CsCuAOs and CsAMADH1 Are Required for Putrescine-Derived γ-Aminobutyric Acid Accumulation in Tea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Determination of GABA and Glutamate Contents
2.3. Determination of the Putrescine Content
2.4. Phylogenetic Tree Construction of Tea Plant CuAO and AMADH Gene Family
2.5. Gene Expression Analysis
2.6. Purification of CsCuAO1, CsCuAO3 and CsAMADH1 In Vitro
2.7. CsCuAOs and CsAMADHs Activities Assay
2.8. Subcellular Localization Analysis of CsCuAO1, CsCuAO3 and CsAMADH1
2.9. Statistical Analysis
3. Results
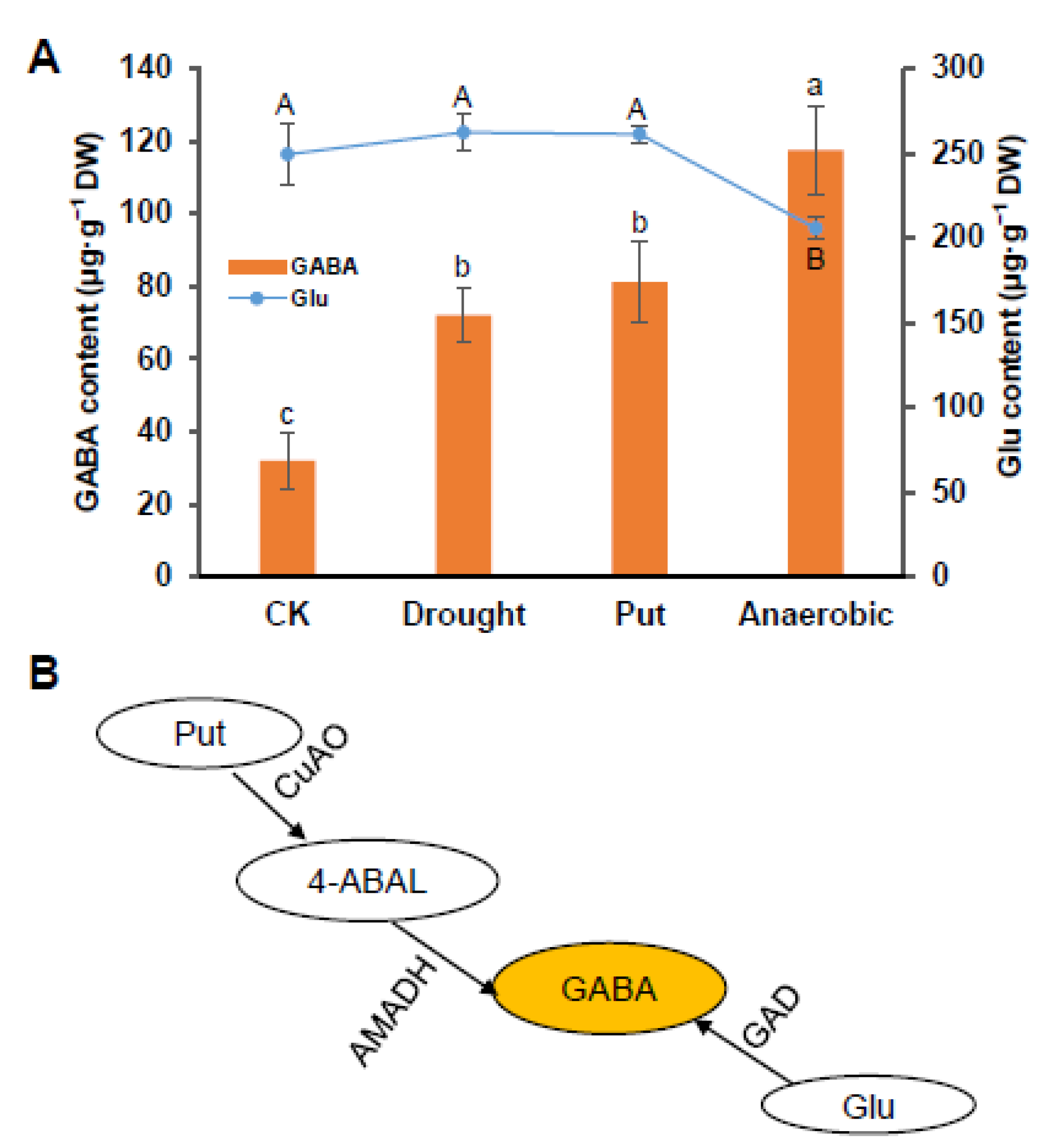
3.1. Changes in GABA, Glutamate, Putrescine Contents under Different Treatments
3.2. Phylogenetic Analysis of Tea Plant CuAO and AMADH Gene Family and Gene Expression
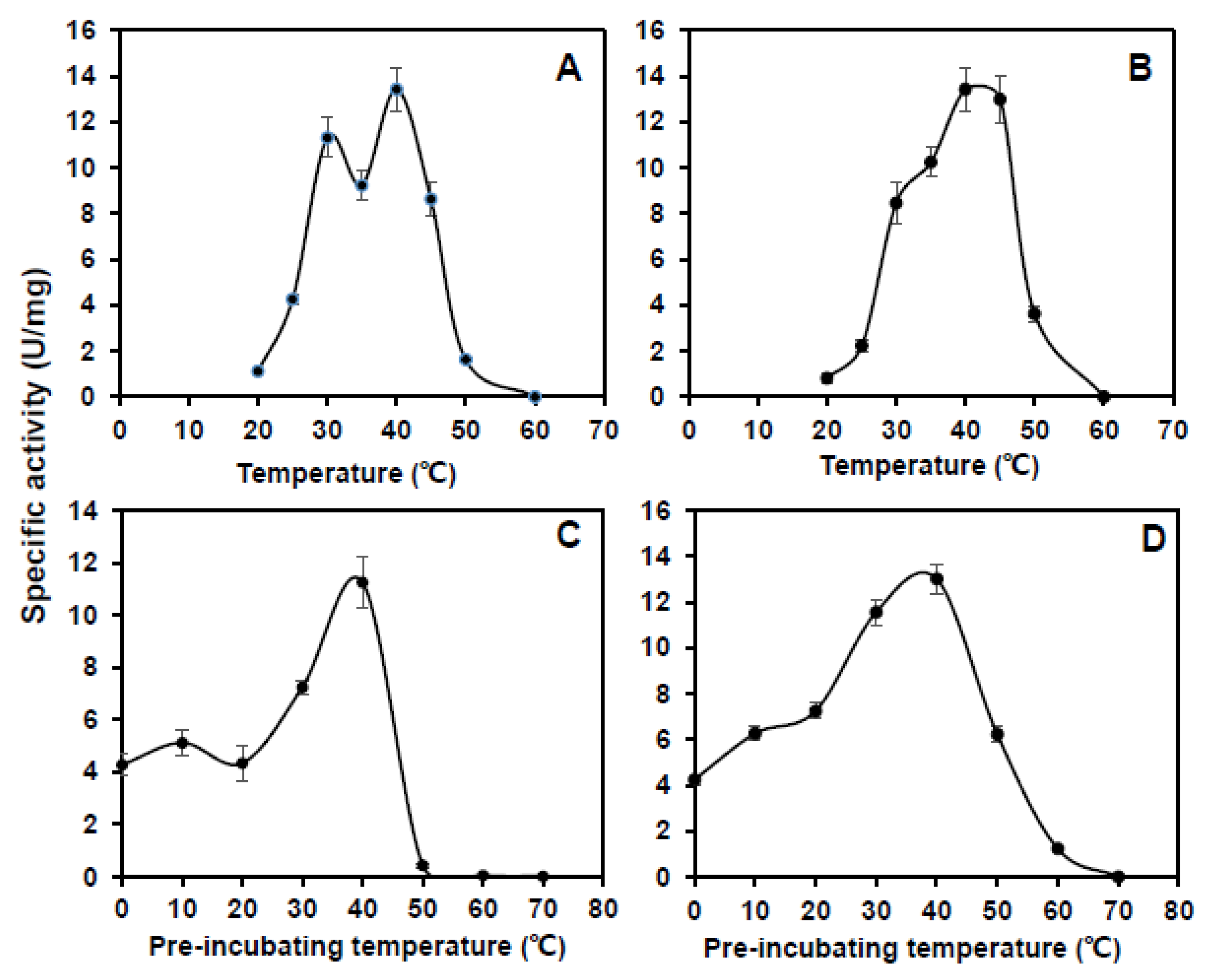
3.3. Purification and Enzyme Kinetics of CsCuAO1, CsCuAO3 and CsAMADH1
3.4. Assays of CsCuAO1, CsCuAO3 and CsAMADH1 Enzyme Activity on GABA Production In Vitro
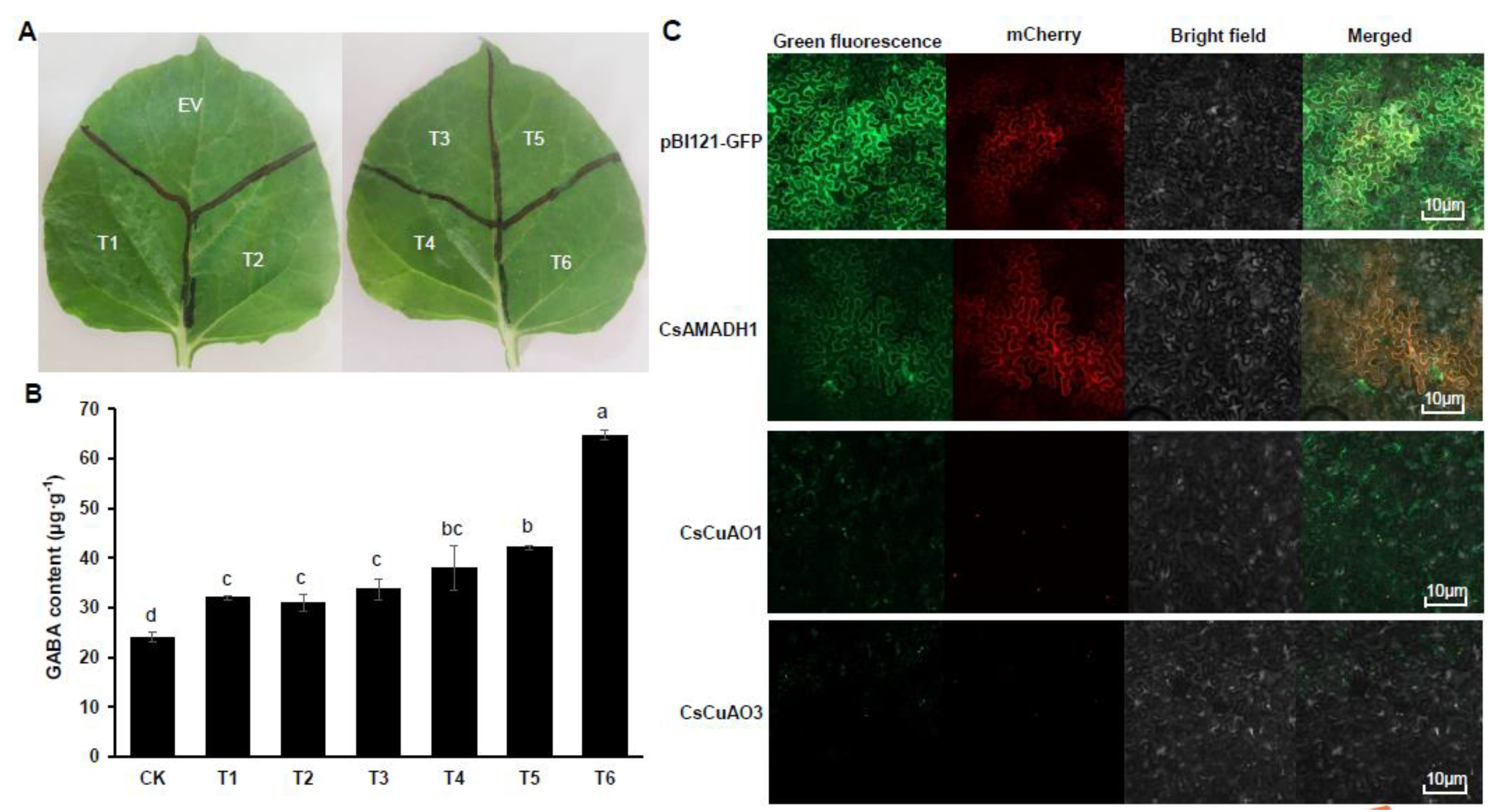
3.5. Transient Transformation Expression in Nicotiana Benthamiana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alharbi, B.; Hunt, J.D.; Dimitrova, S.; Spadafora, N.D.; Cort, A.P.; Colombo, D.; Müller, C.T.; Ghuge, S.A.; Davoli, D.; Cona, A.; et al. Mutation of Arabidopsis Copper-Containing Amine Oxidase Gene AtCuAOδ Alters Polyamines, Reduces Gibberellin Content and Affects Development. Int. J. Mol. Sci. 2020, 21, 7789. [Google Scholar] [CrossRef] [PubMed]
- Bouché, N.; Fromm, H. GABA in plants: Just a metabolite? Trends Plant Sci. 2004, 9, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Pan, H. Role of gamma-Aminobutyric Acid (GABA)A and GABAB receptors in paraventricular nucleus in control of sympathetic vasomotor tone in hypertension. J. Pharmacol. Exp. Ther. 2007, 320, 615–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benson, K.L.; Bottary, R.; Schoerning, L.; Baer, L.; Gonenc, A.; Jensen, J.E.; Winkelman, J.W. 1H MRS Measurement of Cortical GABA and Glutamate in Primary Insomnia and Major Depressive Disorder: Relationship to Sleep Quality and Depression Severity. J. Affect. Disord. 2020, 274, 624–631. [Google Scholar] [CrossRef]
