Abstract
High-throughput DNA sequencing (HTS) was used to study the microbial diversity of commercial traditional Izmir Tulum (IT) and Izmir Brined Tulum (IBT) cheeses from Izmir, Türkiye. Simultaneously, cultivation-dependent methods were used to isolate, identify and characterize bacterial strains displaying probiotic potential. At the phylum level, Firmicutes dominated the microbiota of both cheese types comprising >98% of the population. Thirty genera were observed, with Streptococcus being the most abundant genus and with Streptococcus thermophilus and S. infantarius subsp. infantarius being the most abundant species. Genera, including Bifidobacterium and Chryseobacterium, not previously associated with IT and IBT, were detected. IT cheeses displayed higher operational taxonomic units (OTUs; Richness) and diversity index (Simpson) than IBT cheeses; however, the difference between the diversity of the microbiota of IT and IBT cheese samples was not significant. Three Lacticaseibacillus paracasei strains isolated from IBT cheeses exhibited probiotic characteristics, which included capacity to survive under in vitro simulated gastrointestinal conditions, resistance to bile salts and potential to adhere to HT-29 human intestinal cells. These findings demonstrate that Tulum cheeses harbor bacterial genera not previously reported in this cheese and that some strains display probiotic characteristics.
1. Introduction
Microorganisms play significant roles during both production and ripening of cheese and contribute to the development of flavor, aroma and texture. Type, diversity, number and population dynamics of microorganisms in cheese vary according to the quality of raw milk, whether raw or pasteurized milk is used, starter culture combination, manufacturing technology and hygienic, environmental and ripening conditions [1]. Traditional raw milk cheeses possess a more diverse microbiome and unique flavor profiles compared to industrial cheeses [2]. However, this also exposes traditional raw milk cheeses to quality and safety risks associated with the potential growth of spoilage and/or pathogenic bacteria [3]. On the other hand, the natural microbiota of these cheeses can be a valuable source of bacteria having unique features such as probiotic properties, which can promote health benefits in consumers [4,5,6,7].
Tulum cheese is one of the most widely produced cheese types in Türkiye and takes its name from the goatskin casing used to package cheese during ripening. Its production is common throughout most of Türkiye. However, some differences exist in terms of production method and ripening conditions between regions. This gives rise to many varieties of Tulum cheese, one of the most popular being Izmir Tulum (IT), which is produced in the Aegean region [3,8]. IT is ripened in goatskin bags but because of the lack of sufficient supply of skin bags for industrial production, industrially produced cheese is now ripened in brine in sealed tin cans and is known as Izmir Brined Tulum (IBT) cheese. However, small-scale production of traditional IT in skin bags is still available. IT and IBT cheese also have significant differences in terms of production process. Manufactured from cow’s and/or sheep’s milk without starter culture addition, IBT cheese is acidified for 12 h; the pressed curd is cooked in hot whey (~85 °C) and ripened in 16% brine in sealed tin cans for a minimum of 4 to 6 months (Figure 1b). However, IT is dry salted, filled into skin bags after the draining of whey and does not include any cooking step (Figure 1a).
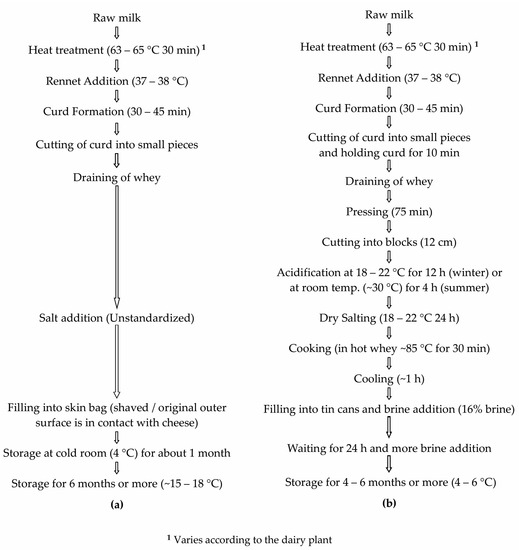
Figure 1.
Production process of (a) Izmir Tulum Cheese, (b) Izmir Brined Tulum Cheese [9].
Defining quality standards for IT and IBT cheeses is very difficult due to variations in manufacturing practices between different processors. Quality defects are reasonably common in IT and especially in IBT cheese, and consumption of the cheeses prior to completion of the required ripening period may pose risks for human health [10]. This generates the need for an accurate and detailed profiling of the microbiome of commercial examples of IT and IBT cheeses, followed by the isolation, selection and characterization of bacterial strains for inclusion in defined starter/adjunct culture blends that will enable the industrial production of cheeses with standardized quality and sensory profiles [11].
The microbiota of IT and IBT cheeses is scarcely described in the literature. Early reports used conventional culture-dependent methods where selective media were used to isolate microorganisms prior to their phenotypic identification [12,13]. However, these methods can underestimate or fail to detect low-abundance or nutritionally fastidious microorganisms [11,14]. In a more recent study, the lactic acid bacteria (LAB) microbiota diversity of IBT cheeses was determined using denaturing gradient gel electrophoresis (DGGE) in combination with real-time qPCR [15]. These molecular techniques have been shown to be valuable tools to profile microbial populations and detect previously unknown bacteria in cheese or raw milk [16,17]. The limitation of these metods is that they are laborious, have low resolution capacity and often reveal only the dominant species present in the samples [2,18].
High-throughput sequencing (HTS) of DNA has become the method of choice to investigate the microbiota and detect dominant and subdominant species of complex microbial ecosystems, including cheese [18,19,20,21,22,23,24]. No prior knowledge of the composition of the microbiome is required when applying HTS, and the fact that it is high-throughput means that many samples can be managed simultaneously in a time-efficient manner [25,26]. The application of HTS to the study of cheese ecosystems has enabled the detection of bacteria not previously associated with particular cheese types [21,27] to unveil the species causing quality defects, such as the pink discoloration defect [28], to identify populations capable of biogenic amine production [29] and to assess the effects of geography, manufacturing process, climatic conditions, seasonal variations and milk heat treatment (raw vs. pasteurized) on the cheese microbiota [1].
The objectives of this study were to:
- ▪
- Obtain an in-depth profile of a selection of commercial IT and IBT cheeses’ microbiomes from different dairies from the Izmir province using culture independent HTS of 16S RNA genes;
- ▪
- Simultaneously use cultivation-dependent methods to isolate, identify and characterize strains demonstrating some probiotic characteristics.
2. Materials and Methods
2.1. Cheese Samples
Commercial examples of five IT and nine IBT cheeses, collected from six different dairy plants in the Izmir Province of Türkiye, were used (for detailed information see Supplementary Material Table S1). Cheese samples (~500 g each) were vacuum-packed into sterile plastic bags, transported to the laboratory under refrigerated conditions and stored in the refrigerator (at +4 °C) until analysis.
2.2. Culture-Independent Profiling of IT and IBT Cheese Microbiota
2.2.1. DNA Extraction from Cheeses
Five IT and five IBT cheeses were selected for the 16S rRNA gene sequence analysis. Genomic DNA was extracted using a PowerFood® microbial DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA, USA), as per manufacturer’s instructions [30]. Before DNA extraction, each cheese sample was prepared and treated according to the method described by O’Sullivan et al. [29]. Quality and purity of the extracted DNA were determined using the NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
2.2.2. HTS of 16S rRNA Gene Amplicons and Bioinformatic Analysis
The V3-V4 variable region of the 16S rRNA genes was amplified using the universal primers:
5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG-3′ (forward) and 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′ (reverse) [31]. The resulting amplicons of 427 bp were purified and sequenced using the Ion Torrent PGM (Thermo Fisher Scientific, Waltham, MA, USA) [29]. The reads were filtered based on sequence quality (removal of low-quality nucleotides at the 3′ end) and length (removal of sequences with less than 250 bp) with PRINSEQ [32].
