Effect of Fortified Inoculation with Indigenous Lactobacillus brevis on Solid-State Fermentation of Light-Flavor Baijiu
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Bacteria and Yeasts
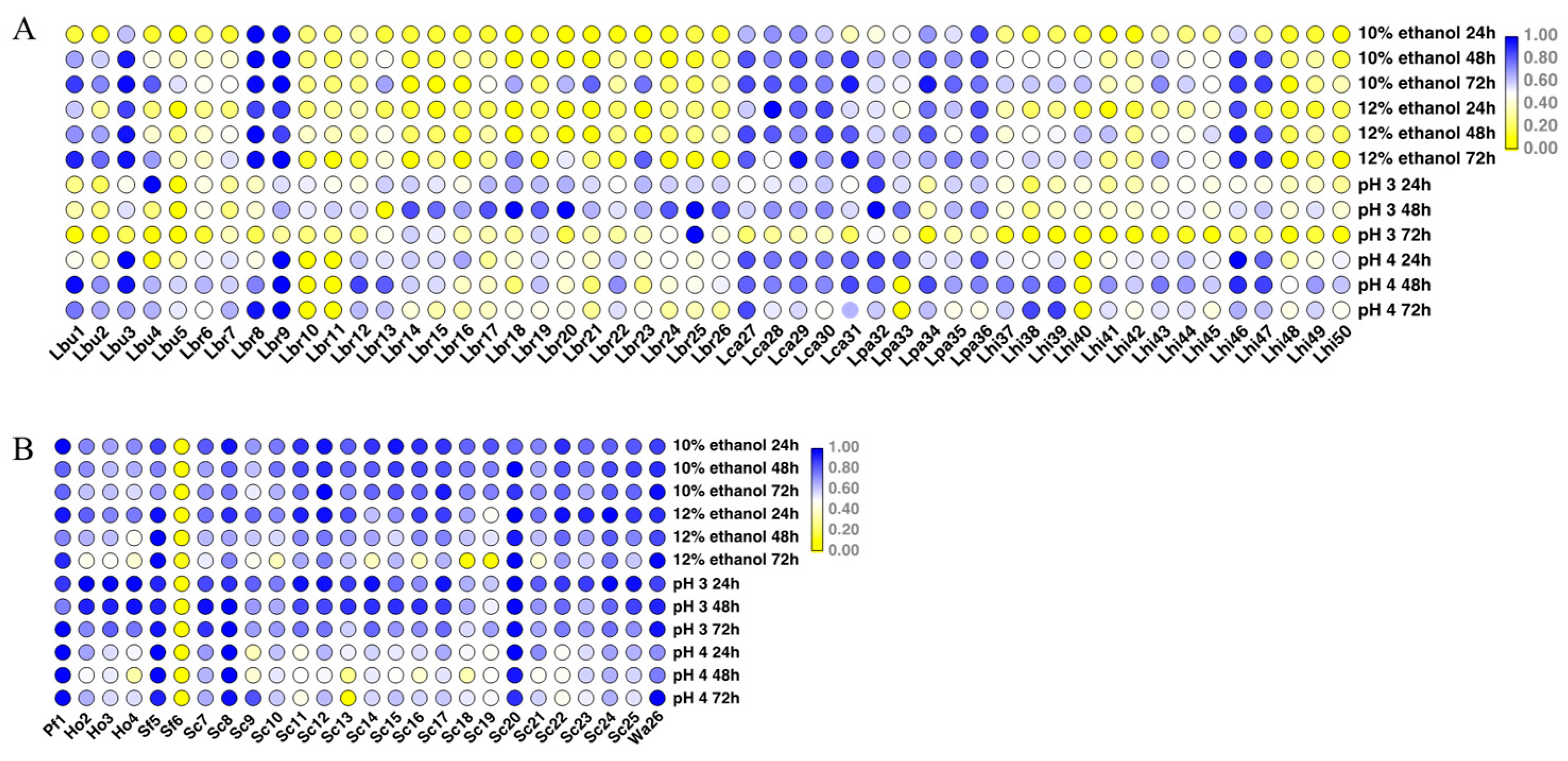
2.2. Environmental Tolerance of Various Species
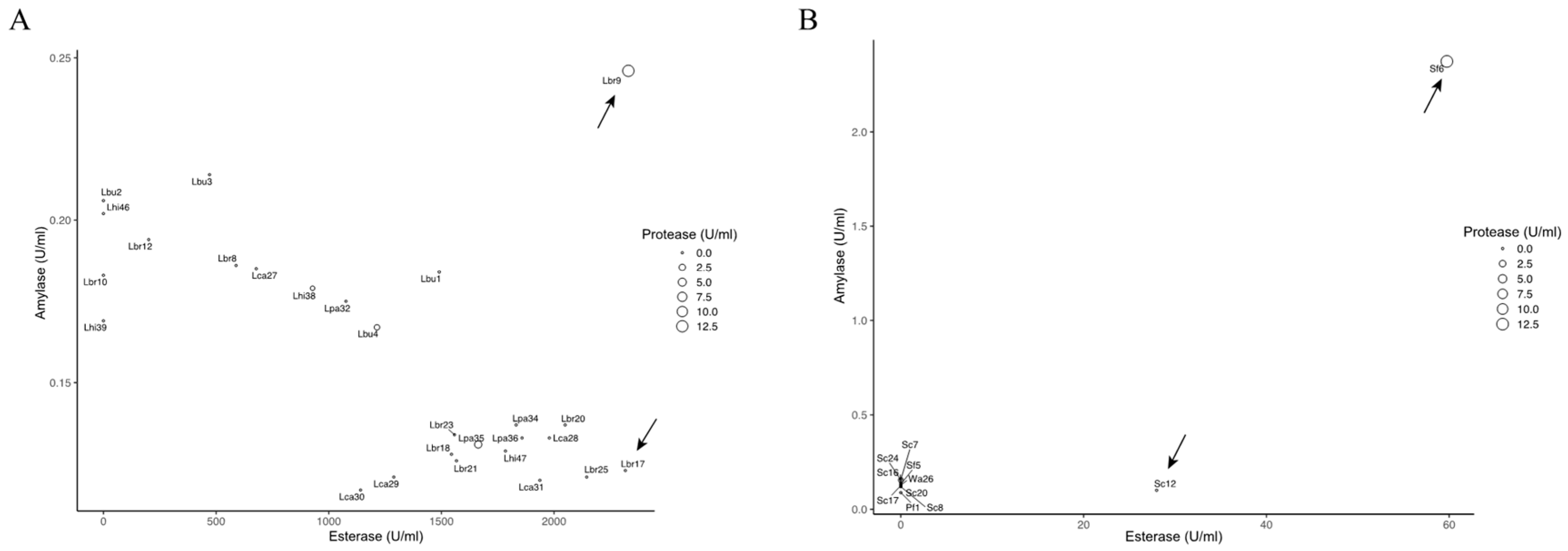
2.3. Enzyme Activities in Various Species
2.4. Fortified Fermentation Inoculated with Promising Candidates
2.5. Analysis of Volatile Compounds in Fermented Grains
2.6. Quantifying Microbial Dynamics during Fermentation
2.7. Physicochemical Properties of Fermented Grains
2.8. Statistical Analysis
3. Results
3.1. Isolation and Identification of LAB and Yeasts from Fermented Grains
3.2. Characterization and Screening of Isolated Strains
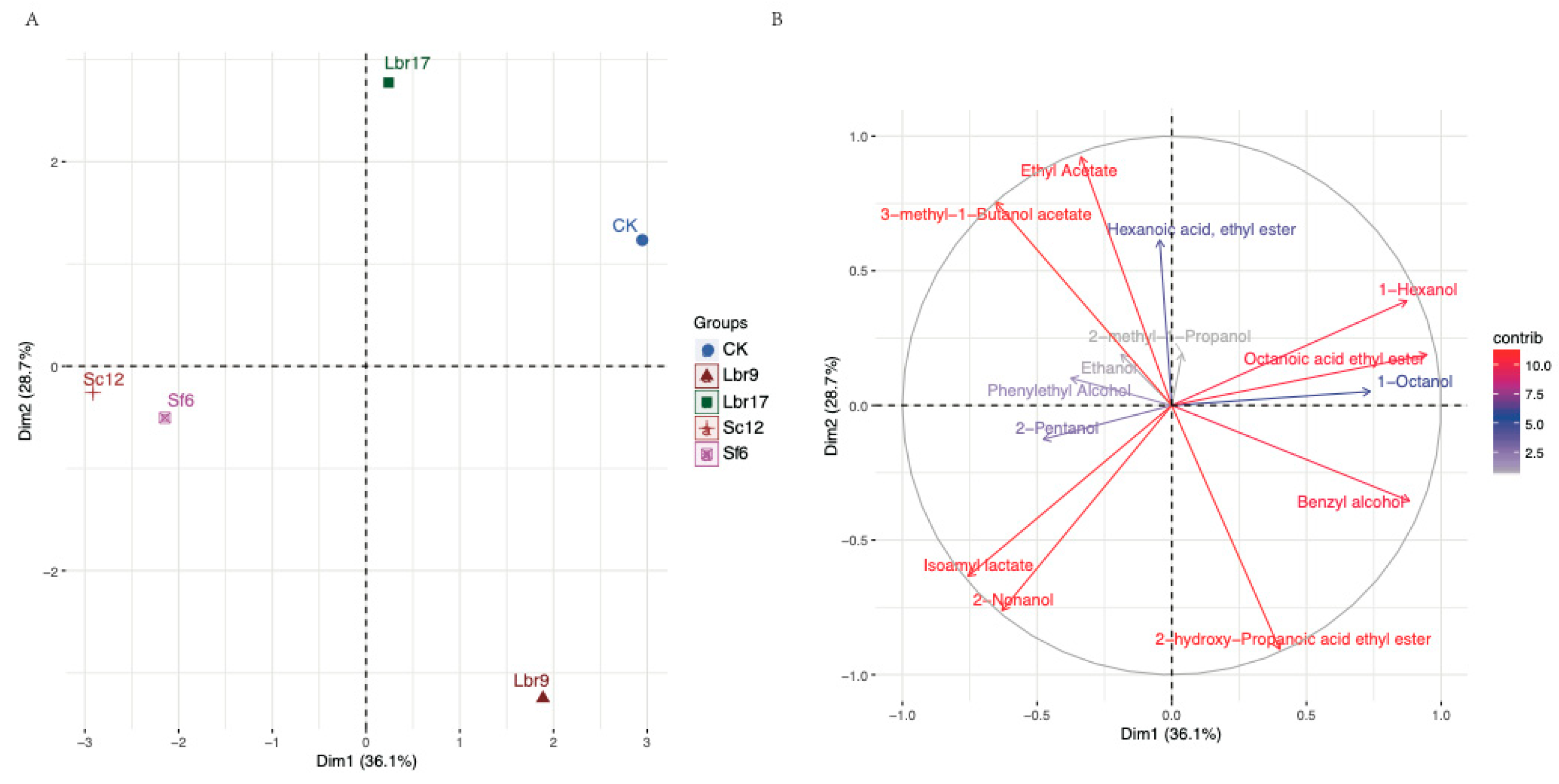
3.3. Fortified Fermentation with Promising Candidate Strains
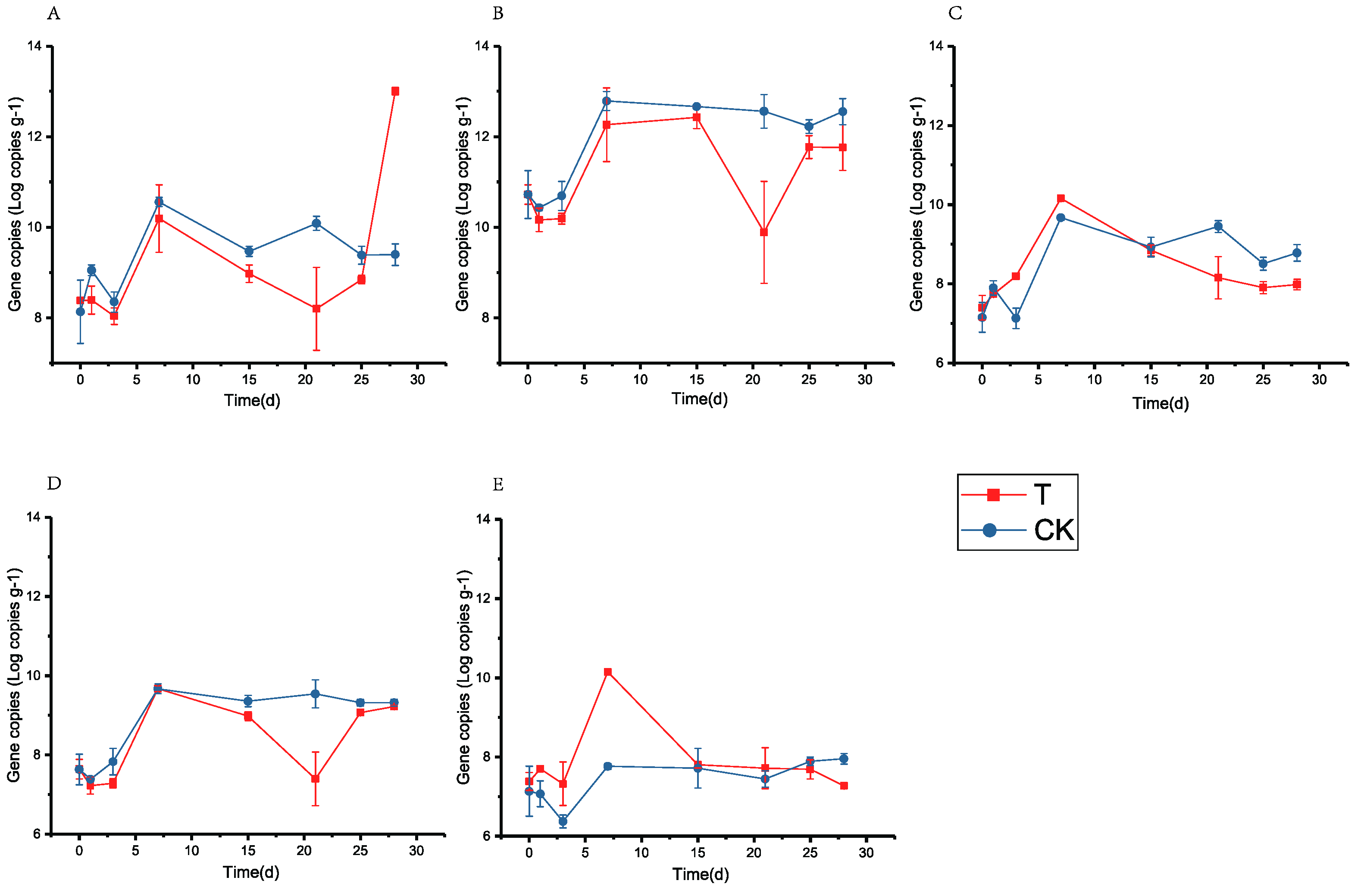
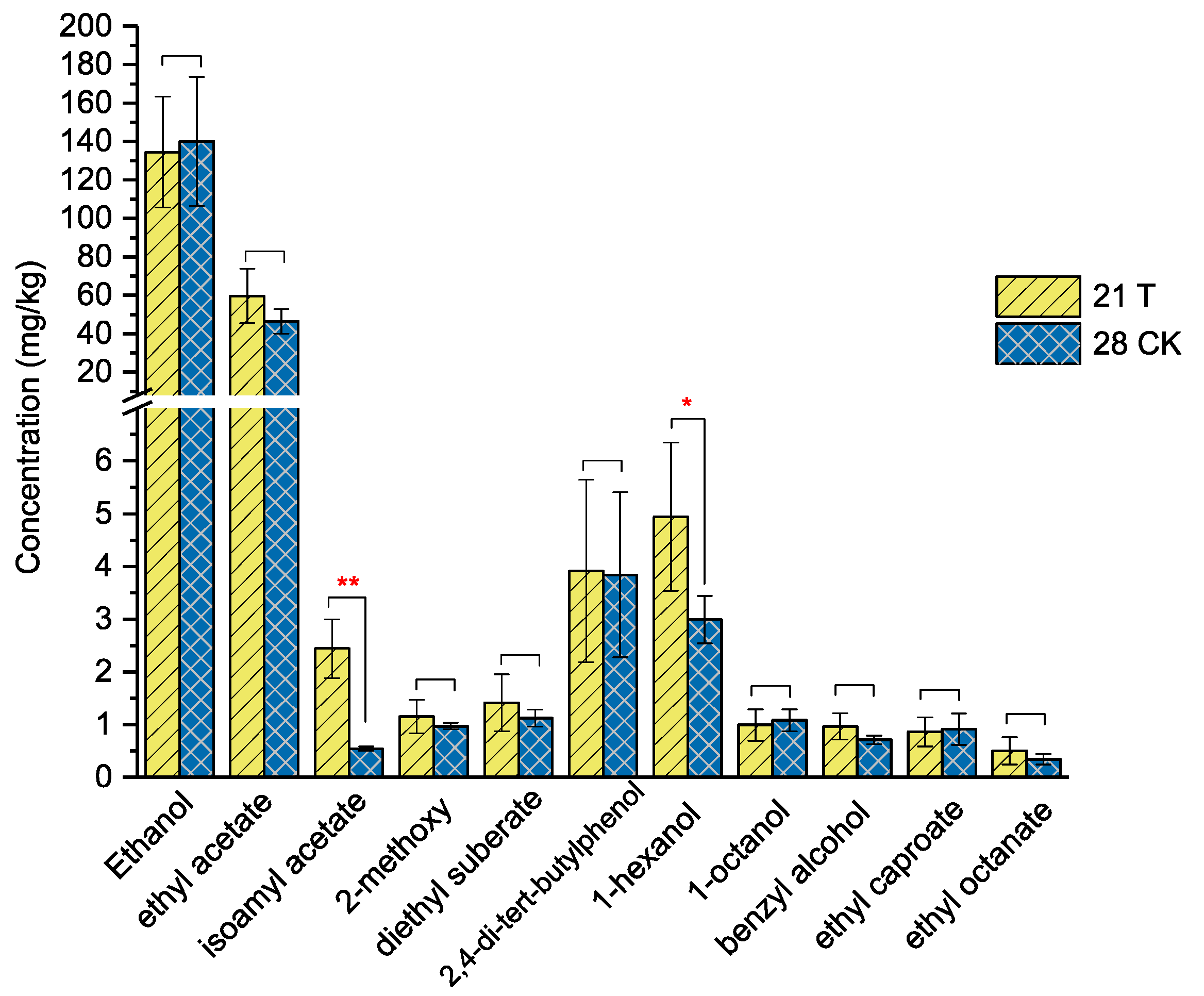
3.4. Effect of Fortified Inoculation with L. brevis Lbr17 on the Fermentation Process
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zheng, X.W.; Han, B.Z. Baijiu (白酒), Chinese liquor: History, classification and manufacture. J. Ethn. Foods 2016, 3, 19–25. [Google Scholar] [CrossRef]
- Kang, J.; Xue, Y.; Chen, X.; Han, B.Z. Integrated multi-omics approaches to understand microbiome assembly in Jiuqu, a mixed-culture starter. Compr. Rev. Food Sci. Food Saf. 2022, 21, 4076–4107. [Google Scholar] [CrossRef]
