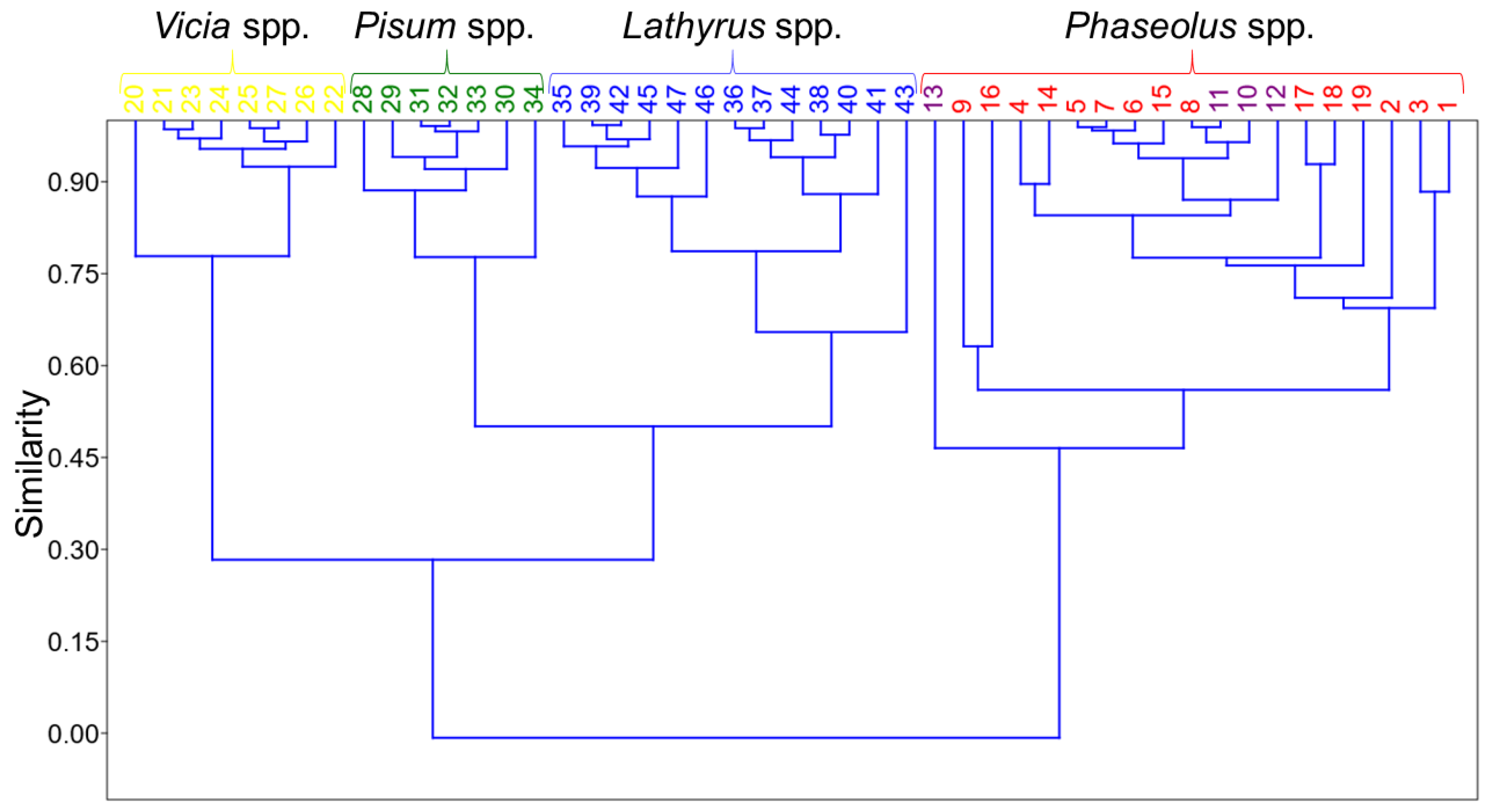
3.2. Hierarchical Cluster Analysis
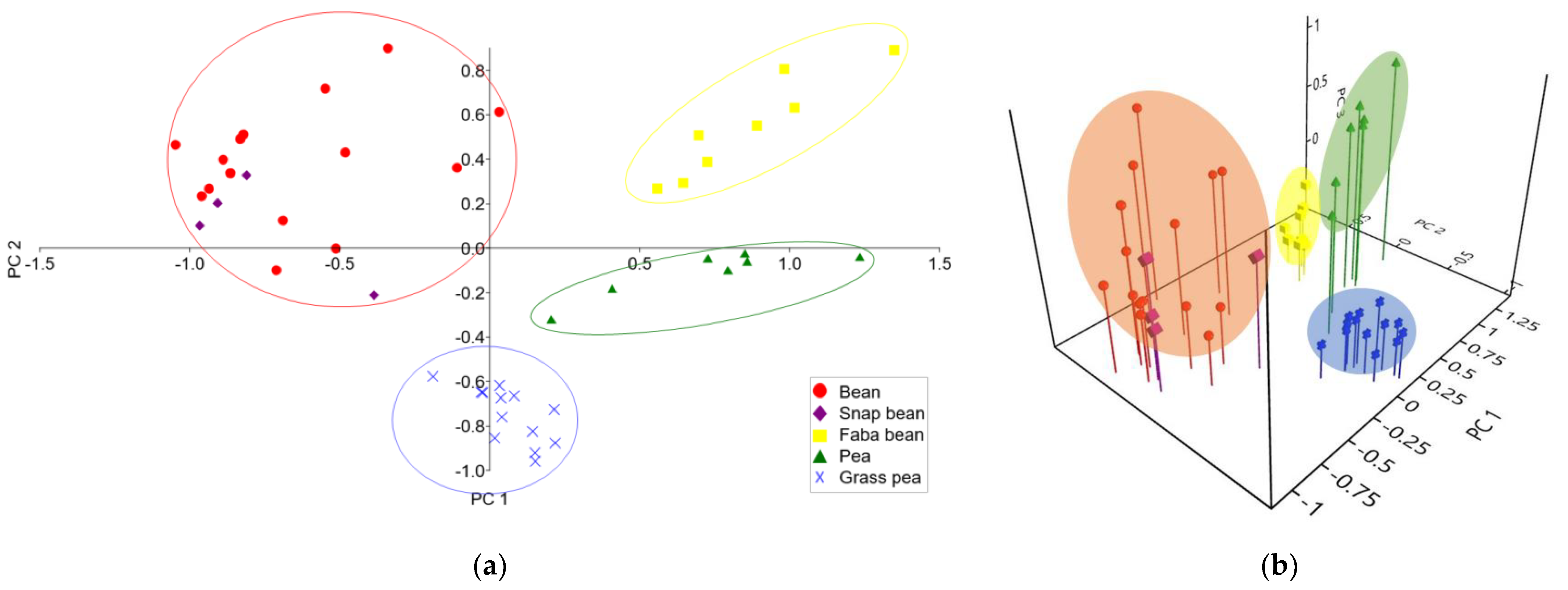
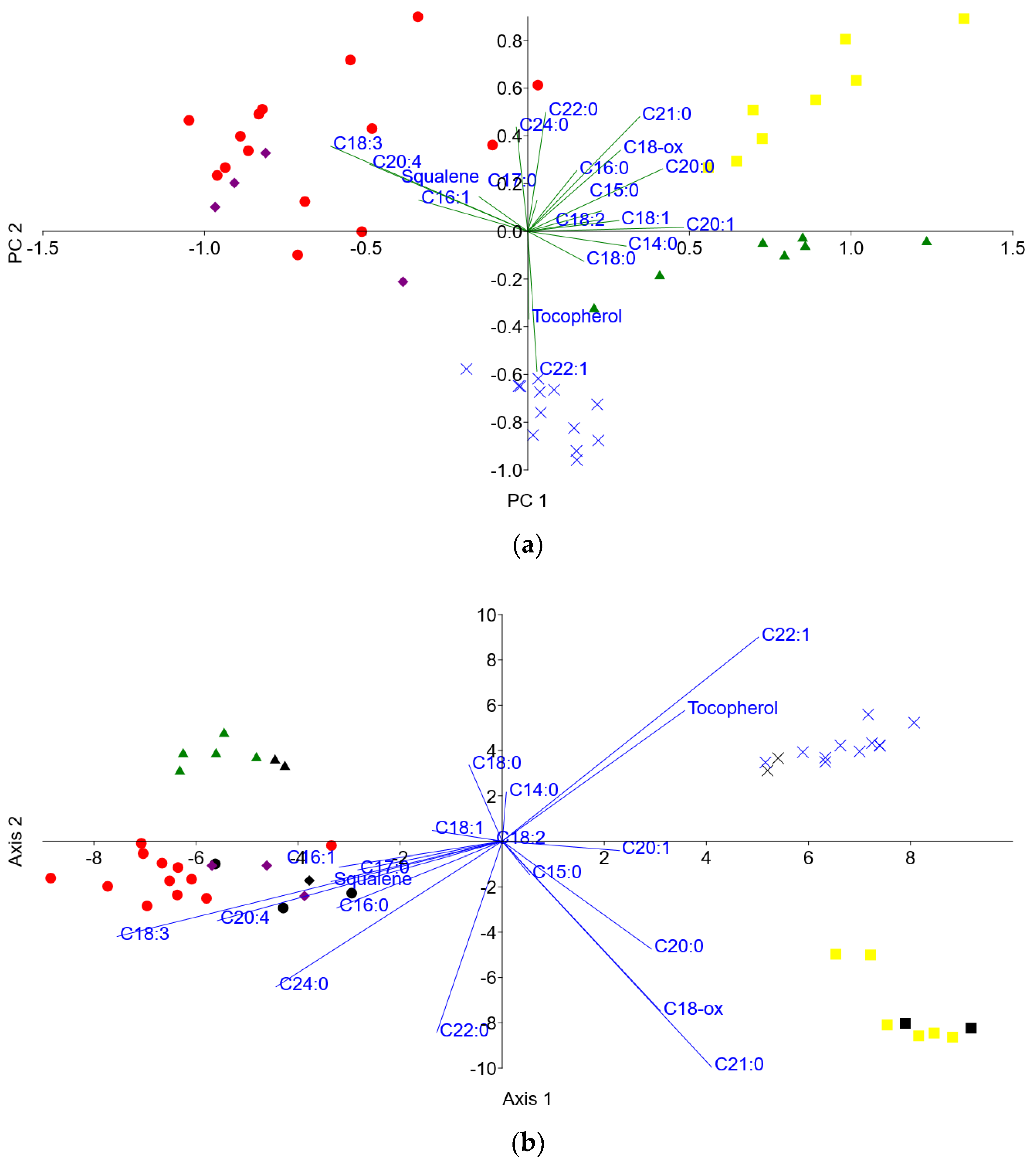
3.3. Principal Component Analysis
3.4. Linear Discriminant Analysis
3.5. Fingerprinting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smykal, P.; Coyne, C.J.; Ambrose, M.J.; Maxted, N.; Schaefer, H.; Blair, M.W.; Berger, J.; Greene, S.J.; Nelson, M.N.; Besharat, N.; et al. Legume crops phylogeny and genetic diversity for science and breeding. CRC Crit. Rev. Plant Sci. 2015, 34, 43–104. [Google Scholar] [CrossRef]