The filtered sequences were clustered at operational taxonomic unit (OTU; with 97% identity level) using UPARSE-OTU algorithm with Usearch v7.0 program [33] and removal of chimeric OTUs against GenomesOnLine database (GOLD). The taxonomic assignment of these OTUs was obtained using the ribosomal database project (RDP) [34]. The β- and α- diversity was determined using R package phyloseq [35], applying statistics by Adonis and ANOVA, respectively. p value greater than 0.05 was not considered significant.
2.3. Culture-Dependent Analysis of IT and IBT Cheeses’ Microbiota
Five grams of each of the 14 cheeses were homogenized in 45 mL of sterile 2% trisodium citrate. Ten-fold serial dilutions were prepared in Maximum Recovery Diluent (Oxoid, Basingstoke, UK), and 1 mL aliquots of appropriate dilutions were plated onto various media for the enumeration of different microbial groups and for the isolation of lactobacilli. M17 (containing lactose) (Merck, Darmstadt, Germany) agar plates incubated at 20 °C for 5 days and at 42 °C for 48 h were used for the enumeration of presumptive lactococci and thermophilic streptococci, respectively. Enterococci were counted on Kanamycin Esculin Azide Agar (KEA) (Merck, Darmstadt, Germany) incubated at 37 °C for 48 h. Overlayed BD™ LBS agar (BD, Heidelberg, Germany) was used for the enumeration and isolation of lactobacilli, incubated at 30 °C for 5 days.
2.4. Selection of Lactobacilli Displaying Probiotic Potential from IBT Cheese
2.4.1. Isolation and Genotyping of Lactobacilli Strains
IBT cheeses from two different dairy plants and two different batches were used to mine for lactobacilli displaying probiotic potential. Colonies were randomly picked from the BD™ LBS agar and purified by streaking onto MRS agar medium twice. After microscopic examination and catalase tests, catalase negative rod-shaped isolates were genotyped at the strain level by pulsed-field gel electrophoresis (PFGE). The protocol described by Simpson et al. [36] was used to extract high molecular weight DNA and to prepare it for digestion. Digestion reactions with AscI restriction endonuclease were performed overnight, according to the supplier’s instructions (New England BioLabs, Hitchin, UK). The restricted DNA was loaded into the wells of a 1% PFGE-grade agarose gel and run in 0.5× Tris-borate buffer using a CHEF-DR® II PFGE apparatus (Bio-Rad, Hercules, CA, USA) and the PFGE conditions stated in Güley et al. [9]. After staining with ethidium bromide, gels were visualized using an AlphaImager system (ProteinSimple, San Jose, CA, USA), and profiles were analyzed using the BioNumerics (Version 7.5) software (hierarchical clustering analysis-UPGMA) (Applied Maths, Sint-Martens-Latem, Belgium).
Isolates showing unique PFGE genotypes were identified to the species level by Sanger sequencing of the 16S rRNA gene. DNA extraction from the strains, PCR amplification of the 16S rRNA gene (primers, PCR reactions etc.), purification of PCR products and 16S rRNA gene sequencing were as described by Güley et al. [9]. Sequencing data were assembled using SeqMan Pro (DNASTAR, Madison, WI, USA) and compared to 16S rRNA gene sequences present in the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov; accessed on 5 January 2023) database using the BLASTN.
2.4.2. Resistance to Simulated Gastrointestinal Tract (GIT) Conditions
The capacity of lactobacilli isolates to transit the upper GIT was evaluated according to Pisano et al. [37], with some minor modifications. Briefly, strains were reactivated twice in 10 mL MRS broth at 37 °C to a final concentration of 108–109 CFU mL−1. Bacterial cells were recovered by centrifugation at 3000× g for 4 min, washed in 5 mL PBS, pH 7.4 (ThermoFisher Scientific, USA), pelletized again and finally resuspended in 10 mL of artificial gastric juice (6.2 g L−1 NaCl, 2.2 g L−1 KCl, 0.22 g L−1 CaCl2, 1.2 g L−1 NaHCO3, pH 3) containing 0.3% pepsin. After incubation at 37 °C for 90 min in a shaking incubator, 18 mL of synthetic duodenum juice (6.4 g L−1 NaHCO3, 0.239 g L−1 KCl, 1.28 g L−1 NaCl, pH 7.2), containing 0.1% pancreatin and 1.5 mL of 10% (w/v) oxgall (Sigma, Ireland), were added to the cell suspension to simulate passage into the upper small intestine. Incubation was continued at 37 °C for 90 min. One mL samples were taken immediately after (i) resuspension in simulated gastric juice, (ii) 90 min exposure to simulated gastric juice and (iii) 90 min exposure to simulated duodenum juice. Samples were serially diluted in MRD, pour-plated using MRS agar and incubated anaerobically at 37 °C for 48 h to enumerate the surviving cells. Experiments were done in triplicate, and results were expressed as the mean log CFU mL−1.
2.4.3. Bile Salt Hydrolase Activity
Bile salt hydrolase (BSH) activity was determined according to the method described by Pisano et al. [37]. Limosilactobacillus reuteri NCIMB 30242 was used as positive control, whereas MRS agar plates without taurodeoxycholic acid sodium salt (TDCA)/glycodeoxycholic acid (GDCA) supplementation were used as negative controls.
2.4.4. Antimicrobial Activity
The ability of the lactobacilli strains to inhibit the growth of food-related pathogens was investigated using the spot-on-lawn method as described by Bolocan et al. [38]. The pathogens used for the test (Escherichia coli O157:H7 P1432* and NCTC 12900‡, Salmonella typhimurium DPC6046* and 3784*, Staphylococcus aureus S17* and Listeria monocytogenes DPC6179*) were obtained from the Teagasc Food Research Center, Moorepark, Culture Collection* and the National Collection of Type Cultures‡ (London, UK). The presence of a distinct inhibition zone around the spots was considered as positive antagonistic effect. The inhibitory activity (IA) was calculated by subtracting the circle diameter (CD, mm) of the lactobacilli colony spreading zone from the inhibition zone diameter observed (IZD, mm) as follows: IA = (IZD − CD)/2 [39].
The inhibitory activity of the lactobacilli strains was also tested using the agar well diffusion method [40]. Cell-free supernatant (CFS) of each strain, grown in sodium acetate-free MRS broth overnight at 30 °C, was obtained by centrifugation at 3000 rpm for 4 min at 4 °C. To assess the contribution of organic acids to the activity observed, the pH of each CFS was adjusted to 6.5–7.0 with 4 mol L−1 NaOH and filter sterilized (0.20 μm cellulose acetate, Sigma-Aldrich, Ireland) before tested. In all assays, the nisin-A producer Lactococcus lactis subsp. lactis NZ9700 was included as a positive control. Analyses were performed in duplicate for each method.
2.5. In-Depth Characterization of GIT-Resistant Lactobacilli Strains Displaying Probiotic Potential
2.5.1. Adhesion Properties
Lactobacilli strains showing resistance to upper GIT conditions, inhibition of pathogens and bile salts resistance were assessed for their capacity to colonize the human intestine by using the human colon adenocarcinoma cell line HT-29 as per Ross et al. [41]. Adhesion rate was calculated as the percentage of bacteria adhered to HT-29 cells compared to the initial number of bacteria added. Experiments were done in triplicate. Lacticaseibacillus rhamnosus GG (ATCC 53103) (LGG) was used in parallel as a positive control.
2.5.2. Biogenic Amines Production
The method described by Bover-Cid et al. [42] was used to screen the strains for the potential to produce the biogenic amines tyramine and histamine.