- Kang, J.; Hu, Y.; Jia, L.; Zhang, M.; Zhang, Z.; Huang, X.; Chen, X.; Han, B.-Z. Response of microbial community assembly and succession pattern to abiotic factors during the second round of light-flavor Baijiu fermentation. Food Res. Int. 2022, 162, 111915. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Fan, Y.; Lu, T.; Kang, J.; Pang, X.; Han, B.; Chen, J. Composition and metabolic functions of the microbiome in fermented grain during light-flavor Baijiu fermentation. Microorganisms 2020, 8, 1281. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Zhu, Y.; Xu, Y. Mystery behind Chinese liquor fermentation. Trends Food Sci. Technol. 2017, 63, 18–28. [Google Scholar]
- Wang, J.; Yan, C.; Ma, C.; Huang, S.; Chang, X.; Li, Z.; Chen, X.; Li, X. Effects of two kinds of Bacillus on flavour formation of Baijiu solid-state fermentation with pure mixed bacteria. Int. J. Food Sci Technol. 2023, 58, 1250–1262. [Google Scholar] [CrossRef]
- Du, H.; Chen, B.; Fu, W.; Yang, F.; Lv, X.; Tan, Y.; Xi, X.; Wang, L.; Xu, Y. Composition and function of viruses in sauce-flavor baijiu fermentation. Int. J. Food Microbiol. 2023, 387, 110055. [Google Scholar] [CrossRef]
- Pang, X.-N.; Han, B.-Z.; Huang, X.-N.; Zhang, X.; Hou, L.-F.; Cao, M.; Gao, L.-J.; Hu, G.-H.; Chen, J.-Y. Effect of the environment microbiota on the flavour of light-flavour Baijiu during spontaneous fermentation. Sci. Rep. 2018, 8, 3396. [Google Scholar] [CrossRef]
