Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological, Biochemical and Physiological Characterization of G. entanii
| Strain Designation | Source and Country of Isolation or Other Features | Reference |
|---|---|---|
| Gluconacetobacter entanii AV429 | Apple cider vinegar (Slovenia) | [15] |
| Gluconacetobacter entanii AV429-2020 | Strain AV429 precultured for 28 months | This study |
| Gluconacetobacter entanii AV429-2022 | Strain AV429 precultured for 43 months | This study |
| Gluconacetobacter entanii FXV2 | Fermented grape must (Portugal) | [4] |
| Gluconacetobacter entanii SI2084 | Apple cider vinegar (Slovenia) | [16] |
| Gluconacetobacter entanii KS542 | Apple cider vinegar (Slovenia) | In-house strain |
| Gluconacetobacter entanii KS544 | Apple cider vinegar (Slovenia) | In-house strain |
| Gluconacetobacter entanii KS545 | Apple cider vinegar (Slovenia) | In-house strain |
2.2. Genome Sequences, Assembly and Annotation
2.3. Phylogenomic Studies
2.4. Comparative Genomic Analysis
3. Results and Discussion
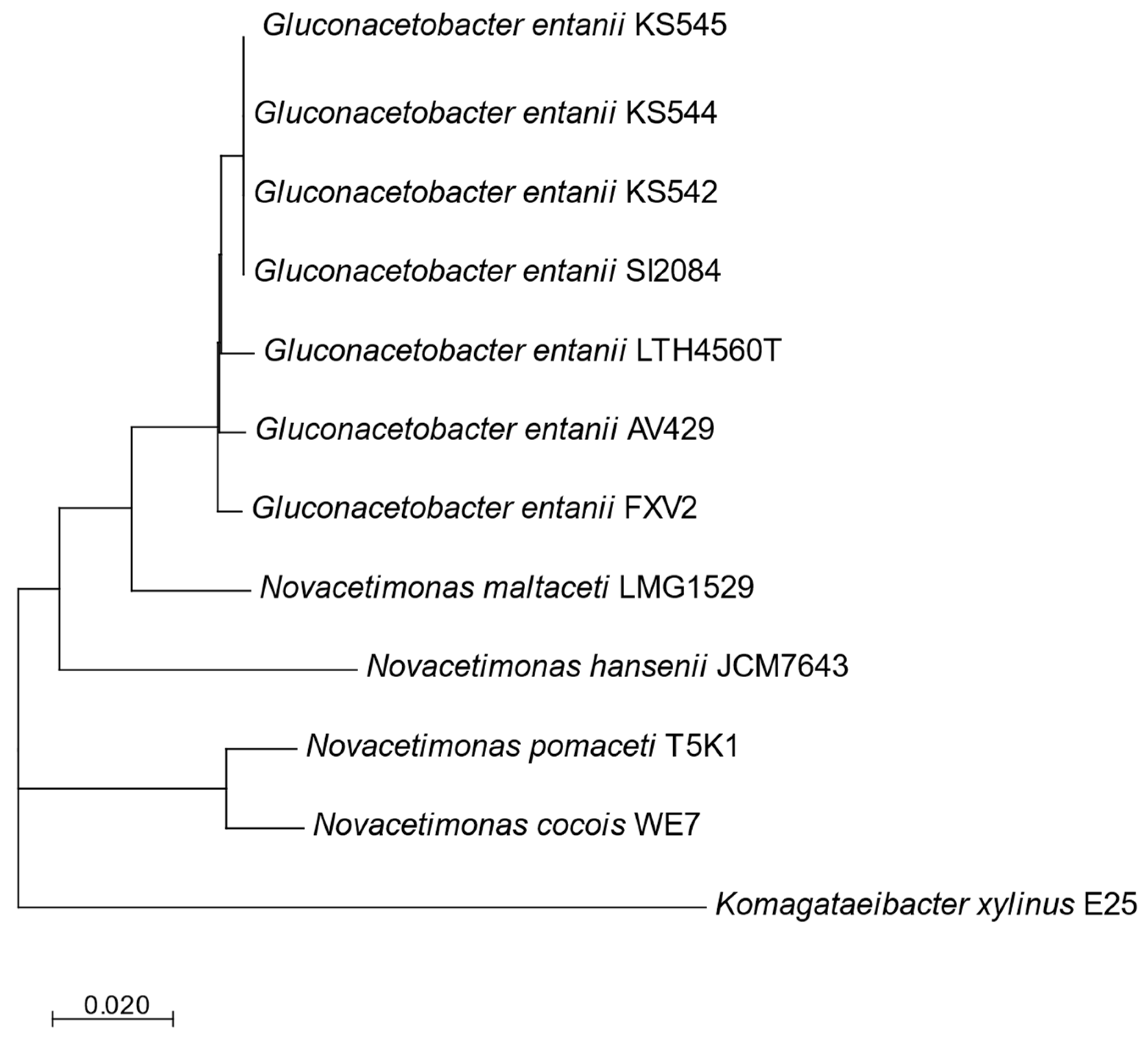
3.1. Phylogenomic Studies
3.2. Morphological, Biochemical and Physiological Characterization of G. entanii
3.3. Basic Genome Characteristics
3.4. Mobilome Analysis
3.5. A Comparative Genomic Functional Analysis
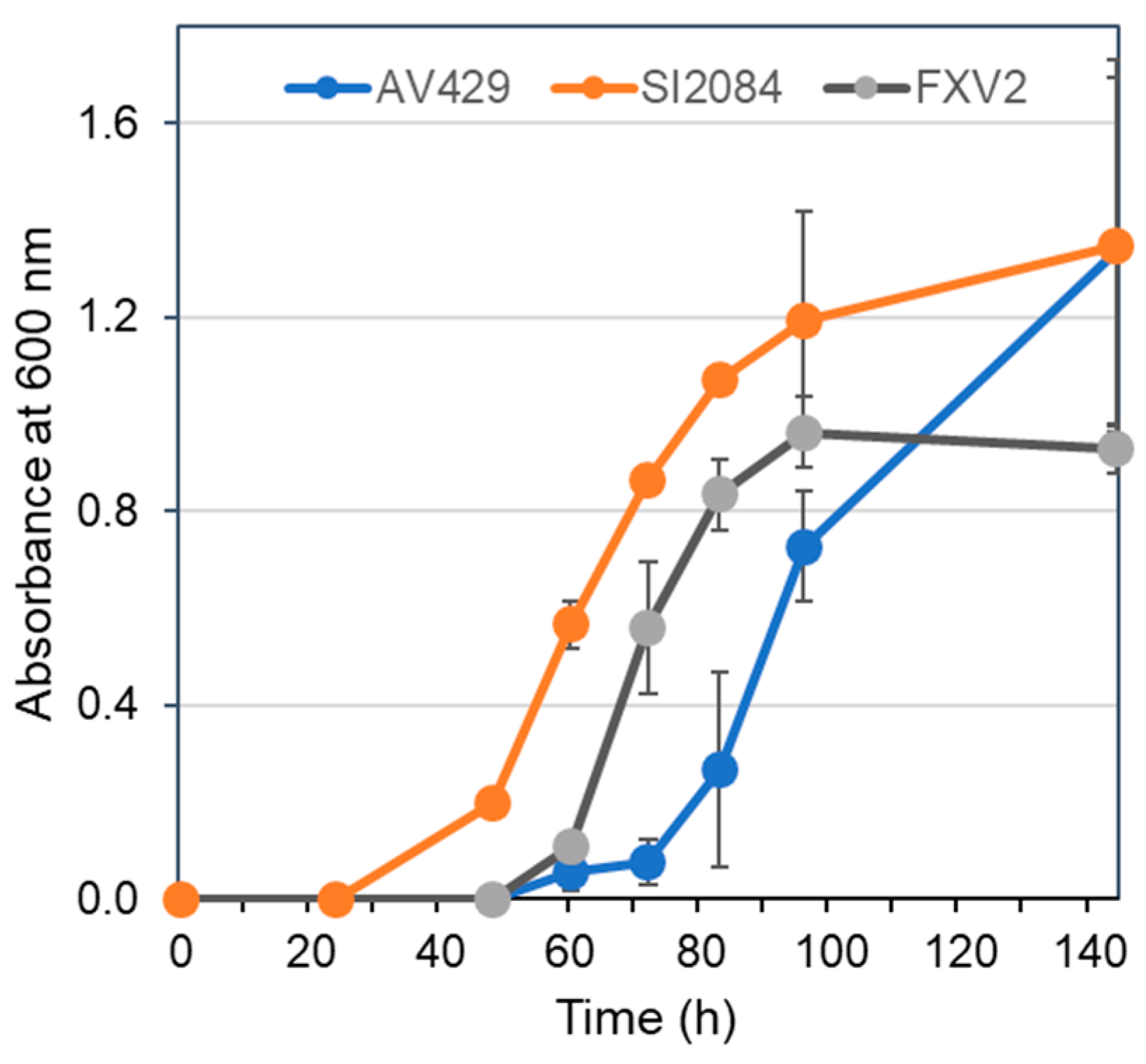
3.6. Genome Stability of G. entanii AV429
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schüller, G.; Hertel, C.; Hammes, W.P. Gluconacetobacter entanii sp. nov., isolated from submerged high-acid industrial vinegar fermentations. Int. J. Syst. Evol. Microbiol. 2000, 50, 2013–2020. [Google Scholar] [CrossRef]
- Sokollek, S.J.; Hammes, W.P. Description of a starter culture preparation for vinegar fermentation. Syst. Appl. Microbiol. 1997, 20, 481–491. [Google Scholar] [CrossRef]
- Trček, J.; Barja, F. Updates on quick identification of acetic acid bacteria with a focus on the 16S-23S rRNA gene internal transcribed spacer and the analysis of cell proteins by MALDI-TOF mass spectrometry. Int. J. Food Microbiol. 2015, 196, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Brandão, P.R.; Crespo, M.T.B.; Nascimento, F.X. Phylogenomic and comparative analyses support the reclassification of several Komagataeibacter species as novel members of the Novacetimonas gen. nov. and bring new insights into the evolution of cellulose synthase genes. Int. J. Syst. Evol. Microbiol. 2022, 72, 005252. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Nakagawa, Y. Subdivision of the genus Gluconacetobacter Yamada, Hoshino and Ishikawa 1998: The proposal of Komagatabacter gen. nov., for strains accommodated to the Gluconacetobacter xylinus group in the α-Proteobacteria. Ann. Microbiol. 2012, 62, 849–859. [Google Scholar] [CrossRef]