- Ting Wong, C.G.; Bottiglieri, T.; Snead, O.C., III. GABA, γ-hydroxybutyric acid, and neurological disease. Ann. Neurol. 2003, 54, S3–S12. [Google Scholar] [CrossRef]
- Chi, Z.; Dai, Y.; Cao, S.; Wei, Y.; Shao, X.; Huang, X.; Xu, F.; Wang, H. Exogenous calcium chloride (CaCl2) promotes γ-aminobutyric acid (GABA) accumulation in fresh-cut pears. Postharvest Biol. Technol. 2020, 174, 111446. [Google Scholar] [CrossRef]
- Lee, J.; Rudell, D.R.; Davies, P.J.; Watkins, C.B. Metabolic changes in 1-methylcyclopropene (1-MCP)-treated ‘Empire’ apple fruit during storage. Metabolomics 2012, 8, 742–753. [Google Scholar] [CrossRef]
- Moschou, P.; Wu, J.; Cona, A.; Tavladoraki, P.; Angelini, R.; Roubelakis-Angelakis, K.A. The polyamines and their catabolic products are significant players in the turnover of nitrogenous molecules in plants. J. Exp. Bot. 2012, 63, 5003–5015. [Google Scholar] [CrossRef] [Green Version]
- Federico, R.; Angelini, R. Polyamine catabolism in plants. In Biochemistry and Physiology of Polyamines in Plants; Slocum, R.D., Flores, H.E., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 41–56. [Google Scholar]
- Fincato, P.; Moschou, P.; Spedaletti, V.; Tavazza, R.; Angelini, R.; Federico, R.; Roubelakis-Angelakis, K.A.; Tavladoraki, P. Functional diversity inside the Arabidopsis polyamine oxidase gene family. J. Exp. Bot. 2010, 62, 1155–1168. [Google Scholar] [CrossRef]
- Podlešáková, K.; Ugena, L.; Spíchal, L.; Doležal, K.; De Diego, N. Phytohormones and polyamines regulate plant stress responses by altering GABA pathway. New Biotechnol. 2018, 48, 53–65. [Google Scholar] [CrossRef]