- Kang, J.; Zheng, X.; Yang, X.; Li, H.; Cheng, J.; Fan, L.; Zhao, H.; Xue, Y.; Ding, Z.; Han, B. Contrasting summer versus winter dynamic microbial communities and their environmental driving factors in the solid-state saccharification process of Fuyu-flavor Baijiu. Food Res. Int. 2022, 154, 111008. [Google Scholar] [CrossRef]
- Li, P.; Lin, W.; Liu, X.; Wang, X.; Gan, X.; Luo, L.; Lin, W. Effect of bioaugmented inoculation on microbiota dynamics during solid-state fermentation of Daqu starter using autochthonous of Bacillus, Pediococcus, Wickerhamomyces and Saccharomycopsis. Food Microbiol. 2017, 61, 83–92. [Google Scholar] [CrossRef]
- Wang, P.; Wu, Q.; Jiang, X.; Wang, Z.; Tang, J.; Xu, Y. Bacillus licheniformis affects the microbial community and metabolic profile in the spontaneous fermentation of Daqu starter for Chinese liquor making. Int. J. Food Microbiol. 2017, 250, 59–67. [Google Scholar] [CrossRef]
- Huang, X.; Fan, Y.; Meng, J.; Sun, S.; Wang, X.; Chen, J.; Han, B.-Z. Laboratory-scale fermentation and multidimensional screening of lactic acid bacteria from Daqu. Food Biosci. 2021, 40, 100853. [Google Scholar] [CrossRef]
- Pang, X.-N.; Chen, C.; Huang, X.-N.; Yan, Y.-Z.; Chen, J.-Y.; Han, B.-Z. Influence of indigenous lactic acid bacteria on the volatile flavor profile of light-flavor Baijiu. LWT 2021, 147, 111540. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Lee, H.J.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of Leuconostoc mesenteroides starter cultures on microbial communities and metabolites during kimchi fermentation. Int. J. Food Microbiol. 2012, 153, 378–387. [Google Scholar] [CrossRef]