- Yamada, Y.; Yukphan, P.; Vu, H.T.L.; Muramatsu, Y.; Ochaikul, D.; Tanasupawat, S.; Nakagawa, Y. Description of Komagataeibacter gen. nov., with proposals of new combinations (Acetobacteraceae). J. Gen. Appl. Microbiol. 2012, 58, 397–404. [Google Scholar] [CrossRef]
- Škraban, J.; Cleenwerck, I.; Vandamme, P.; Fanedl, L.; Trček, J. Genome sequences and description of novel exopolysaccharides producing species Komagataeibacter pomaceti sp. nov. and reclassification of Komagataeibacter kombuchae (Dutta and Gachhui 2007) Yamada et al., 2013 as a later heterotypic synonym of Komagataeibacter hansenii. Syst. Appl. Microbiol. 2018, 41, 581–592. [Google Scholar] [CrossRef]
- Velasco-Bedrán, H.; López-Isunza, F. The unified metabolism of Gluconacetobacter entanii in continuous and batch processes. Process. Biochem. 2007, 42, 1180–1190. [Google Scholar] [CrossRef]
- Dórame-Miranda, R.F.; Gámez-Meza, N.; Medina-Juárez, L.; Ezquerra-Brauer, J.M.; Ovando-Martínez, M.; Lizardi-Mendoza, J. Bacterial cellulose production by Gluconacetobacter entanii using pecan nutshell as carbon source and its chemical functionalization. Carbohydr. Polym. 2019, 207, 91–99. [Google Scholar] [CrossRef]
- Bi, J.C.; Liu, S.X.; Li, C.F.; Li, J.; Liu, L.X.; Deng, J.; Yang, Y.C. Morphology and structure characterization of bacterial celluloses produced by different strains in agitated culture. J. Appl. Microbiol. 2014, 117, 1305–1311. [Google Scholar] [CrossRef]
- Gosselé, F.; Swings, J.; De Ley, J. A rapid, simple and simultaneous detection of 2-keto-, 5-keto-and 2,5-diketogluconic acids by thin-layer chromatography in culture media of acetic acid bacteria. Zentralblatt für Bakteriol. I. Abt. Orig. C Allg. Angew. Ökologische Mikrobiol. 1980, 1, 178–181. [Google Scholar] [CrossRef]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. Isolated from Apple Cider Vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef] [PubMed]
