Development and Validation of a New Method for Detecting Acetic Bacteria in Wine
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Media
2.2. Acetic Acid Analysis
2.3. Quantitative PCR
2.4. Microorganisms
2.5. Olfaction Tests
2.6. Commercial Wines
3. Results
3.1. Development of the Culture Medium
3.2. Validation of the Culture Medium
3.2.1. Growth in the Medium of Acetic Bacteria
3.2.2. Growth in the Medium of Other Oenological Microorganisms
3.2.3. Verification of the Usefulness of the Medium in Commercial Wines
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davis, C.R.; Wibowo, D.J.; Eschenbruch, R.E.; Lee, T.H.; Fleet, G.H. Practical implications of malolactic fermentation: A review. Am. J. Enol. Vitic. 1985, 36, 290–301. [Google Scholar] [CrossRef]
- Bartowsky, E.J.; Henschke, P.A. Acetic acid bacteria spoilage of bottled red wine: A review. Int. J. Food Microbiol. 2008, 125, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Joyeux, A.; Lafon-Lafourcade, S.; Ribéreau-Gayón, P. Evolution of acetic acid bacteria during fermentation and storage of wine. Appl. Environ. Microbiol. 1984, 48, 153–156. [Google Scholar] [CrossRef] [PubMed]
- Guillamón, J.M.; Mas, A. Acetic Acid Bacteria. In Molecular Wine Microbiology; Chapter 9; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Pedraza, R.O. Recent advances in nitrogen-fixing acetic acid bacteria. Int. J. Food Microbiol. 2008, 125, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Millet, V.; Lonvaud-Funel, A. The viable but non-culturable state of wine microorganisms during storage. Lett. Appl. Microbiol. 2000, 30, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Giudici, P.; Zambonelli, C. Biometric and genetic study on acetic acid production for breeding of wine yeast. Am. J. Enol. Vitic. 1992, 43, 370–374. [Google Scholar] [CrossRef]
- Freer, S.N. Acetic acid production by Dekkera/Brettanomyces yeasts. World J. Microbiol. Biotechnol. 2002, 18, 271–275. [Google Scholar] [CrossRef]
- Romano, P.; Suzzi, G.; Comi, G.; Zironi, R. Higher alcohol and acetic acid production by apiculate wine yeasts. J. Appl. Bacteriol. 1992, 73, 126–130. [Google Scholar] [CrossRef]
- Ribérau-Gayon, P.; Dubourdieu, D.; Donèche, B.; Lonvaud, A. Handbook of Enology. The microbiology of Wine and Vinifications; Wiley: West Sussex, UK, 2000; Volume I. [Google Scholar]