- Hidalgo, C.; Torija, M.; Mas, A.; Mateo, E.J. Effect of inoculation on strawberry fermentation and acetification processes using native strains of yeast and acetic acid bacteria. Food Microbiol. 2013, 34, 88–94. [Google Scholar]
- van Rijswijck, I.M.; Wolkers–Rooijackers, J.C.; Abee, T.; Smid, E.J. Performance of non-conventional yeasts in co-culture with brewers’ yeast for steering ethanol and aroma production. Microb. Biotechnol. 2017, 10, 1591–1602. [Google Scholar] [CrossRef]
- Zhang, J.; Hou, Y.; Liu, Q.; Zhang, Y.; Gao, B.; Zou, W.; Zhang, K. Fortified Jiuqu of the Chinese Baijiu: A review on its functional microorganisms, strengthening effects, current challenges, and perspectives. Food Biosci. 2023, 55, 103045. [Google Scholar] [CrossRef]
- Chen, C.; Xiong, Y.; Xie, Y.; Zhang, H.; Jiang, K.; Pang, X.-N.; Huang, M. Metabolic characteristics of lactic acid bacteria and interaction with yeast isolated from light-flavor Baijiu fermentation. Food Biosci. 2022, 50, 102102. [Google Scholar] [CrossRef]
- Erkmen, O. Analysis of Fermented Foods. In Microbiological Analysis of Foods and Food Processing Environments; Erkmen, O., Ed.; Elsevier Inc.: London, UK, 2022. [Google Scholar]
