A Rapid and Accurate Quantitative Analysis of Cellulose in the Rice Bran Layer Based on Near-Infrared Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
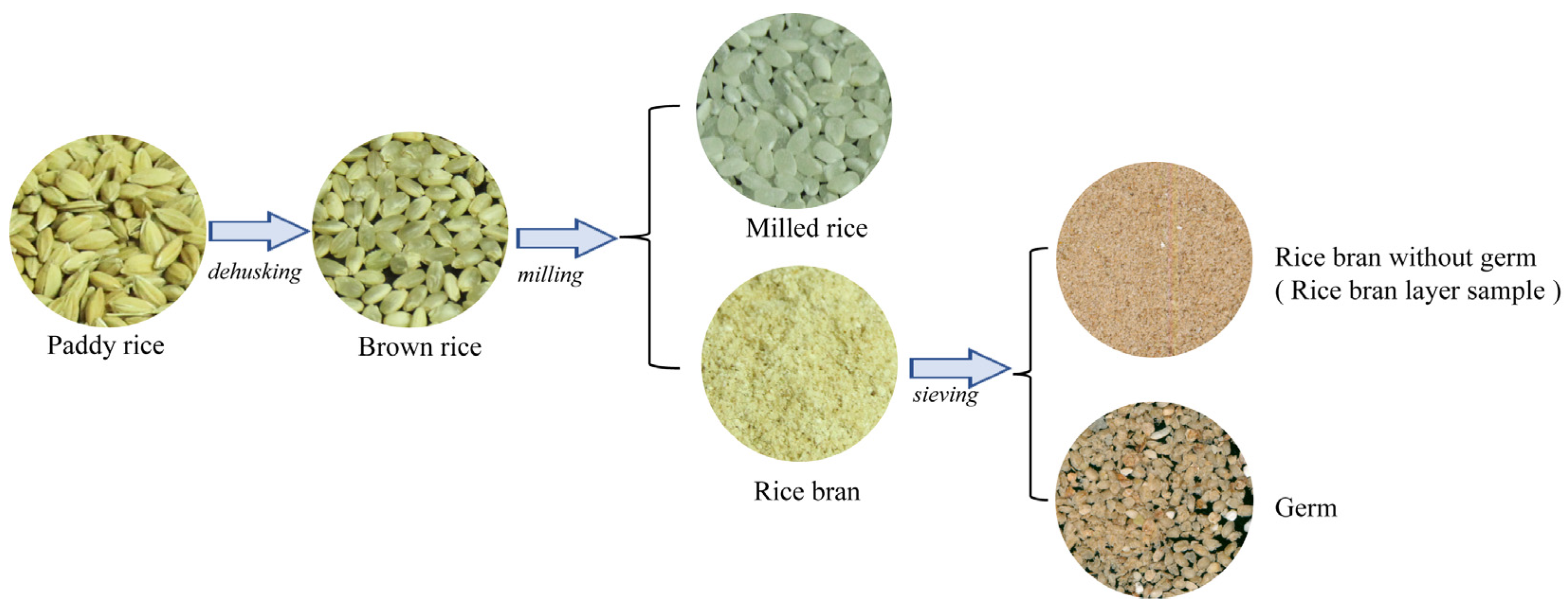
2.1. Materials
2.2. Chemical Analysis
2.2.1. Determination Method of Cellulose
2.2.2. Validation of the Optimized PDO Method
2.3. NIR Spectroscopy Measurement
2.4. The Construction and Evaluation of the NIR Model
2.5. Fusion of NIRdr and NIRdt Spectra
2.6. Statistical Analysis Software
3. Results and Discussion
3.1. Validation of the Optimized PDO Method
3.1.1. Intra-Day Precision and Inter-Day Precision of the Optimized PDO Method
3.1.2. Comparison between the Optimized PDO Method and the Original PDO Method
3.2. Results of Chemical Determination of Rice Bran Layer Sample Set
3.3. Fused Spectra Obtained under Three Different Pretreatment Conditions
3.4. NIR Calibration and Validation Results
3.5. Spectral Ranges of the Optimal Models
3.6. External Prediction of the Models
3.7. Rice Varieties with Low Cellulose Content in Bran Layer Screened by NIRS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mohidem, N.A.; Hashim, N.; Shamsudin, R.; Man, H.C. Rice for Food Security: Revisiting Its Production, Diversity, Rice Milling Process and Nutrient Content. Agriculture 2022, 12, 741. [Google Scholar] [CrossRef]
- Yang, L.; Fan, Y.; Song, S.; Wang, D.; Zhang, Y.; Cao, M. Frictional behavior of brown rice grain during moderate processing. Tribol. Int. 2021, 162, 107123. [Google Scholar] [CrossRef]
- Dhankhar, P.; Hissar, T. Rice milling. IOSR J. Eng. 2014, 4, 34–42. [Google Scholar] [CrossRef]
- Puri, S.; Dhillon, B.; Sodhi, N.S. Effect of degree of milling (Dom) on overall quality of rice—A review. Int. J. Adv. Biotechnol. Res. 2014, 5, 474–489. [Google Scholar]
- Hu, Y.; Willett, W.C.; Manson, J.A.E.; Rosner, B.; Hu, F.B.; Sun, Q. Intake of whole grain foods and risk of coronary heart disease in US men and women. BMC Med. 2022, 20, 192. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Sudha, V.; Wedick, N.M.; RamyaBai, M.; Vijayalakshmi, P.; Lakshmipriya, N.; Gayathri, R.; Kokila, A.; Jones, C.; Hong, B.; et al. Substituting brown rice for white rice on diabetes risk factors in India: A randomised controlled trial. Br. J. Nutr. 2019, 121, 1389–1397. [Google Scholar] [CrossRef]
- Ren, G.Y.; Qi, J.; Zou, Y.L. Association between intake of white rice and incident type 2 diabetes—An updated meta-analysis. Diabetes Res. Clin. Pract. 2021, 172, 108651. [Google Scholar] [CrossRef] [PubMed]
- Heiniö, R.-L.; Noort, M.; Katina, K.; Alam, S.A.; Sozer, N.; De Kock, H.L.; Hersleth, M.; Poutanen, K. Sensory characteristics of wholegrain and bran-rich cereal foods–a review. Trends Food Sci. Technol. 2016, 47, 25–38. [Google Scholar] [CrossRef]
- Roberts, R. Composition and taste evaluation of rice milled to different degrees. J. Food Sci. 1979, 44, 127–129. [Google Scholar] [CrossRef]