- Vidra, A.; Németh, A. Bio-produced acetic acid: A review. Period. Polytech. Chem. Eng. 2018, 62, 245–256. [Google Scholar] [CrossRef]
- Considine, J.A.; Frankish, E. A complete guide to Quality in small-scale wine making. In Chapter 6, Microbiology and Methods; Elsevier: Amsterdam, The Netherlands, 2014; pp. 79–95. ISBN 978-0-12-408081-2. [Google Scholar]
- Lu, Y.; Sun, F.; Wang, W.; Liu, Y.; Wang, J.; Sun, J.; Mu, J.; Gao, Z. Effects of spontaneous fermentation on the microorganisms diversity and volatile compounds during ‘Marselan’ from grape to wine. LWT Food Sci. Technol. 2020, 134, 110–193. [Google Scholar] [CrossRef]
- Mesas, J.M.; Alegre, M.T. El papel de los microorganismos en la elaboración del vino. Cienc. Y Tecnol. Aliment. 1999, 2, 174–183. [Google Scholar] [CrossRef]
- Ultee, A.; Wacker, A.; Kunz, D.; Löwenstein, R.; König, H. Microbial Succession in Spontaneously Fermented Grape Must Before, During and After Stuck Fermentation. S. Afr. J. Enol. Vitic. 2013, 34, 68–78. [Google Scholar] [CrossRef][Green Version]
- Bartowsky, E.J.; Xia, D.; Gibson, R.L.; Fleet, G.H.; Henschke, P.A. Spoilage of bottled red wine by acetic acid bacteria. Lett. Appl. Microbiol. 2003, 36, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Zgardan, D.; Mitina, I.; Mitin, V.; Behta, E.; Rubtov, S.; Boistean, A.; Munteanu, M. Acetic acid bacteria detection in wines by REAL-TIME PCR. Scientific Study & Research. Chem. Chem. Eng. Biotechnol. Food Ind. 2022, 23, 179–188. [Google Scholar]
- Couto, J.A.; Barbosa, A.; Hogg, T. A simple cultural method for the presumptive detection of the yeasts Brettanomyces/Dekkera in wines. Lett. Appl. Microbiol. 2005, 41, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, N.; Gonçalves, G.; Pereira-da-Silva, S.; Malfeito-Ferreira, M.; Loureiro, V. Development and use of a new médium to detect yeasts of the genera Dekkera/Brettanomyces. J. Appl. Microbiol. 2001, 90, 588–599. [Google Scholar] [CrossRef] [PubMed]
- Melo, J. Microbiology of Vinegar: From Isolation, Phenetic Characterization and Detection of Acetic Acid Bacteria to Microbial Profiling of an Industrial Production. Master’s Thesis, Instituto Superior Técnico Lisboa, Lisboa, Portugal, 2016. [Google Scholar]
- ISO 17025:2017; General Requirements for the Competence of Testing and Calibration Laboratories. International Organization for Standardization: Geneva, Switzerland, 2017.