- Renault, H.; Roussel, V.; El Amrani, A.; Arzel, M.; Renault, D.; Bouchereau, A.; Deleu, C. The Arabidopsis pop2-1mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010, 10, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelp, B.J.; Bozzo, G.G.; Trobacher, C.P.; Zarei, A.; Deyman, K.L.; Brikis, C.J. Hypothesis/review: Contribution of putrescine to 4-aminobutyrate (GABA) production in response to abiotic stress. Plant Sci. 2012, 193–194, 130–135. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.G.; Jun, Y.B.; Hau, Z.W.; Liang, L.Y. Higher accumulation of gamma-aminobutyric acid induced by salt stress through stimulating the activity of diamine oxidases in Glycine max (L.) Merr. roots. Plant Physiol. Biochem. 2007, 45, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Zarei, A.; Trobacher, C.P.; Cooke, A.R.; Meyers, A.J.; Hall, J.C.; Shelp, B.J. Apple Fruit Copper Amine Oxidase Isoforms: Peroxisomal MdAO1 Prefers Diamines as Substrates, Whereas Extracellular MdAO2 Exclusively Utilizes Monoamines. Plant Cell Physiol. 2014, 56, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Planas-Portell, J.; Gallart, M.; Tiburcio, A.F.; Altabella, T. Copper-containing amine oxidases contribute to terminal polyamine oxidation in peroxisomes and apoplast of Arabidopsis thaliana. BMC Plant Biol. 2013, 13, 109. [Google Scholar] [CrossRef] [Green Version]
- Cona, A.; Rea, G.; Angelini, R.; Federico, R.; Tavladoraki, P. Functions of amine oxidases in plant development and defence. Trends Plant Sci. 2006, 11, 80–88. [Google Scholar] [CrossRef]
- Flores, H.E.; Filner, P. Polyamine catabolism in higher plants: Characterization of pyrroline dehydrogenase. Plant Growth Regul. 1985, 3, 277–291. [Google Scholar] [CrossRef]
- Shelp, B.J.; Mullen, R.T.; Waller, J. Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 2012, 17, 57–59. [Google Scholar] [CrossRef]
- Díaz-Sanchez, Á.; González-Segura, L.; Mújica-Jiménez, C.; Rudino-Pinera, E.; Montiel, C.; Martínez-Castilla, L.P.; Muñoz-Clares, R.A. Amino Acid Residues Critical for the Specificity for Betaine Aldehyde of the Plant ALDH10 Isoenzyme Involved in the Synthesis of Glycine Betaine. Plant Physiol. 2012, 158, 1570–1582. [Google Scholar] [CrossRef] [Green Version]