- Rubiales, D.; Annicchiarico, P.; Vaz Patto, M.C.; Julier, B. Legumes breeding for the agricultural transition of global agri-food systems: A European perspective. Front. Plant Sci. 2021, 12, 782574. [Google Scholar] [CrossRef]
- Lewis, G.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Royal Botanic Gardens: Kew, UK, 2005. [Google Scholar]
- Khodapanahi, E.; Lefsrud, M.; Orsat, V.; Singh, J.; Warkentin, T.D. Study of pea accessions for development of an oilseed pea. Energies 2012, 5, 3788–3802. [Google Scholar] [CrossRef]
- Dahl, W.J.; Foster, L.M.; Tyler, R.T. Review of the health benefits of peas (Pisum sativum L.). Brit. J. Nutrit. 2012, 108 (Suppl. S1), S3–S10. [Google Scholar] [CrossRef]
- Duc, G.; Bao, S.; Baumc, M.; Redden, B.; Sadiki, M.; Suso, M.J.; Vishniakova, M.; Zong, X. Diversity maintenance and use of Vicia faba L. genetic resources. Field Crop. Res. 2010, 115, 270–278. [Google Scholar] [CrossRef]
- McVicar, R.; Panchuk, D.; Brenzil, C.; Hartley, S.; Pearse, P.; Vandenberg, A. Faba Bean. Gov. Saskatchewan, Agriculture, Crops. 2013. Available online: http://www.agriculture.gov.sk.ca/Default.aspx?DN=a2c9a5ff-8467-4880-9ad7-18a4515eb0d2 (accessed on 1 November 2023).