- Etievant, P. Chapter 14. In Volatile Compounds in Food and Beverage; Maarse, H., Ed.; Routledge: London, UK, 1991; pp. 483–533. [Google Scholar]


| Sample | Region | Variety and/or Type | Cell/mL Acetic Bacteria | Medium GYC | Medium G2 | Medium Kneifel | Medium Wallerstein |
|---|---|---|---|---|---|---|---|
| WINE 1 | Rioja | Tempranillo Red | 2.4 × 104 | No | No | No | Yes |
| WINE 2 | Rioja | Garnacha Red | 2.7 × 103 | No | No | No | Yes |
| WINE 3 | Rioja | Graciano Red | 3.7 × 104 | No | No | No | Yes |
| WINE 4 | Rioja | Tempranillo White | 1.2 × 101 | No | No | No | No |
| WINE 5 | Rioja | Viura White | 4.8 × 102 | No | No | No | No |
| WINE 6 | Ribera Duero | Tempranillo Red | 7.9 × 105 | Yes | Yes | Yes | Yes |
| WINE 7 | Ribera Duero | Tempranillo Red | 4.1 × 104 | No | No | No | Yes |
| WINE 8 | Ribera Duero | Tempranillo Red | 6.9 × 103 | No | No | No | Yes |
| WINE 9 | Ribera Duero | Tempranillo Red | 1.4 × 103 | No | No | No | Yes |
| WINE 10 | Rueda | Verdejo White | <10 | No | No | No | No |
| WINE 11 | Valencia | Bobal Red | 5.1 × 104 | No | No | Yes | Yes |
| WINE 12 | Valencia | Merlot Red | 7.1 × 103 | No | No | No | Yes |
| WINE 13 | Extemadura | Syrah Red | 5.5 × 104 | Yes | No | No | Yes |
| WINE 14 | Extramadura | Malbec Red | 1.8 × 103 | No | No | No | No |
| WINE 15 | Cadiz | Palomino White | 9.6 × 102 | No | No | No | Yes |
| WINE 16 | Cadiz | Palomino White | 5.4 × 103 | No | No | No | Yes |
| WINE 17 | Cadiz | Pedro Ximénez White | 4.8 × 103 | No | No | No | Yes |
| WINE 18 | Priorat | Garnacha Red | 3.6 × 104 | No | No | No | Yes |
| WINE 19 | Priorat | Cabernet Sauvignon Red | 3.9 × 105 | Yes | Yes | Yes | Yes |
| WINE 20 | Navarra | Garnacha Rosé | 4.6 × 103 | No | No | No | Yes |
| WINE 21 | Navarra | Garnacha Rosé | 8.1 × 103 | No | No | No | Yes |
| WINE 22 | Rias Baixas | Albariño | 2.4 × 102 | No | No | No | No |
| WINE 23 | Rias Baixas | Treixadura | 3.1 × 103 | No | No | No | No |
| WINE 24 | Rias Baixas | Albariño | 3.5 × 103 | No | No | No | Yes |
| WINE 25 | Rias Baixas | Godello | 9.1 × 101 | No | No | No | No |
| Components | Quantity |
|---|---|
| Yeast extract | 4 g |
| Peptone | 5 g |
| Glucose | 50 g |
| H2PO4 | 0.55 g |
| CaCl2 | 0.125 g |
| MgSO4 × 7H2O | 0.125 g |
| MnSO4 | 0.0025 g |
| H2O | Complete up to 1000 mL |
| Add before sterilizing 0.5 g of bile salts and 5 microliters of violet crystal. Add after sterilization 0.3 mL of natamycin (pimaricin) (concentration of 66 mg/20 mL) and 0.04 g of cycloheximide. | |