- Navarro, R.R.; Uchimura, T.; Komagata, K. Taxonomic heterogeneity of strains comprising Gluconacetobacter hansenii. J. Gen. Appl. Microbiol. 1999, 45, 295–300. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trcek, J.; Toyama, H.; Czuba, J.; Misiewicz, A.; Matsushita, K. Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 70, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Marič, L. Taksonomska opredelitev ocetnokislinskih bakterij Komagataeibacter sp. AV382, Komagataeibacter sp. AV436 in Komagataeibacter sp. AV429. Master’s Thesis, Faculty of Natural Sciences and Matematics, University of Maribor, Maribor, Slovenia, 2020; 82p. [Google Scholar]
- Vajdič, T. Genetic diversity of exopolysaccharides from acetic acid bacteria isolates originating from apple cider vinegars. Ger. J. Microbiol. 2022, 2, 1–18. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; Dejongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Škraban, J.; Trček, J. Comparative genomics of Acetobacter and other acetic acid bacteria. In Acetic Acid Bacteria: Fundamentals and Food Applications; CRC Press: Boca Raton, FL, USA; ISBN 9781498763707.
- Chen, S.L.; Hung, C.S.; Xu, J.; Reigstad, C.S.; Magrini, V.; Sabo, A.; Blasiar, D.; Bieri, T.; Meyer, R.R.; Ozersky, P.; et al. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc. Natl. Acad. Sci. USA 2006, 103, 5977–5982. [Google Scholar] [CrossRef]
- Mann, S.; Chen, Y.P.P. Bacterial genomic G + C composition-eliciting environmental adaptation. Genomics 2010, 95, 7–15. [Google Scholar] [CrossRef]
- Naya, H.; Romero, H.; Zavala, A.; Alvarez, B.; Musto, H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 2002, 55, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P.C.; Danchin, A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002, 18, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Coucheron, D.H. An Acetobacter xylinum insertion sequence element associated with inactivation of cellulose production. J. Bacteriol. 1991, 173, 5723–5731. [Google Scholar] [CrossRef] [PubMed]
- Dedrick, R.M.; Aull, H.G.; Jacobs-Sera, D.; Garlena, R.A.; Russell, D.A.; Smith, B.E.; Mahalingam, V.; Abad, L.; Gauthier, C.H.; Hatfull, G.F. The prophage and plasmid mobilome as a likely driver of Mycobacterium abscessus diversity. MBio 2021, 12, e03441-20. [Google Scholar] [CrossRef] [PubMed]
- Morgado, S.M.; Vicente, A.C.P. Comprehensive in silico survey of the Mycolicibacterium mobilome reveals an as yet underexplored diversity. Microb. Genom. 2021, 7, 000533. [Google Scholar] [CrossRef]