- Kopečný, D.; Končitíková, R.; Tylichová, M.; Vigouroux, A.; Moskalíková, H.; Soural, M. Plant ALDH10 family: Identifying critical residues for substrate specificity and trapping a thiohemiacetal intermediate. J. Biol. Chem. 2013, 288, 9491–9507. [Google Scholar] [CrossRef] [Green Version]
- Riveros-Rosasa, H.; González-Segura, L.; Julián-Sánchez, A.; Díaz-Sánchez, Á.G.; Muñoz-Clares, R.A. Structural determinants of substrate specificity in aldehyde dehydrogenases. Chem.-Biol. Interact. 2013, 202, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Heim, W.G.; Sykes, K.A.; Hildreth, S.B.; Sun, J.; Lu, R.-H.; Jelesko, J.G. Cloning and characterization of a Nicotiana tabacum methylputrescine oxidase transcript. Phytochem. 2007, 68, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Katoh, A.; Shoji, T.; Hashimoto, T. Molecular Cloning of N-methylputrescine Oxidase from Tobacco. Plant Cell Physiol. 2007, 48, 550–554. [Google Scholar] [CrossRef] [PubMed]
- Weigel, P.; Weretilnyk, E.A.; Hanson, A.D. Betaine Aldehyde Oxidation by Spinach Chloroplasts. Plant Physiol. 1986, 82, 753–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradbury, L.M.T.; Gillies, S.A.; Brushett, D.J.; Waters, D.L.E.; Henry, R.J. Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 2008, 68, 439–449. [Google Scholar] [CrossRef] [Green Version]
- Missihoun, T.D.; Schmitz, J.; Klug, R.; Kirch, H.-H.; Bartels, D. Betaine aldehyde dehydrogenase genes from Arabidopsis with different sub-cellular localization affect stress responses. Planta 2010, 233, 369–382. [Google Scholar] [CrossRef]