2.5.3. Antibiotic Susceptibility
The antibiotic susceptibility of the strains was determined according to the method described by Campedelli et al. [43] using VetMIC™ plates (National Veterinary Institute, Uppsala, Sweden) containing serial 2-fold dilutions of 16 antibiotics. The lowest antibiotic concentration at which no visible growth occurred was defined as the minimum inhibitory concentration (MIC) for each antibiotic. Results were interpreted based on the microbiological cut-off values established by European Food Safety Authority [44] for Lacticaseibacillus paracasei. Cut-off values for antibiotics not covered by EFSA were adopted from Ammor et al. [45] and Danielsen et al. [46]. When the MIC value for a specific antibiotic was higher than the corresponding microbiological cut-off value, the strain was classified as resistant [44]. Tests were performed in triplicate.
2.6. Statistical Analysis
Statistical analysis were carried out using the IBM SPSS® (Version 27.0) software platform for Windows. One Way ANOVA/Tukey’s HSD post hoc test was used to determine statistically significant differences between the control and test strains in adhesion assay. To determine differences between the microbial counts of IT and IBT cheeses, independent samples t test was used. Data were analysed at the significance level of p < 0.05.
3. Results
3.1. HTS Shows That IT and IBT Cheeses Are a Reservoir of Streptococcus and Lactobacillaceae Species
For this study 10 commercial IT (n = 5) and IBT (n = 5) cheeses were used. Following quality filtration and length trimming of the raw data, an average of 44,403 (±8.613 SD) high-quality sequences of 16S rRNA gene was obtained for each sample.
Phylogenetic analysis revealed that Firmicutes dominated both IT and IBT cheeses and contain very low levels of Actinobacteria, Bacteroidetes and Proteobacteria. At the genus level, 30 genera were determined. Bacteria belonging to Streptococcus, Lacticaseibacillus, Lactobacillus, Lactococcus, Enterococcus and Bifidobacterium genera were observed in all samples. Of these, Streptococcus was the predominant genus followed by Lacticaseibacillus, Lactobacillus and Lactococcus. The other genera were always detected at low levels (Table 1).

Table 1.
Relative abundance of OTUs assigned to the phylum and genus level in Izmir Tulum (IT) and Izmir Brined Tulum (IBT) cheeses.
Alpha diversity describes the diversity within an ecosystem, and it has two components: species richness and evenness [47]. IT cheeses displayed higher Operational Taxonomic Units (OTUs; Richness) and diversity index (Simpson) than IBT cheeses; however, the level of α-diversity across the group of samples was not significant in all indexes (Figure 2a). Beta diversity describes the species diversity between two ecosystems, and there are several matrixes to measure β-dversity. The Bray-Curtis dissimilarity index measures the compositional dissimilarity between the microbial communities of two groups and is based on counts on each group [48]. The β-diversity represented by non-metric multidimensional scaling (NMDS) performed using all 16S rRNA gene reads clustering all reads at 97% similarity with the Bray-Curtis distance matrix (Figure 2b). The results showed that the difference between the microbiota of the two cheese types (IT and IBT) was not significant.
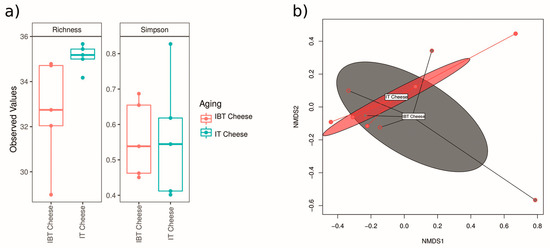
Figure 2.
α-diversity index (a) and β-diversity (b) of the microbiota of IT and IBT cheeses.
The abundance of Streptococcus was very high in both cheese types. However, differences in terms of OTU type and abundance were observed, for example OTU1 (Streptococcus) was highly abundant in both cheese types while OTU8 and OTU86 were only abundant in IT and IBT, respectively (Figure 3). On the other hand, although the differences were not significant statistically, there were inequalities between the populations of Latobacillus (OTUs 7 and 13), Bifidobacterium (OTU 14) and Leuconostoc (OTU 45) of cheese groups (Figure 3).
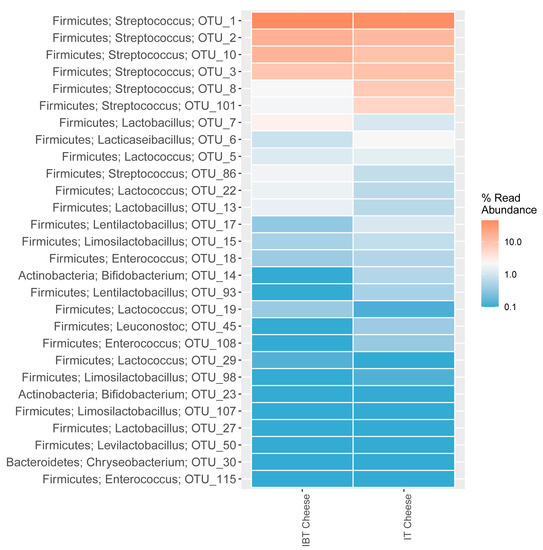
Figure 3.
Distribution of the 28 most abundant OTUs of IT and IBT cheeses, assigned to phylum and genus level.
Taxonomic details up to the species level within the Streptococcus genus revealed that Streptococcus thermophilus was the most abundant OTU in most cheese samples, followed by S. infantarius. In some cheese samples, OTUs belonging to S. infantarius were higher than or equal to the S. thermophilus. Within the lactobacilli Lactobacillus delbrueckii and Lacticaseibacillus paracasei was the most abundant OTUs (Figure 4) observed.
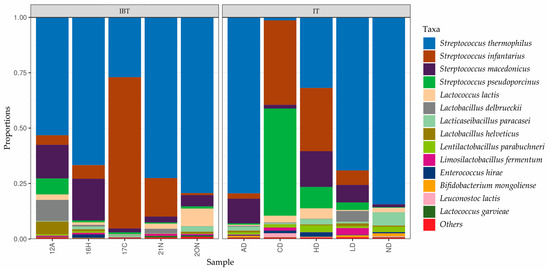
Figure 4.
Relative abundance (%) of sequences identified at species level in IT and IBT Cheeses.
3.2. Microbial Counts of IT and IBT Cheeses
The results of microbial counts in IT and IBT cheeses are presented in Table 2. No statistically significant difference between the bacterial counts of IT and IBT cheeses, in all agar media, was found (p > 0.05). These data suggest that species of the genera Streptococcus, Lactococcus, Enterococcus and lactobacilli are present in IT and IBT cheeses at similar levels and compose the dominant microbiota of these Tulum cheeses.

Table 2.
Microbial counts (log10 CFU g −1) in Izmir Tulum (IT) and Izmir Brined Tulum (IBT) cheeses.
3.3. Isolation and Identification of Lactobacilli
As IBT cheeses are now the most commonly produced Tulum cheeses in the Izmir region, we focused our study on lactobacilli with probiotic characteristics in these cheeses. A total of 73 catalase-negative rod-shaped bacteria were isolated from LBS plates of IBT cheeses, purified and genotyped by PFGE. Clustering analysis of AscI restriction fingerprints revealed 49 unique PFGE pulsotypes (Figure 5). 16S rRNA gene sequencing of a representative of each pulsotype revealed that the isolates belonged to four genera and six different species. Specifically, the 49 PFGE pulsotypes included strains of Lacticaseibacillus paracasei (n = 33), Lacticaseibacillus rhamnosus (n = 11), Lentilactobacillus parabuchneri (n = 2), Lactobacillus delbrueckii subsp. sunkii (n = 1), Schleiferilactobacillus harbinensis (n = 1) and Lacticaseibacillus zeae (n = 1) (Figure 5).