- Qian, C.; Ma, J.; Liang, J.; Zhang, L.; Liang, X. Comprehensive deciphering prophages in genus Acetobacter on the ecology, genomic features, toxin–antitoxin system, and linkage with CRISPR-Cas system. Front. Microbiol. 2022, 13, 951030. [Google Scholar] [CrossRef]
- Chiu, S.F.; Teng, K.W.; Wang, P.C.; Chung, H.Y.; Wang, C.J.; Cheng, H.C.; Kao, M.C. Helicobacter pylori GmhB enzyme involved in ADP-heptose biosynthesis pathway is essential for lipopolysaccharide biosynthesis and bacterial virulence. Virulence 2021, 12, 1610–1628. [Google Scholar] [CrossRef]
- Song, Q.; Wu, H.; Zhang, P.; Tian, K.; Zhu, H.; Qiao, J. LssR plays a positive regulatory role in acid and nisin tolerance response of Lactococcus lactis. J. Dairy Sci. 2022, 105, 6483–6498. [Google Scholar] [CrossRef]
- Shaikhutdinova, R.Z.; Ivanov, S.A.; Dentovskaya, S.V.; Titareva, G.M.; Knirel, Y.A. Characterization of a Transposon Tn5-Generated Mutant of Yersinia pestis Defective in Lipooligosaccharide Biosynthesis. Biochemistry 2019, 84, 398–406. [Google Scholar] [CrossRef]
- Karan, S.; Pratap, B.; Ashish; Saxena, A.K. Low-resolution SAXS and structural dynamics analysis on M. tuberculosis GmhB enzyme involved in GDP-heptose biosynthetic pathway. Int. J. Biol. Macromol. 2019, 136, 676–685. [Google Scholar] [CrossRef]
- Chiba, Y.; Oshima, K.; Arai, H.; Ishii, M.; Igarashi, Y. Discovery and analysis of cofactor-dependent phosphoglycerate mutase homologs as novel phosphoserine phosphatases in hydrogenobacter thermophilus. J. Biol. Chem. 2012, 287, 11934–11941. [Google Scholar] [CrossRef] [PubMed]
- Kawalek, A.; Wawrzyniak, P.; Bartosik, A.A.; Jagura-Burdzy, G. Rules and exceptions: The role of chromosomal ParB in DNA segregation and other cellular processes. Microorganisms 2020, 8, 105. [Google Scholar] [CrossRef] [PubMed]
- Jalal, A.S.B.; Le, T.B.K. Bacterial chromosome segregation by the ParABS system. Open Biol. 2020, 10, 200097. [Google Scholar] [CrossRef] [PubMed]
- Hołówka, J.; Trojanowski, D.; Gind, K.; Wojtaś, B.; Gielniewski, B.; Jakimowicz, D.; Zakrzewska-Czerwińska, J. HupB is a bacterial nucleoid-associated protein with an indispensable eukaryotic-like tail. MBio 2017, 8, e01272-17. [Google Scholar] [CrossRef]
- Hong, Y.G.; Moon, Y.M.; Choi, T.R.; Jung, H.R.; Yang, S.Y.; Ahn, J.O.; Joo, J.C.; Park, K.; Kim, Y.G.; Bhatia, S.K.; et al. Enhanced production of glutaric acid by NADH oxidase and GabD-reinforced bioconversion from l-lysine. Biotechnol. Bioeng. 2019, 116, 333–341. [Google Scholar] [CrossRef]