- Coton, M.; Pawtowski, A.; Taminiau, B.; Burgaud, G.; Deniel, F.; Coulloumme-Labarthe, L.; Fall, A.; Daube, G.; Coton, E. Unraveling microbial ecology of industrial-scale Kombucha fermentations by metabarcoding and culture-based methods. FEMS Microbiol. Ecol. 2017, 93, fix048. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Khaliq, S.; Rajoka, M.I.; Agblevor, F. Solid state fermentation of a raw starch digesting alkaline alpha-amylase from Bacillus licheniformis RT7PE1 and its characteristics. Biotechnol. Res. Int. 2014, 2014, 495384. [Google Scholar] [CrossRef]
- Janssen, P.H.; Monk, C.R.; Morgan, H.W. A thermophilic, lipolytic Bacillus sp., and continuous assay of its p-nitrophenyl-palmitate esterase activity. FEMS Microbiol. Lett. 1994, 120, 195–200. [Google Scholar] [CrossRef]
- Takeuchi, A.; Shimizu-Ibuka, A.; Nishiyama, Y.; Mura, K.; Okada, S.; Tokue, C.; Arai, S.J.B. Purification and characterization of an α-amylase of Pichia burtonii isolated from the traditional starter “Murcha” in Nepal. Biosci. Biotechnol. Biochem. 2006, 70, 3019–3024. [Google Scholar] [CrossRef]
- Classics Lowry, O.; Rosebrough, N.; Farr, A.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Kim, J.; Lim, J.; Lee, C.J.B. Quantitative real-time PCR approaches for microbial community studies in wastewater treatment systems: Applications and considerations. Biotechnol. Adv. 2013, 31, 1358–1373. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Cappello, M.S.; Zapparoli, G.; Logrieco, A.; Bartowsky, E.J. Linking wine lactic acid bacteria diversity with wine aroma and flavour. Int. J. Food Microbiol. 2017, 243, 16–27. [Google Scholar] [CrossRef]