| Microorganism | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 | Day 8 | Day 9 | Day 14 |
|---|---|---|---|---|---|---|---|---|---|---|
| Oenococus oeni | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0.1 | 0.15 |
| Pichia membranafaciens | 0 | 0 | 0 | 0 | 0 | 0 | 0.08 | 0.12 | 0.12 | 0.18 |
| Saccharomyces cerevisae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.08 | 0.10 | 0.15 |
| Brettanomyces spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0.10 | 0.12 | 0.12 | 0.18 |
| Penicillium spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.10 | 0.10 | 0.15 |
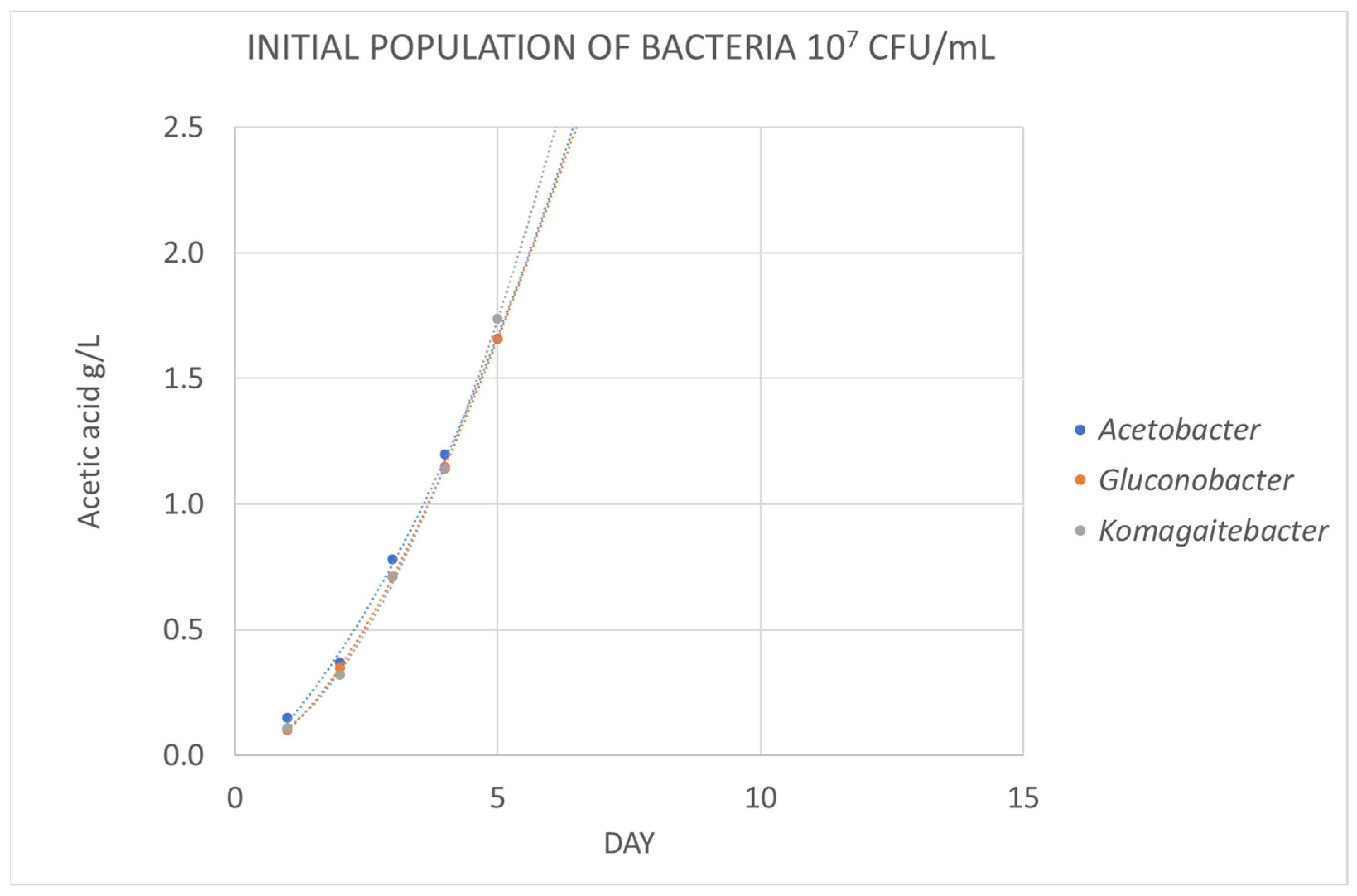
| Acetobacter aceti | 0 | 0.15 | 0.42 | 0.78 | * 1.19 | |||||
| Acetobacter oeni | 0 | 0.1 | 0.38 | 0.77 | 1.15 | |||||
| Gluconobacter oxidans | 0 | 0 | 0 | 0.19 | 0.37 | 0.68 | 1.05 | |||
| Komagataeibacter europaeus | 0 | 0 | 0 | 0.53 | 0.86 | 1.19 | 1.49 |
| Combined Microorganisms | Day 1 | Day 2 | Day 3 | Day 4 | Day 5 | Day 6 | Day 7 |
|---|---|---|---|---|---|---|---|
| Oenococus oeni, Acetobacter aceti | 0 | 0.10 | 0.38 | 0.69 | 1.51 | ||
| Oenococus oeni, Acetobacter oeni | 0 | 0.12 | 0.37 | 0.66 | 1.45 | ||
| Oenococus oeni, Gluconobacter oxidans | 0 | 0 | 0 | 0.15 | 0.34 | 0.62 | 0.98 |
| Oenococus oeni, Komagataeibacter europaeus | 0 | 0 | 0 | 0.46 | 0.78 | 1.09 | |
| Pichia membranafaciens, Acetobacter aceti | 0 | 0.08 | 0.36 | 0.65 | 1.47 | ||
| Pichia membranafaciens, Acetobacter oeni | 0 | 0.10 | 0.39 | 0.67 | 1.56 | ||
| Pichia membranafaciens, Gluconobacter oxidans | 0 | 0 | 0 | 0.14 | 0.36 | 0.65 | 1.01 |
| Pichia membranafaciens, Komagataeibacter europaeus | 0 | 0 | 0 | 0.39 | 0.74 | 1.01 | |
| Saccharomyces cerevisae, Acetobacter aceti | 0 | 0.10 | 0.39 | 0.71 | 1.64 | ||
| Saccharomyces cerevisae, Acetobacter oeni | 0 | 0.11 | 0.36 | 0.67 | 1.67 | ||
| Saccharomyces cerevisae, Gluconobacter oxidans | 0 | 0 | 0 | 0.21 | 0.40 | 0.65 | 1.07 |
| Saccharomyces cerevisae, Komagataeibacter europaeus | 0 | 0 | 0.10 | 0.41 | 0.76 | 1.05 | |
| Brettanomyces spp., Acetobacter aceti | 0 | 0.11 | 0.40 | 0.70 | 1.35 | ||
| Brettanomyces spp., Acetobacter oeni | 0 | 0.10 | 0.38 | 0.67 | 1.42 | ||
| Brettanomyces spp., Gluconobacter oxidans | 0 | 0 | 0 | 0.17 | 0.35 | 0.60 | 0.99 |
| Brettanomyces spp., Komagataeibacter europaeus | 0 | 0 | 0 | 0.29 | 0.70 | 0.99 | |