- Chen, H.H.; Chen, Y.K.; Chang, H.C. Evaluation of physicochemical properties of plasma treated brown rice. Food Chem. 2012, 135, 74–79. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Cheng, M.; Wang, R.; Luo, X.; Li, Y.; Chen, Z. Influence of ultrasonic enzyme treatment on the cooking and eating quality of brown rice. J. Cereal Sci. 2015, 63, 140–146. [Google Scholar] [CrossRef]
- Das, M.; Banerjee, R.; Bal, S. Evaluation of physicochemical properties of enzyme treated brown rice (Part B). LWT-Food Sci. Technol. 2008, 41, 2092–2096. [Google Scholar] [CrossRef]
- Das, M.; Gupta, S.; Kapoor, V.; Banerjee, R.; Bal, S. Enzymatic polishing of rice–A new processing technology. LWT-Food Sci. Technol. 2008, 41, 2079–2084. [Google Scholar] [CrossRef]
- Wattanavanitchakorn, S.; Wansuksri, R.; Chaichoompu, E.; Kamolsukyeunyong, W.; Vanavichit, A. Dietary Fibre Impacts the Texture of Cooked Whole Grain Rice. Foods 2023, 12, 899. [Google Scholar] [CrossRef]
- Casas, G.A.; Laerke, H.N.; Knudsen, K.E.B.; Stein, H.H. Arabinoxylan is the main polysaccharide in fiber from rice coproducts, and increased concentration of fiber decreases in vitro digestibility of dry matter. Anim. Feed Sci. Technol. 2019, 247, 255–261. [Google Scholar] [CrossRef]
- Vansoest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Dampanaboina, L.; Yuan, N.; Mendu, V. Estimation of crystalline cellulose content of plant biomass using the updegraff method. JoVE J. Vis. Exp. 2021, 171, e62031. [Google Scholar]
- Garcia-Iruela, A.; Esteban, L.G.; Fernandez, F.G.; de Palacios, P.; Rodriguez-Navarro, A.B.; Martin-Sampedro, R.; Eugenio, M.E. Changes in cell wall components and hygroscopic properties of Pinus radiata caused by heat treatment. Eur. J. Wood Wood Prod. 2021, 79, 851–861. [Google Scholar] [CrossRef]
- Halliwell, G. A microdetermination of cellulose in studies with cellulase. Biochem. J. 1958, 68, 605–610. [Google Scholar] [CrossRef]
- Fan, S.; Xu, Z.; Cheng, W.; Wang, Q.; Yang, Y.; Guo, J.; Zhang, P.; Wu, Y. Establishment of Non-Destructive Methods for the Detection of Amylose and Fat Content in Single Rice Kernels Using Near-Infrared Spectroscopy. Agriculture 2022, 12, 1258. [Google Scholar] [CrossRef]
- Yu, H.; Guo, L.; Kharbach, M.; Han, W. Multi-Way Analysis Coupled with Near-Infrared Spectroscopy in Food Industry: Models and Applications. Foods 2021, 10, 802. [Google Scholar] [CrossRef]
- Barea-Sepulveda, M.; Ferreiro-Gonzalez, M.; Calle, J.L.P.; Barbero, G.F.; Ayuso, J.; Palma, M. Comparison of different processing approaches by SVM and RF on HS-MS eNose and NIR Spectrometry data for the discrimination of gasoline samples. Microchem. J. 2022, 172, 106893. [Google Scholar] [CrossRef]
- Chang, H.-H.; Chen, Y.-C.; Huang, C.-J.; Kuo, C.-C.; Wang, Y.-M.; Sun, C.-W. Optimization of extracorporeal membrane oxygenation therapy using near-infrared spectroscopy to assess changes in peripheral circulation: A pilot study. J. Biophotonics 2020, 13, e202000116. [Google Scholar] [CrossRef]