- Muñoz-Clares, R.A.; Riveros-Rosas, H.; Garza-Ramos, G.; González-Segura, L.; Mújica-Jiménez, C.; Julián-Sánchez, A. Exploring the evolutionary route of the acquisition of betaine aldehyde dehydrogenase activity by plant ALDH10 enzymes: Implications for the synthesis of the osmoprotectant glycine betaine. BMC Plant Biol. 2014, 14, 149. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Li, Q.; Hu, J.; Wang, M.; Li, X. Molecular Cloning and Characterization of Spermine Synthesis Gene Associated with Cold Tolerance in Tea Plant (Camellia sinensis). Appl. Biochem. Biotechnol. 2015, 177, 1055–1068. [Google Scholar] [CrossRef]
- Tipping, A.J.; McPherson, M. Cloning and Molecular Analysis of the Pea Seedling Copper Amine Oxidase. J. Biol. Chem. 1995, 270, 16939–16946. [Google Scholar] [CrossRef] [Green Version]
- Petřivalský, M.; Brauner, F.; Luhová, L.; Gagneul, D.; Šebela, M. Aminoaldehyde dehydrogenase activity during wound healing of mechanically injured pea seedlings. J. Plant Physiol. 2007, 164, 1410–1418. [Google Scholar] [CrossRef]
- Li, J.; Duan, Y.; Han, Z.; Shang, X.; Zhang, K.; Zou, Z.; Ma, Y.; Li, F.; Fang, W.; Zhu, X. Genome-Wide Identification and Expression Analysis of the NRAMP Family Genes in Tea Plant (Camellia sinensis). Plants 2021, 10, 1055. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Xu, X.; Yang, Z. Characterization of two tea glutamate decarboxylase isoforms involved in GABA production. Food Chem. 2019, 305, 125440. [Google Scholar] [CrossRef] [PubMed]
- Reggiani, R.; Cantu, C.A.; Brambilla, I.; Bertani, A. Accumulation and Interconversion of Amino Acids in Rice Roots under Anoxia. Plant Cell Physiol. 1988, 29, 981–987. [Google Scholar] [CrossRef]
- Liao, J.; Wu, X.; Xing, Z.; Li, Q.; Duan, Y.; Fang, W.; Zhu, X. γ-Aminobutyric Acid (GABA) Accumulation in Tea (Camellia sinensis L.) through the GABA Shunt and Polyamine Degradation Pathways under Anoxia. J. Agric. Food Chem. 2017, 65, 3013–3018. [Google Scholar] [CrossRef]
- Mei, X.; Chen, Y.; Zhang, L.; Fu, X.; Wei, Q.; Grierson, D.; Zhou, Y.; Huang, Y.; Don, G.; Yang, Z. Dual mechanisms regulating glutamate decarboxylases and accumulation of gamma-aminobutyric acid in tea (Camellia sinensis) leaves exposed to multiple stresses. Sci. Rep. 2016, 6, 23685. [Google Scholar] [CrossRef] [Green Version]