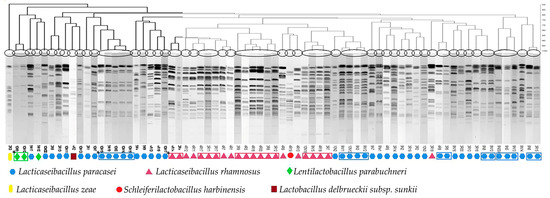
Figure 5.
Dendrogram showing PFGE band patterns of 73 lactobacilli isolates from IBT cheeses, divided into 49 pulsotypes after AscI restriction. * Isolates illustrated within the boxes display similar pulsotypes.
3.4. Investigation of the Probiotic Characteristics of the Lactobacilli Isolates
The representative isolates of the 49 lactobacilli strains were investigated for a range of typical probiotic characteristics. In terms of ability to withstand the harsh GIT conditions, six Lacticaseibacillus paracasei strains and two Lacticaseibacillus rhamnosus strains showed resistance to simulated gastric and duodenum juices with survival levels matching or exceeding those of the well-known probiotic strain Lacticaseibacillus rhamnosus GG, which was used as control (Table 3). These strains were able to grow on MRS supplemented with TDCA, which is indicative of resistance to this bile salt. Weak growth of some strains was also observed on MRS supplemented with GDCA, whereas none of them demonstrated the ability to deconjugate bile salts, but the same was also observed for LGG.

Table 3.
Survival of selected Lactobacillaceae strains and Lacticaseibacillus rhamnosus GG (control) following exposure to simulated GIT conditions (mean ± standard error of three independent experiments).
In terms of desirable antimicrobial activities, most of the strains inhibited a range of food-related pathogens including Staphylococcus aureus, Escherichia coli, Listeria monocytogenes and Salmonella. Of the 8 GIT-resistant strains, Lacticaseibacillus paracasei 4R15, 9N2, 9N14, 3R2 and Lacticaseibacillus rhamnosus 4R11 showed inhibitory activity against all tested pathogens (Table 4). The results of both methodologies used indicated that acid production was responsible for the pathogens’ inhibition.

Table 4.
Inhibitory activity of potential probiotic strains of Lacticaseibacillus paracasei and Lacticaseibacillus rhamnosus against selected pathogens 1 (mean of two independent experiments).
When tested for the ability to adhere to the intestinal epithelium cell line HT-29, three Lacticaseibacillus paracasei strains (4R15, 9N2 and 9N14) were found to possess adhesion properties exceeding those of the probiotic strain LGG (Figure 6). Lacticaseibacillus paracasei 9N2 strain was found to have the highest adherence values that were statistically significantly different (p < 0.05) from LGG and other Lacticaseibacillus paracasei strains (9N14 and 4R15).
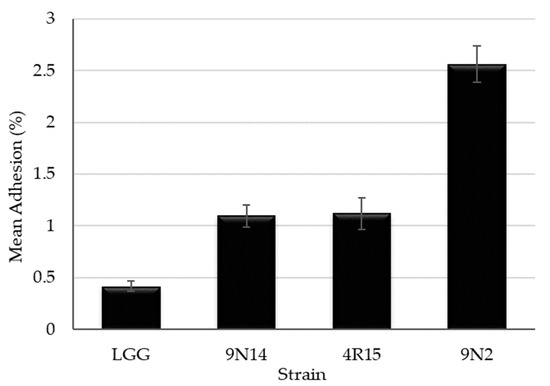
Figure 6.
Adhesion ability to HT-29 human colon cell lines of Lacticaseibacillus paracasei strains with probiotic potential (mean ± standard error of three independent experiments performed in triplicate).
The Lacticaseibacillus paracasei strains 4R15, 9N2 and 9N14 were further characterized for presence of undesirable traits, such as biogenic amine production and resistance to antibiotics. Importantly, none of these strains was found to produce the biogenic amines tyramine and histamine. In terms of antibiotic susceptibility, all three strains were resistant to vancomycin and trimethoprim, and only Lacticaseibacillus paracasei 9N14 was resistant to chloramphenicol. Strains were sensitive to the remaining antibiotics tested (Table 5).

Table 5.
VetMIC™ values (MIC as µg mL−1) * obtained for three Lacticaseibacillus paracasei strains isolated from IBT Cheeses and their comparison with the microbial cut-off values (µg mL−1) established by EFSA a [44], and reported by Ammor et al. b [45] and Danielsen et al. c [46] for Lacticaseibacillus casei/paracasei.
4. Discussion
There are few reports concerning the whole microbiota of IT and IBT cheeses. Species belonging to Enterococcus, Lactobacillus, Leuconostoc and Lactococcus genera were isolated and identified during these studies using conventional culture dependent methods [12,13]. E. durans, E. faecalis, E. faecium and L. casei (now Lacticaseibacillus casei) were found as dominant species. Other Lactobacillus spp., Lactococcus lactis subsp. lactis, L. lactis subsp. cremoris, and Leuconostoc spp. were also identifed in both IT and IBT cheeses [12,13]. Apart from these, more recently, Streptococcus thermophilus, Lactococcus lactis subsp. lactis, Lactobacillus gallinarum, Streptococcus equinus, Streptococcus infantarius subsp. infantarius, E. faecalis, E. faecium and Lactococcus garvieae were found as dominant species using the DGGE method [15].
In the present study, microbiota of both IT and IBT cheeses were investigated using the HTS approach, for the first time, in order to provide insights into the microbiota of these cheeses and to examine if differences between IT and IBT cheese could be identifed (Figure 1). Taxonomic classification of the DNA sequence data identifed mainly four phyla: Firmicutes, Actinobacteria, Bacteroidetes and Proteobacteria in IT and IBT cheeses, which is in agreement with the findings of other works on cheeses [18,20,27]. Similarly, as with the previous reports, at phylum level Firmicutes were the primary microbiota of both cheese types. Besides known genera, the method effectively revealed the presence of a number of other genera not previously associated with IT and IBT cheeses. Different from the former findings on IT and IBT cheeses, Streptococcus was found as the dominant genus in all cheese samples. OTUs belonging to Lacticaseibacillus, Lactobacillus, Enterococcus and Lactococcus genera were also detected in all samples but as relatively low proportions of the overall bacterial population. Besides the identification method used, several factors could be responsible for this difference. It is well established that animal source of milk, pasteurization, raw milk microbiota, production environment and production process parameters, salt content, etc. influence microbial populations present in the resultant cheese [19,27]. Any change in these parameters alter the equilibrium of cheese microbiota. From the time when the original studies were undertaken on these cheeses [8,49] to the present investigation, many changes have occurred to milk production technology, handling and transportation systems, hygiene practices, equipments and even IT and IBT cheese production processes. For example, pasteurization/thermization of milk and immersion of the pressed curd blocks into hot whey (in the region of 85 °C) for 30 min, the so called cooking step, were added to the production process of IBT cheese (Personal communication). Quigley et al. [27] observed a significant difference in the levels of Lactococcus and Lactobacillus between unpasteurized and pasteurized milk cheeses. Also, by comparing the bacterial genera present in artisanal cheeses manufactured from unpasteurized milk relative to those made from pasteurized milk, they detected some genera that were present in raw milk cheeses only, while some others were unique for pasteurized milk cheeses [27].
Taxonomic details up to the species level within the Streptococcus genus revealed that Streptococcus thermophilus and Streptococcus infantarius (S. infantarius subsp. infantarius) were the most abundant OTUs (Figure 4). This is consistent with the findings of recent studies on IBT cheese. Karabey et al. [15] detected Streptococcus thermophilus and Streptococcus infantarius subsp. infantarius amongst the dominant species of IBT cheeses. In a recent study conducted by our research team S. infantarius subsp. infantarius was determined as the species primarily responsible for acid production in IBT cheese [9]. Within the Lacticaseibacillus genus, Lacticaseibacillus paracasei were the most abundant OTU, which is consistent with the results of culture-dependent identification.