- Beppu, T. Genetic organization of Acetobacter for acetic acid fermentation. Antonie Van Leeuwenhoek 1993, 64, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Azuma, Y.; Hosoyama, A.; Matsutani, M.; Furuya, N.; Horikawa, H.; Harada, T.; Hirakawa, H.; Kuhara, S.; Matsushita, K.; Fujita, N.; et al. Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res. 2009, 37, 5768–5783. [Google Scholar] [CrossRef]
- Vandamme, P.; Sutcliffe, I. Out with the old and in with the new: Time to rethink twentieth century chemotaxonomic practices in bacterial taxonomy. Int. J. Syst. Evol. Microbiol. 2021, 71, 5127. [Google Scholar] [CrossRef]


| Species | Accession Number | Number of Bases (Mbp) | G + C% | Genes | Proteins | rRNA | tRNA | Pseudogenes (% of Total Genes) |
|---|---|---|---|---|---|---|---|---|
| Gluconacetobacter entanii LTH4560T | NKUF00000000 | 3.60 | 62.6 | 3444 | 3243 | 5 | 50 | 142 (4.1) |
| Gluconacetobacter entanii AV429 | JABJWD000000000 | 3.75 | 62.6 | 3523 | 3361 | 3 | 46 | 109 (3.1) |
| Gluconacetobacter entanii SI2084 | JAILXQ000000000 | 3.64 | 62.6 | 3560 | 3339 | 3 | 48 | 166 (4.7) |
| Gluconacetobacter entanii FXV2 | WNJT00000000 | 3.61 | 62.5 | 3135 | 2990 | 5 | 48 | 88 (2.9) |
| Carbon Source | LTH 4560T | LTH 4637 | AV429 | SI2084 | FXV2 |
|---|---|---|---|---|---|
| AE (4a/3e) broth | + | + | + | + | + |
| AE (4aa/3e) broth without glucose | − | − | + | W | + |
| AE (4aa/3e) broth; glucose replaced with: | |||||
| Maltose | + | + | W | W | W |
| Sucrose | + | + | + | + | + |
| Sorbitol | W | + | + | + | + |
| Mannitol | − | W | + | + | + |
| Lactate | − | − | + | + | W |
| Gluconate | − | − | + | W | − |
| Fructose | + | + | W | + | − |
| Glycerol | − | − | + | W | + |
| AE (3e) broth | − | − | W | − | W |
| AE (3e) broth; acetic acid replaced with: | |||||
| Lactate | − | − | + | − | − |
| Gluconate | − | − | + | − | − |
| AE (4aa) broth | − | − | + | W | − |
| AE (4aa) broth with 1-propanol | + | + | − | − | − |
| Phenotypic Characteristics | Strain | |||
|---|---|---|---|---|
| LTH 4560T | AV429 | SI2084 | FXV2 | |
| Formation from D-glucose | ||||
| 2-Keto-D-gluconic acid | − | + | + | + |
| 5-Keto-D-gluconic acid | − | + | + | + |
| Growth on carbon sources: | ||||
| D-Ribose | n.k. | W | W | W |
| Sorbitol | n.k. | + | + | + |
| D-Mannitol | n.k. | + | + | + |
| Glycerol | n.k. | + | W | + |
| 1-Propanol | n.k. | W | W | W |