- Wang, N.; Li, L.; Liu, J.; Shi, J.; Li, W. Rapid Detection of Cellulose and HemicelluloseContents of Corn Stover Based on Near-infraredSpectroscopy Combined with Chemometrics. Appl. Opt. 2021, 60, 4282–4290. [Google Scholar] [CrossRef]
- Nielsen, N.S.; Stubbs, T.L.; Garland-Campbell, K.A.; Carter, A.H. Rapid Estimation of Wheat Straw Decomposition Constituents Using Near-Infrared Spectroscopy. Agronomy 2019, 9, 462. [Google Scholar] [CrossRef]
- Nieto-Ortega, B.; Arroyo, J.J.; Walk, C.; Castaares, N.; Canet, E.; Smith, A. Near infrared reflectance spectroscopy as a tool to predict non-starch polysaccharide composition and starch digestibility profiles in common monogastric cereal feed ingredients. Anim. Feed Sci. Technol. 2022, 285, 115214. [Google Scholar] [CrossRef]
- Xu, Z.; Cheng, W.; Fan, S.; Liu, J.; Wang, H.; Li, X.; Liu, B.; Wu, Y.; Zhang, P.; Wang, Q. Data fusion of near-infrared diffuse reflectance spectra and transmittance spectra for the accurate determination of rice flour constituents. Anal. Chim. Acta 2022, 1193, 339384. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.-M.; Liu, B.-M.; Ye, Y.-F.; Tao, L.-Z.; Wu, Y.-J. Screening of Amylose and Protein Mutants and Correlation Analysis of Agronomic Character in Rice. Acta Laser Biol. Sin. 2016, 25, 356–361. (In Chinese) [Google Scholar]
- Cheng, W.-M.; Liu, B.-M.; Ye, Y.-F.; Xu, Z.-P.; Wang, Q.; Liu, J.; Tao, L.-Z.; Wu, Y.-J. Establishment of Rice Amylose and Protein Mutant Lines by Heavy Ion Irradiation. Chin. Agric. Sci. Bull. 2016, 32, 86–90. (In Chinese) [Google Scholar]
- Tian, F.; Tan, F.; Zhu, P. Multi-classification identification of PLS in rice spectra with different pre-treatments and K/S optimisation. Vib. Spectrosc. 2020, 109, 103069. [Google Scholar] [CrossRef]
- Xiong, S.-M.; Zuo, X.-F.; Zhu, Y.-Y. Determination of Cellulose, Hemi-Cellulose and Ligin in Rice Hull. Cereal Feed Ind. 2005, 8, 40–41. (In Chinese) [Google Scholar]
- Niero, G.; Thomas, S.A.; Mouratidou, K.; Visentin, G.; De Marchi, M.; Penasa, M.; Cassandro, M. Lactoferrin concentration in bovine milk: Validation of radial immunodiffusion technique, sources of variation, and association to udder health status. Ital. J. Anim. Sci. 2023, 22, 230–238. [Google Scholar] [CrossRef]
- Niero, G.; Franzoi, M.; Vigolo, V.; Penasa, M.; Cassandro, M.; Boselli, C.; Giangolini, G.; De Marchi, M. Validation of a gold standard method for iodine quantification in raw and processed milk, and its variation in different dairy species. J. Dairy Sci. 2019, 102, 4808–4815. [Google Scholar] [CrossRef] [PubMed]
- Kharouba, M.; Mahmoud, S.H. Development and Validation of a Simple HPLC-UV Assay Method for Determination of Levetiracetam Concentrations in Human Plasma. Analytica 2023, 4, 1–9. [Google Scholar] [CrossRef]