| Penicillium spp., Acetobacter aceti | 0 | 0.14 | 0.49 | 0.72 | 1.41 | ||
| Penicillium spp., Acetobacter oeni | 0 | 0.11 | 0.45 | 0.68 | 1.46 | ||
| Penicillium spp., Gluconobacter oxidans | 0 | 0 | 0 | 0.14 | 0.31 | 0.69 | 0.98 |
| Penicillium spp., Komagataeibacter europaeus | 0 | 0 | 0 | 0.47 | 0.78 | 1.14 |
| Wine | Origin | Type | qPCR Acetobacter Cell/mL | Initial Acetic Acid g/L | Positive Day | Final Acetic Acid g/L | Olfaction |
|---|---|---|---|---|---|---|---|
| 26 | Rioja | Tempranillo Red | 1.8 × 105 | 0.37 | 4 | 0.99 | YES |
| 1 | Rioja | Tempranillo Red | 2.4 × 104 | 0.26 | 4 | 0.73 | YES |
| 6 | Ribera Duero | Tempranillo Red | 7.9 × 105 | 0.30 | 4 | 0.81 | YES |
| 7 | Ribera Duero | Tempranillo Red | 4.1 × 104 | 0.27 | 4 | 0.72 | YES |
| 11 | Valencia | Bobal Red | 5.1 × 104 | 0.24 | 4 | 0.74 | YES |
| 18 | Priorat | Garnacha Red | 3.6 × 104 | 0.25 | 4 | 0.74 | YES |
| 19 | Priorat | Cabernet Sauvignon Red | 3.9 × 105 | 0.24 | 4 | 0.82 | YES |
| 23 | Rias Baixas | Treixadura | 3.1 × 103 | 0.12 | 4 | 0.75 | YES |
| 48 | Extremadura | Red | 2.4 × 105 | 0.4 | 4 | 0.79 | YES |
| 61 | Navarra | Garnacha Rosé | 6.8 × 105 | 0.28 | 4 | 1.16 | YES |
| 67 | Rías Baixas | White | 1.9 × 105 | 0.35 | 4 | 0.81 | YES |
| 78 | La Mancha | White Sweet | 1.5 × 105 | 0.31 | 4 | 1.20 | YES |
| 2 | Rioja | Garnacha Red | 2.7 × 103 | 0.19 | 5 | 1.02 | YES |
| 3 | Rioja | Graciano Red | 3.7 × 104 | 0.22 | 5 | 1.05 | YES |
| 8 | Ribera Duero | Tempranillo Red | 6.9 × 103 | 0.17 | 5 | 1.01 | YES |
| 9 | Ribera Duero | Tempranillo Red | 1.4 × 103 | 0.19 | 5 | 0.95 | YES |
| 12 | Valencia | Merlot Red | 7.1 × 103 | 0.17 | 5 | 1.02 | YES |
| 13 | Extemadura | Syrah Red | 5.5 × 104 | 0.21 | 5 | 1.08 | YES |
| 14 | Extramadura | Malbec Red | 1.8 × 103 | 0.21 | 5 | 0.96 | YES |
| 16 | Cadiz | Palomino White | 5.4 × 103 | 0.13 | 5 | 0.91 | YES |
| 17 | Cadiz | Pedro Ximénez White | 4.8 × 103 | 0.16 | 5 | 0.97 | YES |
| 20 | Navarra | Garnacha Rosé | 4.6 × 103 | 0.18 | 5 | 1.02 | YES |
| 21 | Navarra | Garnacha Rosé | 8.1 × 103 | 0.20 | 5 | 0.91 | YES |
| 24 | Rias Baixas | Albariño | 3.5 × 103 | 0.15 | 5 | 0.94 | YES |
| 40 | Rioja | Red | 6.9 × 104 | 0.18 | 5 | 0.96 | YES |
| 46 | Rioja | Rosé | 1.6 × 104 | 0,2 | 5 | 0.89 | YES |
| 57 | Ribera Duero | Red | 9.9 × 104 | 0.18 | 5 | 0.97 | YES |
| 62 | Navarra | Rosé | 7.1 × 104 | 0.21 | 5 | 0.93 | YES |