The culture-independent approach used in this study identified other genera for the first time in mature IT and IBT cheeses. Among these, Bifidobacteria and Chryseobacterium were detected in all samples while others were observed in some of them.
Members of the Chryseobacterium genus are gram negative, non-spore-forming, proteolytic, psychrotrophic bacteria that are widely distributed in a variety of environments, such as fresh water, sewage, soil and foods [50]. Chryseobacterium spp. have been frequently described in raw milk and in cheese among subdominant genera [18,27,51]. Chrysobacterium have the capacity to produce hydrolytic thermostable enzymes that lead to formation of undesirable aroma and flavor compounds during milk storage and cheese ripening, and thus, there is concern that they have the potential to cause spoilage defects in dairy products [52]. Further investigations are required to determine the source of these bacteria in IT and IBT cheeses, the species present in the cheese and their contribution to the maturation of the cheeses in order to understand whether they impact on key quality attributes of these cheeses.
The presence of bifidobacteria and Bifidobacterium mongoliense in some traditional and artisanal raw milk cheeses was also reported previously [19,27,53]. Bifidobacteria in cheese probably originates from raw milk contaminated with animal feces and may contribute to some organoleptic and technological characteristics [53]. The growth characteristics and nutrient requirements of bifidobacteria are different from most LAB. They have low proteolytic activity and usually require an anaerobic environment to survive. Their growth and long-term survival in cheese is supported by LAB metabolism, which alters cheese pH, limits oxygen levels and provides growth promoters [54]. It has been shown that S. thermophilus strains with high oxygen consumption ability enhance the viability of bifidobacteria [54]. In the present study, the presence of Bifidobacteria in all IT and IBT cheese samples may be linked to the high levels of Streptococcus thermophilus found in the cheese. Bifidobacterium ssp. are widely used as probiotic microorganisms. Bifidobacterium mongoliense strains showing in vitro resistance to gastric and pancreatic juices, and bile salts, and having ability to digest milk oligosaccharides and produce antivirulent metabolites, have been reported [53,55]. Therefore, efforts should be made in the future to recover and investigate the probiotic potential of bifidobacteria from IBT and IT cheeses.
Culture dependent microbial counts of cheeses exhibited different profile than metagenomic analysis. No statistical differences were observed between the bacterial counts of both IT and IBT across the four growth conditions tested implying that Streptococcus, Lactococcus, Enteroccocus and lactobacilli are present at similar levels and constitute the dominant LAB microbiota of IT and IBT cheeses. However, HTS data indicated that the microbiome of all cheeses were dominated by Streptococcus. It is well known that M17 and LBS agar (also known as Rogosa Agar) media are not very selective, and species of enterococci, lactobacilli, streptococci and lactococci are able to grow on both of these media [56,57]. KEA agar is used as a selective medium for the enumeration of enterococci. Growth of some Lactobacillus and Pediococcus species on KEA agar, with colony structure similar to enterococci, has been observed [58]. Therefore, the lack of selectivity could partly explain the broadly similar populations of bacteria observed under each of the four growth conditions tested. On the other hand, HTS analysis measures samples’ total DNA, which in the case of ripened cheeses certainly includes DNA originating from live, injured, dead and possibly the so-called viable-but-not-culturable cells [59]. Therefore, as we previousely demonstrated, Streptococcus is primarialy responsible for acid production during cheese manufacture [9] and thus present as the dominant component of the microbiome during cheese manufacture. However, by the end of ripening it is possible that many of these are no longer viable while their DNA would still be present in the cheese and being detected by HTS. Even so, the abundance of the genus Streptococcus in IT and IBT cheeses microbiota is obvious.
Although it has been reported that the use of different packaging materials (skinbag versus plastic, and wooden box) for Tulum cheeses affected the microbial composition of the cheeses [60], in present study no significant difference was determined between the microbiota of IT and IBT cheeses.
Traditional fermented foods, especially cheeses, can be a good source of microorganisms possessing probiotic properties [4,5,6,7]. In this respect, mesophilic lactobacilli are particularly sought after as they constitute a significant fraction of Non-Starter LAB microbiota of ripened cheeses and can find wide applications as starters, adjunct cultures and probiotics [61,62]. Therefore, when seeking to determine if bacteria from Tulum cheese may express characteristics associated with probiotics, we focused our study on mesophilic lactobacilli.
In this study, strains of six different species belonging to four genera from the Lactobacillaceae family were isolated from IBT cheeses, with Lacticaseibacillus paracasei being the most commonly encountered followed by Lacticaseibacillus rhamnosus. Selected strains of Lacticaseibacillus casei, Lacticaseibacillus paracasei and Lacticaseibacillus rhamnosus have applications as probiotics and are claimed to have beneficial effects on human health [63].
In this study, the overall lactobacilli population of IBT cheese were investigated for their probiotic characteristics for the first time. Tolerance to GIT conditions and ability to adhere to intestinal cells are mandatory features of bacteria displaying probiotic potential [4,64]. Amongst 49 tested strains, six Lacticaseibacillus paracasei and two Lacticaseibacillus rhamnosus showed ability to remain viable following simulated GIT conditions, with survival levels matching or exceeding those of the well known probiotic strain LGG. The strains Lacticaseibacillus paracasei 4R15, 9N2 and 9N14, and Lacticaseibacillus rhamnosus 4R11 exhibited the best survival rates with less than 2.5 log reduction. Good survival ability of several Lacticaseibacillus paracasei [37], Lacticaseibacillus casei and Lacticaseibacillus rhamnosus strains [5] have been reported by other researchers.
Another important function of a probiotic bacteria is to protect the host gastrointestinal tract from pathogen infection through the production of inhibitory compounds such as organic acids (e.g., lactic acid, acetic acid), hydrogen peroxide, and bacteriocins [65]. Here, the GIT-resistant strains of Lacticaseibacillus paracasei (4R15, 9N2, 9N14, 3R2) and Lacticaseibacillus rhamnosus (4R11) showed ability to inhibit all tested pathogens, and the inhibition was due to organic acids. In agreement with our findings, the antagonistic effects of Lacticaseibacillus paracasei and Lacticaseibacillus rhamnosus strains against pathogens have been associated with production of organic acids [37].
Although its contribution is not fully described, BSH activity is thought to be a necessary feature for probiotic strains to withstand the toxicity of conjugated bile salts in the duodenum and survive in the highly competitive environment of the human intestinal tract. However, some reports suggest that resistance to bile salts in lactobacilli is not always related to hydrolase activity [66]. Lactobacilli strains tested in this study were able to grow in the presence of conjugated bile salts but did not hydrolyze GDCA and TDCA. Similar characteristics were observed in lactobacilli strains isolated from fermented foods [37,66].
Adhesion to and colonization to the gastrointestinal tract of the host is another important trait of probiotic bacteria. Therefore, the ability of the strains to adhere to the human intestinal cell lines is an important criterion while evaluating the probiotic potential of novel strains [39,67]. Adhesion ability is a strain and matrix specific trait [64] with levels of adhesion ranging from 3 to 20% for various Lacticaseibacillus casei and Lacticaseibacillus paracasei strains being reported [37,68]. In our study, three Lacticaseibacillus paracasei strains were able to adhere to the human intestinal cell line HT-29 at levels varying from 1% to 2.6% and higher than that observed for the reference strain LGG. Similar to our findings, low adhesion levels of LGG to HT-29 cells were reported by other authors [69].
