Investigating Differential Expressed Genes of Limosilactobacillus reuteri LR08 Regulated by Soybean Protein and Peptides
Abstract
1. Introduction
2. Materials and Methods
2.1. The Preparation and Properties of Soybean Protein and Peptide Hydrolysates
2.2. Activation and Growth Condition of Strains
2.3. Library Construction for Transcriptome Sequencing
2.4. Reverse Transcription of RNA
2.5. Fluorescence Quantitative PCR Reaction
2.6. Statistical Analysis
3. Results and Discussion
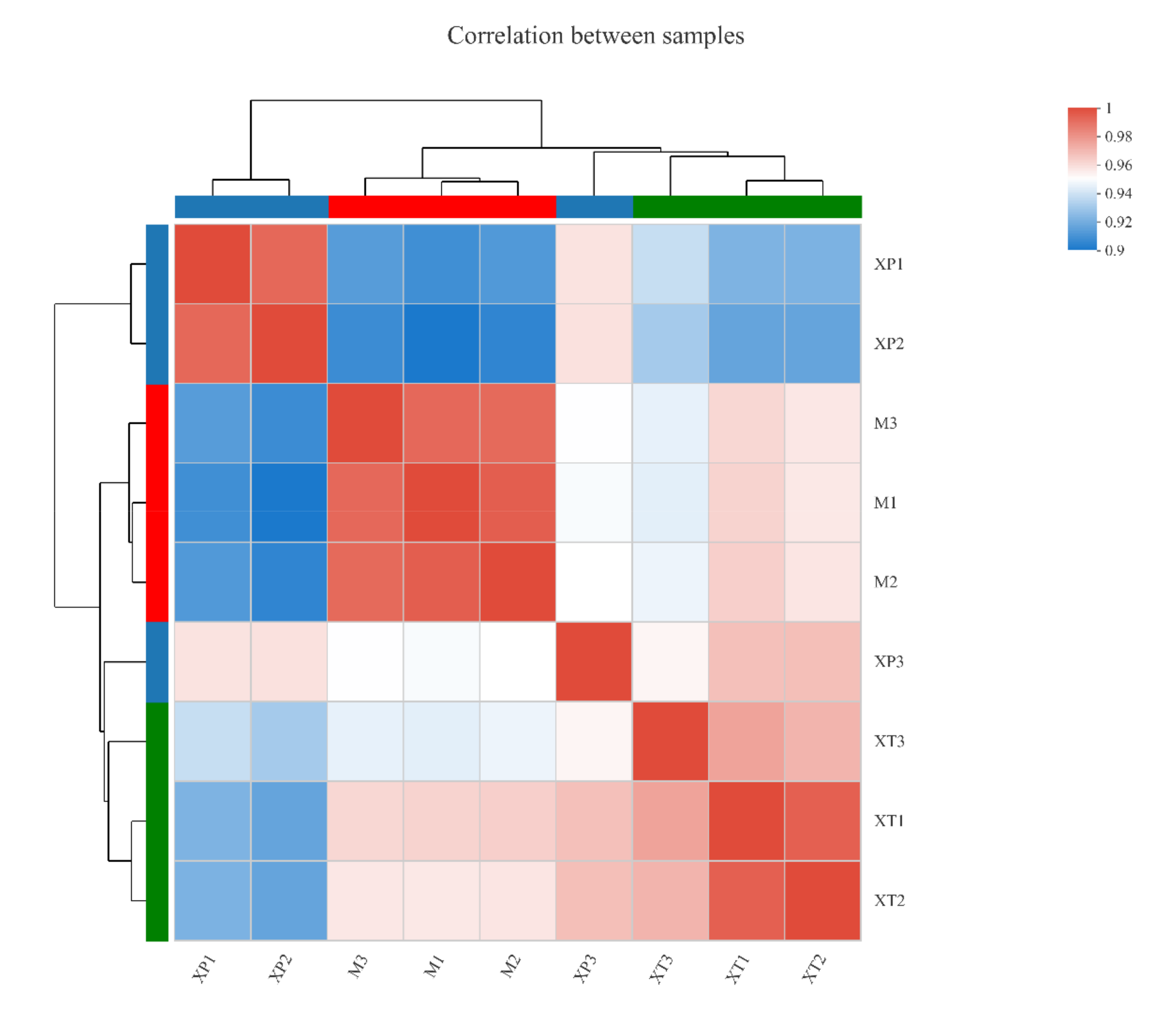
3.1. Overview of the L. reuteri LR08 Transcriptomic Changes Response to dpep and dpro