- Apaydin, H.; Ertan, S.; Uzekmekci, S. Broad bean (Vicia faba)—A natural source of L-DOPA—Prolongs “on” periods in patenients with parkinson’s disease who have “on-off” fluctuations. Move. Disord. 2000, 15, 164–166. [Google Scholar] [CrossRef]
- Caracuta, V.; Vardi, J.; Paz, Y.; Boaretto, E. Farming legumes in the pre-pottery Neolithic: New discoveries from the site of Ahihud (Israel). PLoS ONE 2017, 12, e0177859. [Google Scholar] [CrossRef] [PubMed]
- Barrow, M.V.; Simpson, C.F.; Miller, E.J. Lathyrism: A Review. Quart. Review Biol. 1974, 49, 101–128. [Google Scholar] [CrossRef] [PubMed]
- De Ron, A.; Gonzalez, A.; Rodino, A.; Santalla, M.; Godoy, L.; Papa, R. History of the common bean crop: Its evolution beyond its areas of origin and domestication. Arbor 2016, 192, a317. [Google Scholar] [CrossRef]
- Munoz, C.; Duque, M.C.; Debouck, D.; Blair, M. Taxonomy of tepary bean (Phaseolus acutifolius) and wild relatives as determined by amplified fragment length polymorphism (AFLP) markers. Crop Sci. 2006, 46, 1744–1754. [Google Scholar] [CrossRef]
- Bitocchi, E.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a Model for Understanding Crop Evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef]
- Huschek, G.; Bönick, J.; Merkel, D.; Huschek, D.; Rawel, H. Authentication of leguminous-based products by targeted biomarkers using high resolution time of flight mass spectrometry. LWT 2018, 90, 164–171. [Google Scholar] [CrossRef]
- Madesis, P.; Ganopoulos, I.; Anagnostis, A.; Tsaftaris, A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control 2012, 25, 576–582. [Google Scholar] [CrossRef]
- Pastor, K.; Vujasinovic, V.; Marjanovic Jeromela, A.; Vujic, D.; Jovanovic, D.; Acanski, M. Gas chromatography—Mass spectrometry system applied to determine botanical origin of various types of edible vegetable oils. J. Serbian Chem. Soc. 2019, 84, 1019–1025. [Google Scholar]
- Yan, J.; Oey, S.B.; van Leeuwen, S.P.J.; van Ruth, S.M. Discrimination of processing grades of olive oil and other vegetable oils by monochloropropanediol esters and glycidyl esters. Food Chem. 2018, 248, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Ilić, M.; Pastor, K.; Romanić, R.; Vujić, Đ.; Ačanski, M. A New Challenge in Food Authenticity: Application of a Novel Mathematical Model for Rapid Quantification of Vegetable Oil Blends by Gas Chromatography—Mass Spectrometry (GC-MS). Anal. Lett. 2022, 55, 2752–2763. [Google Scholar] [CrossRef]
- Ilić, M.; Pastor, K.; Romanić, R.; Vujić, Đ.; Ačanski, M. A GC-MS based Fatty Acid Profiling Approach for Uncovering the Composition of Edible Oil Blends. Food Anal. Methods 2023, 16, 1149–1155. [Google Scholar] [CrossRef]
- Pastor, K.; Ilić, M.; Kojić, J.; Ačanski, M.; Vujić, D. Classification of Cereal Flour by Gas Chromatography—Mass Spectrometry (GCMS) Liposoluble Fingerprints and Automated Machine Learning. Anal. Lett. 2022, 55, 2220–2226. [Google Scholar] [CrossRef]
- Pastor, K.; Pezo, L.; Vujić, Đ.; Jovanović, Đ.; Ačanski, M. Discriminating cereal and pseudocereal species using binary system of GC/MS data—Pattern recognition approach. J. Serbian Chem. Soc. 2018, 83, 317–329. [Google Scholar] [CrossRef]
- Pastor, K.; Zorlu, G.; Osman, S.; Öztürk Sevdik, Y.; Kojić, J.; Nastić, N.; Senyuva, H. Chemometric fatty acidomics to distinguish between yeast and sourdough breads from Serbia and Turkey. J. Food Nutr. Res. 2022, 61, 339–351. [Google Scholar]