As they may contribute to the transfer of antibiotic resistance genes, all bacteria intended for use as probiotics or starter cultures must be assessed for their sensitivity/resistance to antibiotics of human and veterinary importance and must comply with the guidelines set out by the EFSA [44]. In the present study, all tested strains were resistant to vancomycin and trimethoprim. Vancomycin resistance is well characterized in lactobacilli and has also been reported for Lacticaseibacillus paracasei strains [37,39]. It is attributed to the synthesis of peptidoglycan precursors terminating with D-alanyl-D-lactate conferring vancomycin resistance. Such resistance is intrinsic, chromosomally encoded and not transferable [70]. Lactobacilli, including the Lacticaseibacillus species, have also been reported as being intrinsically resistant to trimethoprim [43]. In LAB, trimethoprim resistance is associated with the absence of the folic acid synthesis pathway, which is the target of this antibiotic, and it has been described as an intrinsic feature [71]. Intrinsic and acquired resistance by mutation are assumed to have a low potential of horizontal transfer [44]. Lactobacilli are generally known as susceptible to chloramphenicol; however, in recent years transferable chloramphenicol resistant genes as well as phenotypic resistance have been observed in lactobacilli [43,45]. Lacticaseibacillus paracasei 9N14 strain showed resistance to chloramphenicol. The strains tested in our study, except 9N14, do not seem to represent a source for transferable resistance genes since they were phenotypically susceptible to the remaining 14 antibiotics. But the absence of acquired or transferable resistance factors must be proven genotypically in these strains, especially in the strain 9N14, before used as probiotics.
Biogenic amines occur in foods as a result of amino acid decarboxylation by decarboxylase positive microorganisms and can cause toxicological effects (e.g., hypertension, headaches, palpitations and vomiting) in humans. Cheese can contain potentially harmful levels of biogenic amines, especially histamine and tyramine [72]. The EFSA regards histamine and tyramine as the most important biogenic amines from a toxicological standpoint [73]. Lactobacilli species are amongst the major biogenic amines producers in cheese, with Lacticaseibacillus paracasei strains being reported to produce tyramine [74]. It is important to note that the probiotic candidates in our study do not produce biogenic amines. This is a desirable trait for food grade microorganisms.
These data, taken together, demonstrate that strains of lactobacilli from Tulum cheese encode a range of characteristics required of a probiotic. They also survive in the cheese during ripening and are among the dominant strains present. These all support the view that these strains could be applied during cheese manufacture with a view to enhancing the health-promoting properties of cheese. However, the in vivo potential of these strains as probiotics would first need to be confirmed.
5. Conclusions
The application of HTS of DNA gave detailed new information about the microbiota of commercial examples of IT and IBT cheeses. This approach:
- Highlighted the dominance of the genus Streptococcus and, within the genus, the abundance of the species S. thermophilus and S. infantarius subsp. infantarius.
- It unveiled the presence of genera, including Bifidobacteria and Chryseobacterium, that have not been reported in these cheese types before.
Results from the culture-dependent approach confirmed Streptococcus as a key microbial population in IT and IBT cheeses but also demonstrated that Lactococcus, Enterococcus and lactobacilli are present in large populations.
This information will provide the base for further comprehensive studies to solve the quality problems and to create appropriate starter/adjunct cultures for these cheeses, in order to produce a Tulum cheese of standardized quality.
The investigations also gave valuable information about the Lactobacillaceae microbiota of the IBT cheeses. Evaluation of their in vitro probiotic properties displayed the presence of a potentially highly beneficial microbiota in these traditional cheeses. Three Lacticaseibacillus paracasei strains exhibited favorable in vitro probiotic properties including: Potential to survive passage through the GIT, inhibition of selected pathogens, adhesion ability to human colon cells, antibiotic sensitivity, absence of biogenic amine production.
Therefore these strains could be candidates for inclusion as starter/adjunct cultures in the manufacture of Tulum cheeses or of probiotic-containing fermented foods. This is the first study that the overall lactobacilli population from IBT cheese were investigated for their probiotic characteristics. Additional studies are required to confirm their in vivo probiotic properties and technological attributes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/foods12183482/s1, Table S1: Cheese samples used in the study.
Author Contributions
Conceptualization, Z.G., V.F. and T.B.; Methodology, Z.G. and V.F.; Investigation, Z.G., D.O., M.M., S.S. and V.P.; Formal analysis, R.C.-R. and Z.G.; Writing—Original Draft, Z.G.; Visualization, Z.G.; Writing—Review & Editing, Z.G., V.F. and T.B.; Supervision, T.B.; Resources, T.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data is contained within the article or supplementary material. Extra data will be provided on request.
Acknowledgments
Ziba Güley gratefully acknowledges receipt of a fellowship from the Scientific and Technological Research Council of Türkiye (TÜBİTAK) BİDEB 2219. Authors especially thank Paul D. Cotter, Rita Hickey and Kieran Jordan for providing opportunity to make use of the facilities in their laboratories during this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yeluri Jonnala, B.R.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D. Sequencing of the cheese microbiome and its relevance to industry. Front. Microbiol. 2018, 9, 1020. [Google Scholar] [CrossRef]
- Masoud, W.; Takamiya, M.; Vogensen, F.K.; Lillevang, S.; Al-Soud, W.A.; Sørensen, S.J.; Jakobsen, M. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int. Dairy J. 2011, 21, 142–148. [Google Scholar] [CrossRef]
- Hayaloglu, A.A.; Fox, P.F.; Guven, M.; Cakmakci, S. Cheeses of Turkey: 1. Varieties ripened in goat-skin bags. Lait 2007, 87, 79–95. [Google Scholar] [CrossRef]
- Leeuwendaal, N.; Stanton, C.; O’Toole, P.W.; Beresford, T.P. The potential of non-starter lactic acid bacteria from Cheddar cheese to colonise the gut. J. Funct. Foods. 2021, 83, 104425. [Google Scholar] [CrossRef]
- Solieri, L.; Bianchi, A.; Mottolese, G.; Lemmetti, F.; Giudici, P. Tailoring the probiotic potential of non-starter Lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol. 2014, 38, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Tulumoğlu, Ş.; Kaya, H.I.; Şimşek, Ö. Probiotic characteristics of Lactobacillus fermentum strains isolated from tulum cheese. Anaerobe 2014, 30, 120–125. [Google Scholar] [CrossRef]
- Mohammed, S.; Çon, A.H. Isolation and characterization of potential probiotic lactic acid bacteria from traditional cheese. LWT 2021, 152, 112319. [Google Scholar] [CrossRef]
- Koca, N. The Effects of the Different Starter Culture Combinations on the Properties of Izmir Tulum (Canned) Cheese. Master’s Thesis, Ege University, Izmir, Türkiye, 1996. Available online: https://tez.yok.gov.tr/UlusalTezMerkezi/tezSorguSonucYeni.jsp (accessed on 21 May 2023).