| Growth in the presence of 30% D-Glucose | − | + | − | + |
| Utilization of ammoniacal nitrogen in: | ||||
| Hoyer–Frateur medium with | ||||
| D-Glucose | n.k. | + | + | + |
| D-Mannitol | n.k. | − | W | − |
| Ethanol | n.k. | + | + | W |
| Asai medium with | ||||
| D-Glucose | n.k. | + | − | − |
| D-Mannitol | n.k. | + | − | − |
| Ethanol | n.k. | + | + | W |
| Growth without acetic acid | − | + | + | W |
| Growth on RAE medium in the presence of 1% ethanol and acetic acid at: | ||||
| 4% | n.k. | + | + | + |
| 5% | n.k. | W | W | + |
| 6% | n.k. | − | W | + |
| 7% | n.k. | − | W | − |
| Growth on RAE medium in the presence of 3% ethanol and acetic acid at: | ||||
| 4% | n.k. | + | + | + |
| 5% | n.k. | − | + | + |
| 6% | n.k. | − | + | − |
| 7% | n.k. | − | + | − |
| IS-Element (Length) | G. entanii LTH4560 | G. entanii AV429 | G. entanii SI2084 | G. entanii FXV2 |
|---|---|---|---|---|
| ISGxy1 (1313 bp) | 4 | 2 | - | 1 |
| IS1452 (1411 bp) | 2 | 1 | 1 | - |
| IS1031A (930 bp) | 4 | - | - | - |
| IS1031C or D (930 bp) | 3 | - | - | - |
| ISGdi13 (1452 bp) | 2 | - | 1 | - |
| Tn5393 (5470 bp) | 1 | - | 1 | - |
| IS1032 (916 bp) | - | 1 | - | 1 |
| ISGdi8 (1356 bp) | - | - | 1 | - |
| ISPpa1 (1376 bp) | - | - | 1 | - |
| ISGdi11 (1200 bp) | - | - | - | 1 |
| Strain Designation | Intact | Incomplete | Questionable |
|---|---|---|---|
| Gluconacetobacter entanii AV429 | 0 | 2 | 2 |
| Gluconacetobacter entanii AV429-28 | 1 | 2 | 0 |
| Gluconacetobacter entanii AV429-43 | 1 | 5 | 0 |
| Gluconacetobacter entanii FXV2 | 0 | 2 | 0 |
| Gluconacetobacter entanii SI2084 | 2 | 0 | 0 |
| Gluconacetobacter entanii KS542 | 2 | 0 | 0 |
| Gluconacetobacter entanii KS544 | 1 | 0 | 1 |
| Gluconacetobacter entanii KS545 | 1 | 0 | 1 |
| Bacterial Host Strain | Node Length (kbp)/Node no. | Total Proteins of the Node/Phage Specific Proteins | Two Most Common Phage Species (acc. no.) | NCBI BlastN Similarity Results (acc.no.): Query Cover (%)/nt-Identity (%) |
|---|---|---|---|---|
| Gluconacetobacter entanii AV429-28 | 27.3/(node 3) | 32/13 | Escherichia coli phage Stx2a_F451 (NC_049924) | Komagataeibacter phage phiKM1 (LC644974.1): 43/95.7 |
| Escherichia coli phage Stx2_1717 (NC_011357) | - | |||
| Gluconacetobacter entanii AV429-43 | 26.0/(node 4) | 30/13 | Erwinia phage ENT90 (NC_019932) | Komagataeibacter phage phiKM1 (LC644974.1): 45/95.7 |
| Burkholderia phage phi644-2 (NC_009235) | - | |||