- Angelini, R.; Cona, A.; Federico, R.; Fincato, P.; Tavladoraki, P.; Tisi, A. Plant amine oxidases “on the move”: An update. Plant Physiol. Biochem. 2010, 48, 560–564. [Google Scholar] [CrossRef]
- Reumann, S.; Quan, S.; Aung, K.; Yang, P.; Manandhar-Shrestha, K.; Holbrook, D.; Linka, N.; Switzenberg, R.; Wilkerson, C.G.; Weber, A.P.; et al. In-Depth Proteome Analysis of Arabidopsis Leaf Peroxisomes Combined with in Vivo Subcellular Targeting Verification Indicates Novel Metabolic and Regulatory Functions of Peroxisomes. Plant Physiol. 2009, 150, 125–143. [Google Scholar] [CrossRef] [Green Version]
- Moller, S.; McPherson, M. Developmental expression and biochemical analysis of theArabidopsis atao1gene encoding an H2O2-generating diamine oxidase. Plant J. 1998, 13, 781–791. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, K.; Duan, Y.; Cao, Y.; Chen, Y.; Zou, Z.; Li, F.; Shen, Q.; Yang, X.; Ma, Y.; Fang, W.; et al. CsCuAOs and CsAMADH1 Are Required for Putrescine-Derived γ-Aminobutyric Acid Accumulation in Tea. Foods 2022, 11, 1356. https://doi.org/10.3390/foods11091356
Zhang K, Duan Y, Cao Y, Chen Y, Zou Z, Li F, Shen Q, Yang X, Ma Y, Fang W, et al. CsCuAOs and CsAMADH1 Are Required for Putrescine-Derived γ-Aminobutyric Acid Accumulation in Tea. Foods. 2022; 11(9):1356. https://doi.org/10.3390/foods11091356
Chicago/Turabian StyleZhang, Kexin, Yu Duan, Yu Cao, Yiwen Chen, Zhongwei Zou, Fang Li, Qiang Shen, Xiaowei Yang, Yuanchun Ma, Wanping Fang, and et al. 2022. "CsCuAOs and CsAMADH1 Are Required for Putrescine-Derived γ-Aminobutyric Acid Accumulation in Tea" Foods 11, no. 9: 1356. https://doi.org/10.3390/foods11091356
APA StyleZhang, K., Duan, Y., Cao, Y., Chen, Y., Zou, Z., Li, F., Shen, Q., Yang, X., Ma, Y., Fang, W., & Zhu, X. (2022). CsCuAOs and CsAMADH1 Are Required for Putrescine-Derived γ-Aminobutyric Acid Accumulation in Tea. Foods, 11(9), 1356. https://doi.org/10.3390/foods11091356