- Zheng, X.-W.; Yan, Z.; Han, B.-Z.; Zwietering, M.H.; Samson, R.A.; Boekhout, T.; Nout, M.R. Complex microbiota of a Chinese “Fen” liquor fermentation starter (Fen-Daqu), revealed by culture-dependent and culture-independent methods. Food Microbiol. 2012, 31, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Huang, X.; Chen, J.; Han, B. Formation of a mixed-species biofilm is a survival strategy for unculturable lactic acid bacteria and Saccharomyces cerevisiae in Daqu, a Chinese traditional fermentation starter. Front. Microbiol. 2020, 11, 138. [Google Scholar] [CrossRef]
- Huffer, S.; Clark, M.E.; Ning, J.C.; Blanch, H.W.; Clark, D.S. Role of alcohols in growth, lipid composition, and membrane fluidity of yeasts, bacteria, and archaea. Appl. Environ. Microbiol. 2011, 77, 6400–6408. [Google Scholar] [CrossRef]
- Yang, X.; Teng, K.; Zhang, J.; Wang, F.; Zhang, T.; Ai, G.; Han, P.; Bai, F.; Zhong, J. Transcriptome responses of Lactobacillus acetotolerans F28 to a short and long term ethanol stress. Sci. Rep. 2017, 7, 2650. [Google Scholar] [CrossRef]
- Fang, R.; Syahputra, J.C.; Airhunmwunde, O.; Wu, Y.; Lv, C.; Huang, J.; Xiao, G.; Chen, Q.J.L. Improving the enzyme property of ornithine transcarbamylase from Lactobacillus brevis through site-directed mutation. LWT 2020, 133, 109953. [Google Scholar] [CrossRef]
- Wang, C.L.; Shi, D.J.; Gong, G.L. Microorganisms in Daqu: A starter culture of Chinese Maotai-flavor liquor. World J. Microb. Biot. 2008, 24, 2183–2190. [Google Scholar] [CrossRef]