| 63 | Madrid | Red | 4.2 × 104 | 0.25 | 5 | 1.25 | YES |
| 68 | Rías Baixas | White | 6.7 × 104 | 0.15 | 5 | 0.96 | YES |
| 76 | La Mancha | Red | 8.0 × 104 | 0.23 | 5 | 1.07 | YES |
| 80 | Yecla | Red | 3.6 × 104 | 0.2 | 5 | 0.77 | YES |
| 5 | Rioja | Viura White | 4.8 × 102 | 0.16 | 6 | 1.05 | YES |
| 15 | Cadiz | Palomino White | 9.6 × 102 | 0.12 | 6 | 0.96 | YES |
| 22 | Rias Baixas | Albariño | 2.4 × 102 | 0.13 | 6 | 0.95 | YES |
| 41 | Rioja | Red | 4.3 × 104 | 0.19 | 6 | 1.13 | YES |
| 44 | Rioja | White | 3.8 × 103 | 0.1 | 6 | 0.86 | YES |
| 45 | Rioja | Rosé | 1.3 × 104 | 0.24 | 6 | 0.95 | YES |
| 47 | Rioja | Rosé | 2.3 × 103 | 0.17 | 6 | 0.91 | YES |
| 49 | Ribera Duero | Red | 7.0 × 103 | 0.32 | 6 | 0.88 | YES |
| 51 | Ribera Duero | Red | 2.5 × 103 | 0.32 | 6 | 0.95 | YES |
| 53 | Ribera Duero | Red | 3.5 × 103 | 0.35 | 6 | 1.14 | YES |
| 69 | Rías Baixas | White | 3.1 × 103 | 0.21 | 6 | 0.83 | YES |
| 70 | Rías Baixas | White | 3.7 × 103 | 0.22 | 6 | 0.93 | YES |
| 75 | La Mancha | Red | 7.8 × 103 | 0.2 | 6 | 0.94 | YES |
| 43 | Rioja | Red | 4.5 × 102 | 0.11 | 7 | 0.95 | YES |
| 52 | Ribera Duero | Red | 4.3 × 102 | 0.2 | 7 | 0.83 | YES |
| 54 | Rioja | Red | 1.4 × 103 | 0.16 | 7 | 0.91 | YES |
| 56 | Toro | Red | 1.7 × 102 | 0.21 | 7 | 0.93 | YES |
| 58 | Navarra | Red | 2.7 × 102 | 0.13 | 7 | 0.89 | YES |
| 59 | Navarra | Rosé | 3.5 × 102 | 0.15 | 7 | 1.08 | YES |
| 72 | Ribeiro | White | 2.8 × 102 | 0.15 | 7 | 0.84 | YES |
| 77 | La Mancha | White | 6.9 × 102 | 0.14 | 7 | 1.02 | YES |
| 4 | Rioja | Tempranillo White | 1.2 × 101 | 0.08 | 8 | 0.89 | YES |
| 25 | Rias Baixas | Godello | 9.1 × 101 | 0.09 | 8 | 0.93 | YES |
| 50 | Ribera Duero | VRed | 5.7 × 102 | 0.17 | 8 | 0.98 | YES |
| 71 | Rías Baixas | White | 1.2 × 101 | 0.14 | 11 | 0.98 | YES |
| 73 | Ribeiro | Red | 3.6 × 101 | 0.15 | 11 | 0.94 | YES |
| 10 | Rueda | Verdejo White | <10 | 0.06 | 14 | 0.12 | NO |
| 27 | Rioja | Tempranillo Red | <10 | 0.23 | 14 | 1.01 | YES |
| 28 | Rias Baixas | Albariño White | <10 | 0.09 | 14 | 0.17 | NO |
| 29 | Rias Baixas | Albariño White | <10 | 0.14 | 14 | 0.21 | NO |
| 30 | Rias Baixas | Albariño White | <10 | 0.14 | 14 | 0.23 | NO |
| 31 | Rioja | Tempranillo White | <10 | 0.06 | 14 | 0.10 | NO |
| 32 | Cariñena | Garnacha Red | <10 | 0.12 | 14 | 0.41 | NO |
| 33 | Rias Baixas | Albariño White | <10 | 0.08 | 14 | 0.15 | NO |