- Caligiani, A.; Nocetti, M.; Lolli, V.; Marseglia, A.; Palla, G. Development of a quantitative GC-MS method for the detection of cyclopropane fatty acids in cheese as new molecular markers for Parmigiano Reggiano authentication. J. Agric. Food Chem. 2016, 64, 4158–4164. [Google Scholar] [CrossRef] [PubMed]
- Husak, R.; Sebranek, J.; Bregendahl, K. A survey of commercially available broilers marketed as organic, free-range, and conventional broilers for cooked meat yields, meat composition, and relative value. Poult. Sci. 2008, 87, 2367–2376. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Seong, P.N.; Cho, S.H.; Kim, J.H.; Lee, J.M.; Jo, C.; Lim, D.G. Fatty acid composition and meat quality traits of organically reared Korean native black pigs. Livest. Sci. 2009, 120, 96–102. [Google Scholar] [CrossRef]
- Çayan, F.; Deveci, E.; Tel-Çayan, G.; Duru, M.E. Chemometric approaches for the character-ization of the fatty acid composition of seventeen mushroom species. Anal. Lett. 2020, 53, 2784–2798. [Google Scholar] [CrossRef]
- Caprioli, G.; Giusti, F.; Ballini, R.; Sagratini, G.; Vila-Donat, P.; Vittori, S.; Fiorini, D. Lipid nutritional value of legumes: Evaluation of different extraction methods and determination of fatty acid composition. Food Chem. 2016, 192, 965–971. [Google Scholar] [CrossRef]
- Los, F.G.B.; Zielinski, A.A.F.; Wojeicchowski, J.P.; Nogueira, A.; Demiate, I.M. Beans (Phaseolus vulgaris L.): Whole seeds with complex chemical composition. Curr. Opin. Food Sci. 2018, 19, 63–71. [Google Scholar] [CrossRef]
- Chaurasia, S. Chapter 17—Green Beans, in Nutritional Composition and Antioxidant Properties of Fruits and Vegetables; Jaiswal, A.K., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 289–300. [Google Scholar]
- Llorach, R.; Favari, C.; Alonso, D.; Garcia-Aloy, M.; Andres-Lacueva, C.; Urpi-Sarda, M. Comparative metabolite fingerprinting of legumes using LC-MS-based untargeted metabolomics. Food Res. Int. 2019, 126, 108666. [Google Scholar] [CrossRef]
- Sayre-Chavez, B.; Baxter, B.; Broeckling, C.D.; Muñoz-Amatriaín, M.; Manary, M.; Ryan, E.P. Non-targeted metabolomics of cooked cowpea (Vigna unguiculata) and pigeon pea (Cajanus cajan) from Ghana using two distinct and complementary analytical platforms. Food Chem. Mol. Sci. 2022, 4, 100087. [Google Scholar] [CrossRef]
- Longobardi, F.; Innamorato, V.; Di Gioia, A.; Ventrella, A.; Lippolis, V.; Logrieco, A.F.; Catucci, L.; Agostiano, A. Geographical origin discrimination of lentils (Lens culinaris Medik.) using 1H NMR fingerprinting and multivariate statistical analyses. Food Chem. 2017, 237, 743–748. [Google Scholar] [CrossRef]
- Foschi, M.; D’Archivio, A.A.; Rossi, L. Geographical discrimination and authentication of lentils (Lens culinaris Medik.) by ICP-OES elemental analysis and chemometrics. Food Control 2020, 118, 107438. [Google Scholar] [CrossRef]
- Grembecka, M.; Szefer, P. Elemental Profiles of Legumes and Seeds in View of Chemometric Approach. Appl. Sci. 2022, 12, 1577. [Google Scholar] [CrossRef]
- Lioi, L.; Galasso, I.; Santantonio, M.; Lanave, C.; Bollini, R.; Sparvoli, F. Lectin gene sequences and species relationships among cultivated legumes. Genet. Resour. Crop Evol. 2006, 53, 1615–1623. [Google Scholar] [CrossRef]
- Omar, G.I.; Saqer, M.M.; Adwan, G.M. Phylogenetic Relationship among Some species of the Genera Lens, Vicia, Lathyrus and Pisum (Leguminosae) in Palestine. Jordan J. Biol. Sci. 2019, 12, 289–296. [Google Scholar]