- Güley, Z.; Fallico, V.; Cabrera-Rubio, R.; Cotter, P.D.; Beresford, T. Identification of Streptococcus infantarius subsp. infantarius as the species primarily responsible for acid production in Izmir Brined Tulum cheese from the Aegean region of Türkiye. Food Res. Int. 2022, 160, 111707. [Google Scholar] [CrossRef]
- Jansen, W.; Linard, C.; Noll, M.; Nöckler, K.; Al Dahouk, S. Brucella-positive raw milk cheese sold on the inner European market: A public health threat due to illegal import? Food Control 2019, 100, 130–137. [Google Scholar] [CrossRef]
- Mayo, B.; Rachid, C.T.; Alegria, A.; Leite, A.M.; Peixoto, R.S.; Delgado, S. Impact of next generation sequencing techniques in food microbiology. Curr. Genomics 2014, 15, 293–309. [Google Scholar] [CrossRef]
- Gökovalı, T. Salamuralı Tulum Peynirinin Olgunlaşması Sırasında Meydana Gelen Mikrobiyolojik Değişiklikler Üzerinde Araştırma (A Research on Microbiological Changes during Ripening of Brined Tulum Cheese). Ph.D. Thesis, Ege University, Izmir, Türkiye, 1980. (In Turkish). [Google Scholar]
- Kılıç, S.; Gönç, S. Izmir Tulum peynirinin olgunlasmasinda rol oynayan mikroorganizma gruplarinin belirlenmesi uzerine bir arastirma (A research on the determination of microorganism groups that play a role in the ripening of Izmir Tulum cheese). Ege Üniv. Ziraat Fak. Derg. 1992, 29, 71–78. [Google Scholar]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. Molecular approaches to analysing the microbial composition of raw milk and raw milk cheese. Int. J. Food Microbiol. 2011, 150, 81–94. [Google Scholar] [CrossRef]
- Karabey, B.; Eroglu, D.; Vural, C.; Ozdemir, G.; Yerlikaya, O.; Kinik, O. Determination of the microbial flora in traditional İzmir Tulum cheeses by Denaturing Gradient Gel Electrophoresis. J. Food Sci. Technol. 2018, 55, 956–963. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, M.; Terzic-Vidojevic, A.; Jovcic, B.; Begovic, J.; Golic, N.; Topisirovic, L. Characterization of lactic acid bacteria isolated from Bukuljac, a homemade goat’s milk cheese. Int. J. Food Microbiol. 2008, 122, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, C.L.; Torriani, S.; Akkermans, A.D.L.; de Vos, W.M.; Vaughan, E.E. Diversity, dynamics, and activity of bacterial communities during production of an Artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 2002, 68, 1882–1892. [Google Scholar] [CrossRef]
- Dalmasso, A.; Soto del Rio, M.d.l.D.; Civera, T.; Pattono, D.; Cardazzo, B.; Bottero, M.T. Characterization of microbiota in Plaisentif cheese by high-throughput sequencing. LWT—Food Sci. Technol. 2016, 69, 490–496. [Google Scholar] [CrossRef]
- Alegría, Á.; Szczesny, P.; Mayo, B.; Bardowski, J.; Kowalczyk, M. Biodiversity in Oscypek, a traditional Polish cheese, determined by culture-dependent and -independent approaches. Appl. Environ. Microbiol. 2012, 78, 1890–1898. [Google Scholar] [CrossRef]
- Biolcati, F.; Ferrocino, I.; Bottero, M.T.; Dalmasso, A. Short communication: High-throughput sequencing approach to investigate Italian artisanal cheese production. J. Dairy Sci. 2020, 103, 10015–10021. [Google Scholar] [CrossRef]
- Coelho, M.C.; Malcata, F.X.; Silva, C.C.G. Distinct bacterial communities in São Jorge cheese with protected designation of origin (PDO). Foods 2023, 12, 990. [Google Scholar] [CrossRef]
- Kamilari, E.; Anagnostopoulos, D.A.; Papademas, P.; Kamilaris, A.; Tsaltas, D. Characterizing Halloumi cheese’s bacterial communities through metagenomic analysis. LWT 2020, 126, 109298. [Google Scholar] [CrossRef]
- Yurt, M.N.Z.; Ersoy Omeroglu, E.; Tasbasi, B.B.; Acar, E.E.; Altunbas, O.; Ozalp, V.C.; Sudagidan, M. Bacterial and fungal microbiota of mould-ripened cheese produced in Konya. Int. J. Dairy Technol. 2023, 76, 627–637. [Google Scholar] [CrossRef]
- Walsh, A.M.; Macori, G.; Kilcawley, K.N.; Cotter, P.D. Meta-analysis of cheese microbiomes highlights contributions to multiple aspects of quality. Nat. Food 2020, 1, 500–510. [Google Scholar] [CrossRef]
- Westaway, J.A.F.; Huerlimann, R.; Miller, C.M.; Kandasamy, Y.; Norton, R.; Rudd, D. Methods for exploring the faecal microbiome of premature infants: A review. Matern. Health Neonatol. Perinatol. 2021, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D. High-Throughput Sequencing and Metagenomics: Moving Forward in the Culture-Independent Analysis of Food Microbial Ecology. Appl. Environ. Microbiol. 2013, 79, 3148–3155. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; Cotter, P.D. High-Throughput Sequencing for detection of subpopulations of bacteria not previously associated with Artisanal cheeses. Appl. Environ. Microbiol. 2012, 78, 5717–5723. [Google Scholar] [CrossRef] [PubMed]
- Quigley, L.; O’Sullivan, D.J.; Daly, D.; O’Sullivan, O.; Burdikova, Z.; Vana, R.; Beresford, T.P.; Ross, R.P.; Fitzgerald, G.F.; McSweeney, P.L.H.; et al. Thermus and the pink discoloration defect in cheese. mSystems 2016, 1, e00023-16. [Google Scholar] [CrossRef]
- O’Sullivan, D.J.; Fallico, V.; O’Sullivan, O.; McSweeney, P.L.H.; Sheehan, J.J.; Cotter, P.D.; Giblin, L. High-throughput DNA sequencing to survey bacterial histidine and tyrosine decarboxylases in raw milk cheeses. BMC Microbiol. 2015, 15, 266. [Google Scholar] [CrossRef]
- Quigley, L.; O’Sullivan, O.; Beresford, T.P.; Paul Ross, R.; Fitzgerald, G.F.; Cotter, P.D. A comparison of methods used to extract bacterial DNA from raw milk and raw milk cheese. J. Appl. Microbiol. 2012, 113, 96–105. [Google Scholar] [CrossRef]
- Illumina 16S Metagenomics Sequencing Workflow. Available online: https://www.illumina.com/content/dam/illumina-marketing/documents/products/other/16s-metagenomics-faq-1270-2014-003.pdf (accessed on 24 June 2021).