| 34 | Ribeiro | Red | <10 | 0.16 | 14 | 0.27 | NO |
| 35 | Ribeiro | Red | <10 | 0.16 | 14 | 0.26 | NO |
| 36 | Cadiz | Oloroso | <10 | 0.42 | 14 | 0.44 | NO |
| 37 | Sevilla | White | <10 | 0.13 | 14 | 0.15 | NO |
| 38 | Campo Borja | Rosé semisweet | <10 | 0.12 | 14 | 0.18 | NO |
| 39 | Rioja | Red | <10 | 0.21 | 14 | 0.28 | NO |
| 42 | Rioja | Red | <10 | 0.24 | 14 | 0.28 | NO |
| 55 | Hungría | White | <10 | 0.1 | 14 | 0.20 | NO |
| 60 | Navarra | Rosé | <10 | 0.08 | 14 | 0.21 | NO |
| 64 | Priorat | Red | <10 | 0.06 | 14 | 0.13 | NO |
| 65 | Priorat | Red | <10 | 0.12 | 14 | 0.17 | NO |
| 66 | Rías Baixas | White | <10 | 0.06 | 14 | 0.21 | NO |
| 74 | Bierzo | Red | <10 | 0.07 | 14 | 0.21 | NO |
| 79 | Jerez | White Sweet | <10 | 0.08 | 14 | 0.18 | NO |
| Days Necessary for the Appearance of the Odour | Estimated Population of Acetic Bacteria | What to Do? |
|---|---|---|
| >12 days/No appearance | Absence in 20 mL | Control in 1 month |
| 10 days | Very weak (about 100 bacteria/mL) | Control in 15 days |
| 8 days | Weak (100 to 1000 bacteria/mL) | Control in 1 week |
| 6–7 days | Media (1000 to 10,000 bacteria/mL) | 2 controls: immediate and after 5 days |
| 4 days | Significant: Danger (10,000 to 100,000 bacteria/mL) | ACT: Filtration/Centrifugation /Flash-pasteurization Add SO2 After a few days return to control |
| 3 days | Strong: A lot of danger (+1,000,000 bacteria/mL) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra, A.; Ovejas, A.; González-Arenzana, L.; Gutiérrez, A.R.; López-Alfaro, I. Development and Validation of a New Method for Detecting Acetic Bacteria in Wine. Foods 2023, 12, 3734. https://doi.org/10.3390/foods12203734
Parra A, Ovejas A, González-Arenzana L, Gutiérrez AR, López-Alfaro I. Development and Validation of a New Method for Detecting Acetic Bacteria in Wine. Foods. 2023; 12(20):3734. https://doi.org/10.3390/foods12203734
Chicago/Turabian StyleParra, Alejandro, Aroa Ovejas, Lucía González-Arenzana, Ana Rosa Gutiérrez, and Isabel López-Alfaro. 2023. "Development and Validation of a New Method for Detecting Acetic Bacteria in Wine" Foods 12, no. 20: 3734. https://doi.org/10.3390/foods12203734
APA StyleParra, A., Ovejas, A., González-Arenzana, L., Gutiérrez, A. R., & López-Alfaro, I. (2023). Development and Validation of a New Method for Detecting Acetic Bacteria in Wine. Foods, 12(20), 3734. https://doi.org/10.3390/foods12203734