- Yildizdogan, Z.; Ikten, C.; Mutlu, N.; Toker, C. Genetic relationships among the genera Cicer L., Lathyrus L., Lens Mill., and Vicia L., together with similarity of Lens taxa based on morphological and AFLP markers. Turk. J. Bot. 2016, 40, 566–575. [Google Scholar] [CrossRef]

| Compound | Abbr. * | Retention Time [min] | Match Range [%] |
|---|---|---|---|
| Tetradecanoic acid, methyl ester | C14:0 | 8.74 | 81.2–95.2 |
| Pentadecanoic acid, methyl ester | C15:0 | 9.44 | 80.7–92.2 |
| 9-Hexadecenoic acid, methyl ester | C16:1 | 10.00 | 80.0–89.9 |
| Hexadecanoic acid, methyl ester | C16:0 | 10.13 | 92.5–97.3 |
| Heptadecanoic acid, methyl ester | C17:0 | 10.79 | 80.1–86.5 |
| 9,12-Octadecadienoic acid, methyl ester | C18:2 | 11.25 | 89.6–96.7 |
| 9,12,15-Octadecatrienoic acid, methyl ester | C18:3 | 11.29 | 82.5–95.6 |
| 9-Octadecenoic acid, methyl ester | C18:1 | 11.30 | 80.0–93.6 |
| Stearic acid, methyl ester | C18:0 | 11.42 | 91.4–95.9 |
| 5,11,14,17-Eicosatetraenoic acid, methyl ester | C20:4 | 12.36 | 81.8–84.4 |
| Oxiraneoctanoic acid, 3-octyl-, methyl ester | C18-ox | 12.44 | 81.9–92.6 |
| 11-Eicosenoic acid, methyl ester | C20:1 | 12.48 | 80.6–90.2 |
| Eicosanoic acid, methyl ester | C20:0 | 12.61 | 81.7–93.0 |
| Heneicosanoic acid, methyl ester | C21:0 | 13.20 | 80.5–83.4 |
| 13-Docosenoic acid, methyl ester | C22:1 | 13.71 | 80.7–87.9 |
| Docosanoic acid, methyl ester | C22:0 | 13.86 | 80.1–90.5 |
| Tetracosanoic acid, methyl ester | C24:0 | 15.50 | 80.2–88.2 |
| Squalene | Squalene | 16.41 | 81.0–91.7 |
| γ-Tocopherol, O-methyl- | Tocopherol | 18.40 | 87.0–90.9 |
| Groups * | Bean | Snap Bean | Faba Bean | Pea | Grass Pea | Total |
|---|---|---|---|---|---|---|
| Bean | 12 | 0 | 0 | 0 | 0 | 12 |
| Snap bean | 0 | 3 | 0 | 0 | 0 | 3 |
| Faba bean | 0 | 0 | 6 | 0 | 0 | 6 |
| Pea | 0 | 0 | 0 | 5 | 0 | 5 |
| Grass Pea | 0 | 0 | 0 | 0 | 11 | 11 |
| Total | 12 | 3 | 6 | 5 | 11 | 37 |
| Groups * | Bean | Snap Bean | Faba Bean | Pea | Grass Pea | Total |
|---|---|---|---|---|---|---|
| Bean | 14 | 1 | 0 | 0 | 0 | 15 |
| Snap bean | 0 | 4 | 0 | 0 | 0 | 4 |
| Faba bean | 0 | 0 | 8 | 0 | 0 | 8 |
| Pea | 0 | 0 | 0 | 7 | 0 | 7 |
| Grass Pea | 0 | 0 | 0 | 0 | 13 | 13 |
| Total | 14 | 5 | 8 | 7 | 13 | 47 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ilić, M.; Pastor, K.; Ilić, A.; Vasić, M.; Nastić, N.; Vujić, Đ.; Ačanski, M. Legume Fingerprinting through Lipid Composition: Utilizing GC/MS with Multivariate Statistics. Foods 2023, 12, 4420. https://doi.org/10.3390/foods12244420
Ilić M, Pastor K, Ilić A, Vasić M, Nastić N, Vujić Đ, Ačanski M. Legume Fingerprinting through Lipid Composition: Utilizing GC/MS with Multivariate Statistics. Foods. 2023; 12(24):4420. https://doi.org/10.3390/foods12244420
Chicago/Turabian StyleIlić, Marko, Kristian Pastor, Aleksandra Ilić, Mirjana Vasić, Nataša Nastić, Đura Vujić, and Marijana Ačanski. 2023. "Legume Fingerprinting through Lipid Composition: Utilizing GC/MS with Multivariate Statistics" Foods 12, no. 24: 4420. https://doi.org/10.3390/foods12244420
APA StyleIlić, M., Pastor, K., Ilić, A., Vasić, M., Nastić, N., Vujić, Đ., & Ačanski, M. (2023). Legume Fingerprinting through Lipid Composition: Utilizing GC/MS with Multivariate Statistics. Foods, 12(24), 4420. https://doi.org/10.3390/foods12244420