- Vander Heyden, Y.; Massart, D. Review of the use of robustness and ruggedness in analytical chemistry. Data Handl. Sci. Technol. 1996, 19, 79–147. [Google Scholar]
- Xiang, D.; LoBrutto, R.; Cheney, J.; Wabuyele, B.W.; Berry, J.; Lyon, R.; Wu, H.; Khan, M.A.; Hussain, A.S. Evaluation of transmission and reflection modalities for measuring content uniformity of pharmaceutical tablets with near-infrared spectroscopy. Appl. Spectrosc. 2009, 63, 33–47. [Google Scholar] [CrossRef]
- Fearn, T.; Riccioli, C.; Garrido-Varo, A.; Guerrero-Ginel, J.E. On the geometry of SNV and MSC. Chemom. Intell. Lab. Syst. 2009, 96, 22–26. [Google Scholar] [CrossRef]
- Yun, Y.-H.; Liang, Y.-Z.; Xie, G.-X.; Li, H.-D.; Cao, D.-S.; Xu, Q.-S. A perspective demonstration on the importance of variable selection in inverse calibration for complex analytical systems. Analyst 2013, 138, 6412–6421. [Google Scholar] [CrossRef] [PubMed]
- Miao, X.; Miao, Y.; Liu, Y.; Tao, S.; Zheng, H.; Wang, J.; Wang, W.; Tang, Q. Measurement of nitrogen content in rice plant using near infrared spectroscopy combined with different PLS algorithms. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2023, 284, 121733. [Google Scholar] [CrossRef]
- Wang, N.; Feng, J.R.; Li, L.W.; Liu, J.M.; Sun, Y. Rapid Determination of Cellulose and Hemicellulose Contents in Corn Stover Using Near-Infrared Spectroscopy Combined with Wavelength Selection. Molecules 2022, 27, 3373. [Google Scholar] [CrossRef]
- Liu, J.M.; Jin, S.; Bao, C.H.; Sun, Y.; Li, W.Z. Rapid determination of lignocellulose in corn stover based on near-infrared reflectance spectroscopy and chemometrics methods. Bioresour. Technol. 2021, 321, 124449. [Google Scholar] [CrossRef] [PubMed]








| Time | The Parallel Measured Value of the Rice Bran Layer Cellulose Content, % | Mean, % | RSDr | RSDR | |||||
|---|---|---|---|---|---|---|---|---|---|
| Value 1 | Value 2 | Value 3 | Value 4 | Value 5 | Value 6 | ||||
| Day1 | 3.86 | 3.89 | 3.80 | 3.91 | 3.86 | 3.86 | 3.86 | 0.96% | 0.70% |
| Day2 | 3.84 | 3.86 | 3.82 | 3.83 | 3.87 | 3.86 | 3.84 | 0.51% | |
| Day3 | 3.88 | 3.84 | 3.81 | 3.82 | 3.84 | 3.84 | 3.84 | 0.63% | |
| Sample | Original PDO Method | Optimized PDO Method | ||||
|---|---|---|---|---|---|---|
| Mean (%) | SD (%) | RSD | Mean (%) | SD (%) | RSD | |
| A | 3.57 | 0.097 | 2.73% | 3.38 | 0.058 | 1.73% |
| B | 3.62 | 0.161 | 4.45% | 3.54 | 0.057 | 1.61% |
| C | 3.73 | 0.144 | 3.86% | 3.57 | 0.075 | 2.10% |
| D | 3.69 | 0.061 | 1.65% | 3.74 | 0.054 | 1.45% |
| E | 4.58 | 0.051 | 1.12% | 4.38 | 0.061 | 1.39% |
| F | 4.30 | 0.155 | 3.60% | 4.52 | 0.011 | 0.25% |
| G | 4.63 | 0.092 | 1.99% | 4.75 | 0.080 | 1.69% |
| H | 3.00 | 0.116 | 3.85% | 2.98 | 0.030 | 1.00% |
| I | 4.16 | 0.087 | 2.10% | 4.08 | 0.065 | 1.60% |
| J | 3.59 | 0.046 | 1.28% | 3.60 | 0.070 | 1.95% |
| K | 4.33 | 0.125 | 2.88% | 4.41 | 0.059 | 1.33% |