- Wang, S.; Wu, Q.; Nie, Y.; Wu, J.; Xu, Y. Construction of synthetic microbiota for reproducible flavor compound metabolism in Chinese light-aroma-type liquor produced by solid-state fermentation. Appl. Environ. Microbiol. 2019, 85, e03090-18. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-R.; Ma, E.-B.; Yan, L.-Z.; Meng, H.; Du, X.-W.; Zhang, S.-W.; Quan, Z.-X. Bacterial and fungal diversity in the traditional Chinese liquor fermentation process. Int. J. Food Microbiol. 2011, 146, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Tomita, S.; Nakamura, T. Aggregation of Lactobacillus brevis associated with decrease in pH by glucose fermentation. Biosci. Biotechnol. Biochem. 2019, 83, 1523–1529. [Google Scholar] [CrossRef]
- Kang, J.; Jia, L.; Zhang, Z.; Zhang, M.; Huang, X.; Chen, X.; Han, B.-Z. Comparison of physicochemical characteristics and microbiome profiles of low-temperature Daqu with and without adding tartary buckwheat. Food Biosci. 2022, 49, 101931. [Google Scholar] [CrossRef]
- Xu, W.; Huang, Z.Y.; Zhang, X.J.; Li, Q.; Lu, Z.M.; Shi, J.S.; Xu, Z.H.; Ma, Y.H. Monitoring the microbial community during solid-state acetic acid fermentation of Zhenjiang aromatic vinegar. Food Microbiol. 2011, 28, 1175–1181. [Google Scholar] [CrossRef]
- Duniere, L.; Jin, L.; Smiley, B.; Qi, M.; Rutherford, W.; Wang, Y.; Mcallister, T. Impact of adding Saccharomyces strains on fermentation, aerobic stability, nutritive value, and select lactobacilli populations in corn silage. J. Anim. Sci. 2015, 93, 82–87. [Google Scholar] [CrossRef]