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Titus Brown, C.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Simpson, P.J.; Stanton, C.; Fitzgerald, G.F.; Ross, R.P. Genomic diversity within the genus Pediococcus as revealed by randomly amplified polymorphic DNA PCR and pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 2002, 68, 765–771. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pisano, M.B.; Viale, S.; Conti, S.; Fadda, M.E.; Deplano, M.; Melis, M.P.; Monica, D.; Cosentino, S. Preliminary evaluation of probiotic properties of Lactobacillus strains isolated from Sardinian dairy products. Biomed. Res. Int. 2014, 2014, 286390. [Google Scholar] [CrossRef]
- Bolocan, A.S.; Pennone, V.; O’Connor, P.M.; Coffey, A.; Nicolau, A.I.; McAuliffe, O.; Jordan, K. Inhibition of Listeria monocytogenes biofilms by bacteriocin-producing bacteria isolated from mushroom substrate. J. Appl. Microbiol. 2017, 122, 279–293. [Google Scholar] [CrossRef]
- Verdenelli, M.C.; Ghelfi, F.; Silvi, S.; Orpianesi, C.; Cecchini, C.; Cresci, A. Probiotic properties of Lactobacillus rhamnosus and Lactobacillus paracasei isolated from human faeces. Eur. J. Nutr. 2009, 48, 355–363. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Ross, S.A.; Lane, J.A.; Kilcoyne, M.; Joshi, L.; Hickey, R.M. Defatted bovine milk fat globule membrane inhibits association of enterohaemorrhagic Escherichia coli O157:H7 with human HT-29 cells. Int. Dairy J. 2016, 59, 36–43. [Google Scholar] [CrossRef]
- Bover-Cid, S.; Holzapfel, W.H. Improved screening procedure for biogenic amine production by lactic acid bacteria. Int. J. Food Microbiol. 1999, 53, 33–41. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Clarke, S.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O’Toole, P.W. Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef]
- EFSA. EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP); Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Ammor, M.S.; Belén Flórez, A.; Mayo, B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007, 24, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Thukral, A.K. A review on measurement of Alpha diversity in biology. Agric. Res. J. 2017, 54, 1–10. [Google Scholar] [CrossRef]
- Kers, J.G.; Saccenti, E. The power of microbiome studies: Some considerations on which alpha and beta metrics to use and how to report results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Yaygın, H. Salamuralı Tulum peynirinin yapılışı ve özellikleri üzerinde araştırmalar (Studies on the production and properties of brined Tulum cheese). Ege Üniv. Ziraat Fak. Derg. 1971, 8, 91–124. [Google Scholar]
- Bernardet, J.F.; Hugo, C.; Bruun, B. The Genera Chryseobacterium and Elizabethkingia. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 638–676. ISBN 978-0-387-25497-5. [Google Scholar] [CrossRef]
- Franciosi, E.; De Sabbata, G.; Gardini, F.; Cavazza, A.; Poznanski, E. Changes in psychrotrophic microbial populations during milk creaming to produce Grana Trentino cheese. Food Microbiol. 2011, 28, 43–51. [Google Scholar] [CrossRef]
- Tsôeu, L.I.; Jooste, P.J.; Charimba, G.; Hugo, C.J. Spoilage potential of a novel group of bacteria isolated from dairy products. S. Afr. J. Sci. 2016, 112, 1–8. [Google Scholar] [CrossRef][Green Version]
- Delcenserie, V.; Taminiau, B.; Gavini, F.; de Schaetzen, M.A.; Cleenwerck, I.; Theves, M.; Mahieu, M.; Daube, G. Detection and characterization of Bifidobacterium crudilactis and B. mongoliense able to grow during the manufacturing process of French raw milk cheeses. BMC Microbiol. 2013, 13, 239. [Google Scholar] [CrossRef]
- Boylston, T.D.; Vinderola, C.G.; Ghoddusi, H.B.; Reinheimer, J.A. Incorporation of bifidobacteria into cheeses: Challenges and rewards. Int. Dairy J. 2004, 14, 375–387. [Google Scholar] [CrossRef]
- Bondue, P.; Milani, C.; Arnould, E.; Ventura, M.; Daube, G.; LaPointe, G.; Delcenserie, V. Bifidobacterium mongoliense genome seems particularly adapted to milk oligosaccharide digestion leading to production of antivirulent metabolites. BMC Microbiol. 2020, 20, 111. [Google Scholar] [CrossRef] [PubMed]
- Guley, Z.; Uysal, H.R.; Kilic, S. Lactic acid bacteria flora of Konya Kuflu cheese: A traditional cheese from Konya province in Turkey. J. Microbiol. Biotechnol. Food Sci. 2015, 4, 238–242. [Google Scholar] [CrossRef]
- Jackson, M.S.; Bird, A.R.; McOrist, A.L. Comparison of two selective media for the detection and enumeration of Lactobacilli in human faeces. J. Microbiol. Methods 2002, 51, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.; Domig, K.J.; Kneifel, W. Comparison of selective media for the enumeration of probiotic enterococci from animal feed. Food Technol. Biotechnol. 2005, 43, 147–155. [Google Scholar]
- Bellali, S.; Lagier, J.C.; Million, M.; Anani, H.; Haddad, G.; Francis, R.; Yimagou, E.K.; Khelaifia, S.; Levasseur, A.; Raoult, D.; et al. Running after ghosts: Are dead bacteria the dark matter of the human gut microbiota? Gut Microbes 2021, 13, e1897208. [Google Scholar] [CrossRef]
- Sengul, M.; Turkoglu, H.; Cakmakci, S.; Con, A.H. Effects of casing materials and ripening period on some microbiological properties of Tulum cheese. Pak. J. Biol. Sci. 2001, 4, 854–857. [Google Scholar] [CrossRef]
- Beresford, T.P.; Fitzsimons, N.A.; Brennan, N.L.; Cogan, T.M. Recent advances in cheese microbiology. Int. Dairy J. 2001, 11, 259–274. [Google Scholar] [CrossRef]
- Settanni, L.; Moschetti, G. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 2010, 27, 691–697. [Google Scholar] [CrossRef]
- Lavermicocca, P.; Dekker, M.; Russo, F.; Valerio, F.; Di Venere, D.; Sisto, A. Lactobacillus paracasei-enriched vegetables containing health promoting molecules. In Probiotics, Prebiotics, and Synbiotics: Bioactive Foods in Health Promotion, 1st ed.; Watson, R., Preedy, V.R., Eds.; Elsevier Academic Press: Cambridge, MA, USA, 2016; pp. 361–370. ISBN 978-0-12-802189-7. [Google Scholar]
- Jensen, H.; Grimmer, S.; Naterstad, K.; Axelsson, L. In vitro testing of commercial and potential probiotic lactic acid bacteria. Int. J. Food Microbiol. 2012, 153, 216–222. [Google Scholar] [CrossRef]
- Kanmani, P.; Satish Kumar, R.; Yuvaraj, N.; Paari, K.A.; Pattukumar, V.; Arul, V. Probiotics and its functionally valuable products—A review. Crit. Rev. Food Sci. Nutr. 2013, 53, 641–658. [Google Scholar] [CrossRef]
- Moser, S.A.; Savage, D.C. Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 2001, 67, 3476–3480. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO (Food and Agriculture Organization of the United Nations/World Health Organization). Probiotics in Food. Health and Nutritional Properties and Guidelines for Evaluation; Food and Nutrition Paper 85; FAO: Rome, Italy, 2006. Available online: http://www.fao.org/3/a0512e/a0512e.pdf (accessed on 2 July 2023).
- Caggia, C.; De Angelis, M.; Pitino, I.; Pino, A.; Randazzo, C.L. Probiotic features of Lactobacillus strains isolated from Ragusano and Pecorino Siciliano cheeses. Food Microbiol. 2015, 50, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Garriga, M.; Rubio, R.; Aymerich, T.; Ruas-Madiedo, P. Potentially probiotic and bioprotective lactic acid bacteria starter cultures antagonise the Listeria monocytogenes adhesion to HT29 colonocyte-like cells. Benef. Microbes 2015, 6, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Handwerger, S.; Pucci, M.J.; Volk, K.J.; Liu, J.; Lee, M.S. Vancomycin-resistant Leuconostoc mesenteroides and Lactobacillus casei synthesize cytoplasmic peptidoglycan precursors that terminate in lactate. J. Bacteriol. 1994, 176, 260–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katla, A.K.; Kruse, H.; Johnsen, G.; Herikstad, H. Antimicrobial susceptibility of starter culture bacteria used in Norwegian dairy products. Int. J. Food Microbiol. 2001, 67, 147–152. [Google Scholar] [CrossRef]
- Linares, D.M.; Del Río, B.; Ladero, V.; Martínez, N.; Fernández, M.; Martín, M.C.; Álvarez, M.A. Factors influencing biogenic amines accumulation in dairy products. Front. Microbiol. 2012, 3, 180. [Google Scholar] [CrossRef]
- EFSA. EFSA Panel on Biological Hazards (BIOHAZ); Scientific Opinion on risk based control of biogenic amine formation in fermented foods. EFSA J. 2011, 9, 2393. [Google Scholar] [CrossRef]
- Carafa, I.; Nardin, T.; Larcher, R.; Viola, R.; Tuohy, K.; Franciosi, E. Identification and characterization of wild lactobacilli and pediococci from spontaneously fermented Mountain Cheese. Food Microbiol. 2015, 48, 123–132. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).