| L | 4.19 | 0.155 | 3.71% | 4.15 | 0.047 | 1.14% |
| M | 4.06 | 0.114 | 2.80% | 4.16 | 0.035 | 0.84% |
| N | 4.08 | 0.110 | 2.70% | 4.04 | 0.041 | 1.01% |
| O | 4.36 | 0.145 | 3.33% | 4.41 | 0.059 | 1.33% |
| Analyte | Calibration Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Range | Mean | SE | SD | N | Range | Mean | SE | SD | |
| Cellulose content (%) | 99 | 2.80–4.92 | 3.89 | 0.054 | 0.55 | 42 | 2.81–4.89 | 3.83 | 0.086 | 0.57 |
| Spectral Type | Pretreatment | Spectral Range (cm−1) | Cross-Validation | Validation Set | LVs | ||
|---|---|---|---|---|---|---|---|
| R2CV | RMSECV/% | R2v | RMSEV/% | ||||
| NIRdr | SNV | 7506–5446; 4605–4242 | 0.852 | 0.211 | 0.829 | 0.239 | 8 |
| NIRdt | 1st der + SNV | 7752.9–6094 | 0.879 | 0.191 | 0.850 | 0.224 | 8 |
| NIRdr-NIRdt | 1st der + SNV | 7583.1–6109.7; 8732–7259; 11,671.7–10,198.3 | 0.895 | 0.178 | 0.906 | 0.177 | 9 |
| NIRdr-NIRdt (NM) | 1st der + SNV | 7583.1–6109.7; 8732–7259 | 0.890 | 0.182 | 0.903 | 0.18 | 8 |
| NIRdr-NIRdt (1st der) | MSC | 7583.1–4636.2; 8732–5793 | 0.898 | 0.176 | 0.914 | 0.169 | 9 |
| Model Type | Sample Number | R2p | RMSEP/% |
|---|---|---|---|
| NIRdr | 20 | 0.748 | 0.344 |
| NIRdt | 20 | 0.815 | 0.257 |
| NIRdr-NIRdt | 20 | 0.838 | 0.278 |
| NIRdr-NIRdt (NM) | 20 | 0.855 | 0.229 |
| NIRdr-NIRdt (1st der) | 20 | 0.903 | 0.213 |
| Cellulose Content Level | Sample Name | Reference Cellulose Content (%) | Predicted Cellulose Content (%) |
|---|---|---|---|
| Low | O1 | 3.15 | 3.42 |
| O2 | 3.10 | 3.28 | |
| O5 | 3.19 | 3.30 | |
| O9 | 3.35 | 3.51 | |
| O17 | 3.22 | 3.48 | |
| Normal | O7 | 4.20 | 4.13 |
| O13 | 4.26 | 4.37 | |
| W7 | 4.26 | 4.14 | |
| High | O19 | 4.74 | 4.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Qin, C.; Xu, Z.; Wang, Q.; Yang, Y.; Ni, X.; Cheng, W.; Zhang, P.; Zhan, Y.; Tao, L.; et al. A Rapid and Accurate Quantitative Analysis of Cellulose in the Rice Bran Layer Based on Near-Infrared Spectroscopy. Foods 2023, 12, 2997. https://doi.org/10.3390/foods12162997
Fan S, Qin C, Xu Z, Wang Q, Yang Y, Ni X, Cheng W, Zhang P, Zhan Y, Tao L, et al. A Rapid and Accurate Quantitative Analysis of Cellulose in the Rice Bran Layer Based on Near-Infrared Spectroscopy. Foods. 2023; 12(16):2997. https://doi.org/10.3390/foods12162997
Chicago/Turabian StyleFan, Shuang, Chaoqi Qin, Zhuopin Xu, Qi Wang, Yang Yang, Xiaoyu Ni, Weimin Cheng, Pengfei Zhang, Yue Zhan, Liangzhi Tao, and et al. 2023. "A Rapid and Accurate Quantitative Analysis of Cellulose in the Rice Bran Layer Based on Near-Infrared Spectroscopy" Foods 12, no. 16: 2997. https://doi.org/10.3390/foods12162997
APA StyleFan, S., Qin, C., Xu, Z., Wang, Q., Yang, Y., Ni, X., Cheng, W., Zhang, P., Zhan, Y., Tao, L., & Wu, Y. (2023). A Rapid and Accurate Quantitative Analysis of Cellulose in the Rice Bran Layer Based on Near-Infrared Spectroscopy. Foods, 12(16), 2997. https://doi.org/10.3390/foods12162997