- Wu, S.H.; Xu, S.J.; Chen, X.; Sun, H.S.; Hu, M.L.; Bai, Z.H.; Zhuang, G.Q.; Zhuang, X.L. Bacterial communities changes during food waste spoilage. Sci. Rep. 2018, 8, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Katharina, Z.; Olivier, C.; Patrick, M.L.; Joana, C.; Aline, L.; Isabelle, M.P. Characterization of the yeast ecosystem in grape must and wine using real-time PCR. Food Microbiol. 2010, 27, 559–567. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Huang, X.; Sun, S.; Han, B. Effect of Fortified Inoculation with Indigenous Lactobacillus brevis on Solid-State Fermentation of Light-Flavor Baijiu. Foods 2023, 12, 4198. https://doi.org/10.3390/foods12234198
Chen X, Huang X, Sun S, Han B. Effect of Fortified Inoculation with Indigenous Lactobacillus brevis on Solid-State Fermentation of Light-Flavor Baijiu. Foods. 2023; 12(23):4198. https://doi.org/10.3390/foods12234198
Chicago/Turabian StyleChen, Xiaoxue, Xiaoning Huang, Shanfeng Sun, and Beizhong Han. 2023. "Effect of Fortified Inoculation with Indigenous Lactobacillus brevis on Solid-State Fermentation of Light-Flavor Baijiu" Foods 12, no. 23: 4198. https://doi.org/10.3390/foods12234198
APA StyleChen, X., Huang, X., Sun, S., & Han, B. (2023). Effect of Fortified Inoculation with Indigenous Lactobacillus brevis on Solid-State Fermentation of Light-Flavor Baijiu. Foods, 12(23), 4198. https://doi.org/10.3390/foods12234198