| Gluconacetobacter entanii SI2084 | 22.9/(node 145) | 29/21 | Shigella phage SfII (NC_021857) | Komagataeibacter phage phiKM1 (LC644974.1): 94/93.7 |
| Enterobacteria phage SfI (NC_027339) | - | |||
| 39.9/(node 166) | 57/32 | Vibrio phage martha 12B12 (NC_021070) | - | |
| Escherichia phage D108 (NC_013594) | - | |||
| Gluconacetobacter entanii KS542 | 42.0/(node 7) | 63/35 | Vibrio phage martha 12B12 (NC_021070) | - |
| Escherichia phage D108 (NC_013594) | - | |||
| 24.0/(node 9) | 30/21 | Enterobacteria phage SfI (NC_027339) | Komagataeibacter phage phiKM1 (LC644974.1): 95/95.5 | |
| Shigella phage SfII (NC_021857) | - | |||
| Gluconacetobacter entanii KS544 | 39.4/(node 30) | 56/31 | Vibrio phage martha 12B12 (NC_021070) | - |
| Escherichia phage D108 (NC_013594) | - | |||
| Gluconacetobacter entanii KS545 | 42.1/(node 7) | 63/35 | Vibrio phage martha 12B12 (NC_021070) | - |
| Escherichia phage D10 (NC_013594) | - |
| KO | Definition |
|---|---|
| K03273 | gmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] |
| K09732 | K09732; uncharacterized protein |
| K22305 | psp; phosphoserine phosphatase [EC:3.1.3.3] |
| KO | Definition |
|---|---|
| K03273 | GmhB; D-glycero-D-manno-heptose 1,7-bisphosphate phosphatase [EC:3.1.3.82 3.1.3.83] |
| K07733 | AlpA; prophage regulatory protein |
| K09732 | K09732; uncharacterized protein |
| K14414 | RtcR; transcriptional regulatory protein RtcR |
| K22305 | Psp; phosphoserine phosphatase [EC:3.1.3.3] |
| K23123 | PxpB; 5-oxoprolinase (ATP-hydrolysing) subunit B [EC:3.5.2.9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jelenko, K.; Cepec, E.; Nascimento, F.X.; Trček, J. Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars. Foods 2023, 12, 214. https://doi.org/10.3390/foods12010214
Jelenko K, Cepec E, Nascimento FX, Trček J. Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars. Foods. 2023; 12(1):214. https://doi.org/10.3390/foods12010214
Chicago/Turabian StyleJelenko, Karin, Eva Cepec, Francisco X. Nascimento, and Janja Trček. 2023. "Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars" Foods 12, no. 1: 214. https://doi.org/10.3390/foods12010214
APA StyleJelenko, K., Cepec, E., Nascimento, F. X., & Trček, J. (2023). Comparative Genomics and Phenotypic Characterization of Gluconacetobacter entanii, a Highly Acetic Acid-Tolerant Bacterium from Vinegars. Foods, 12(1), 214. https://doi.org/10.3390/foods12010214