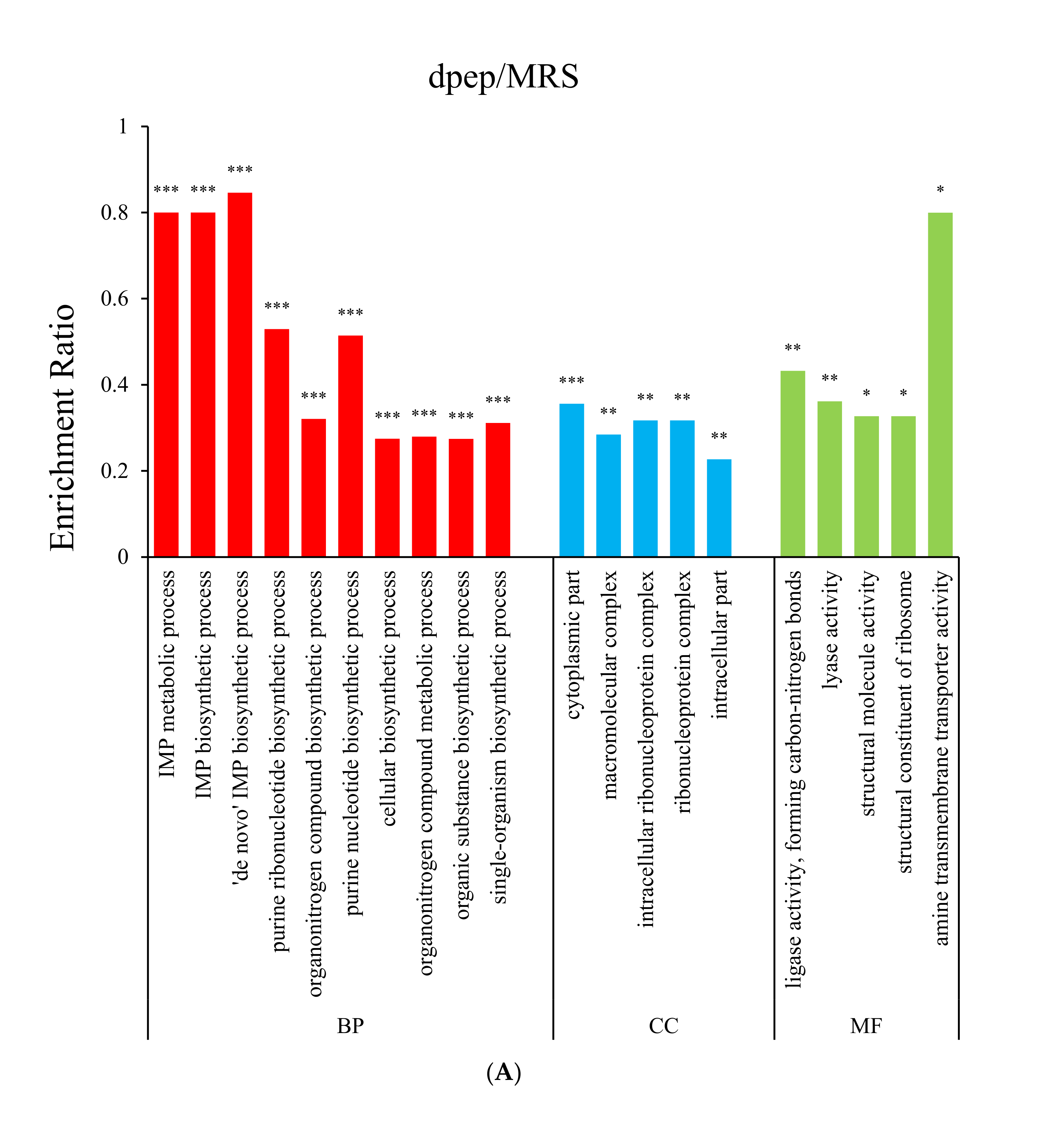
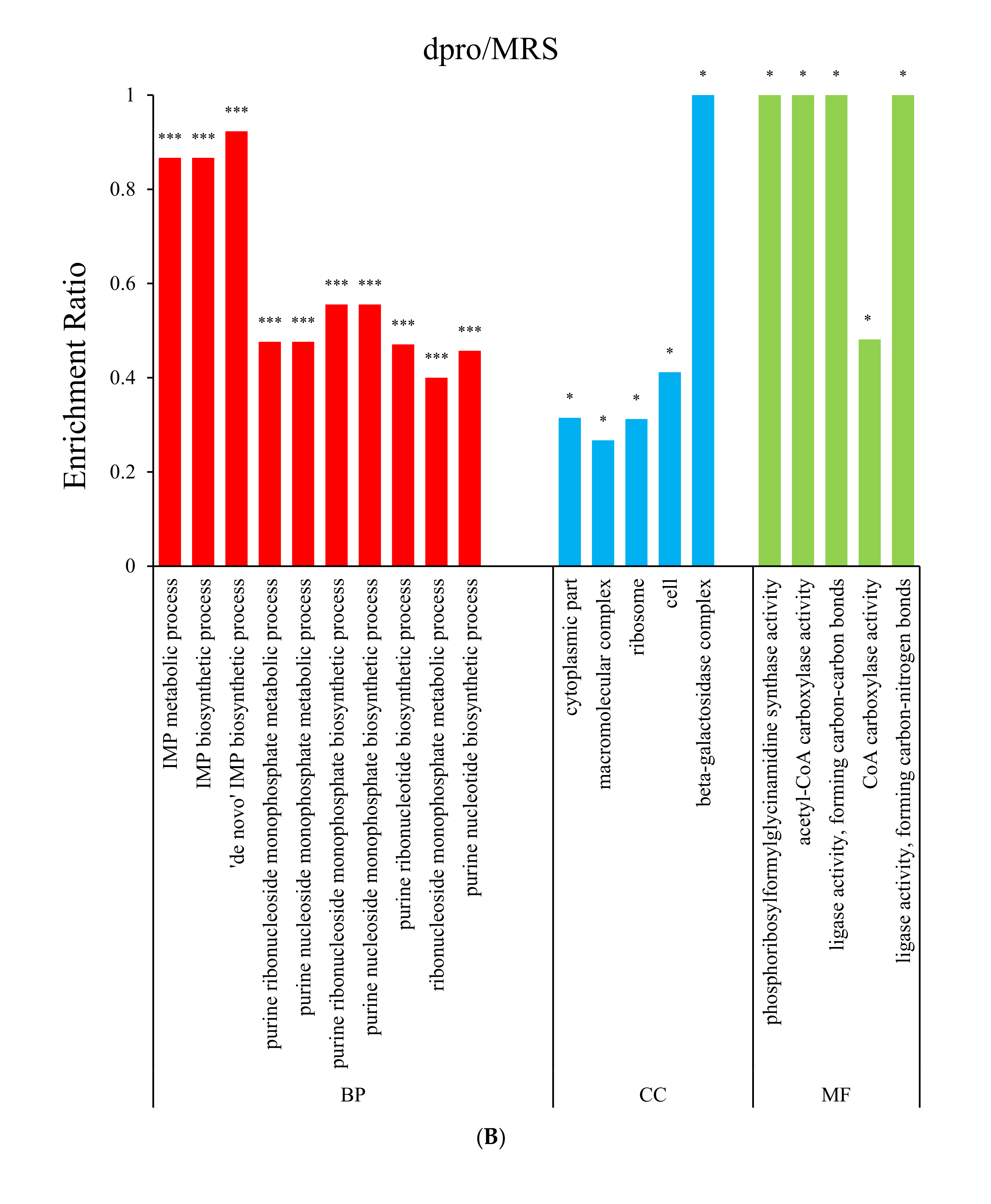
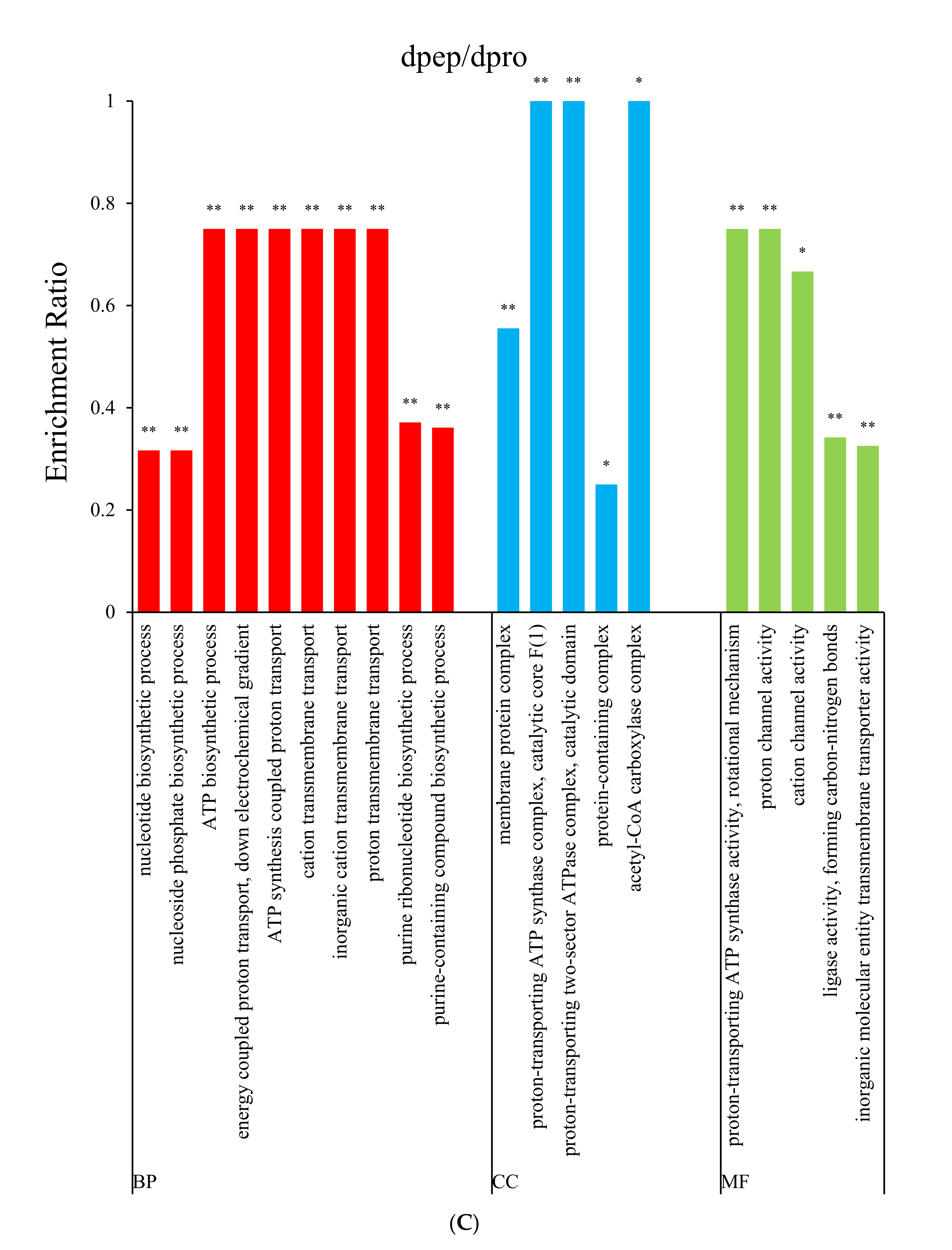
3.2. Gene Ontology (GO) Enrichment Analysis
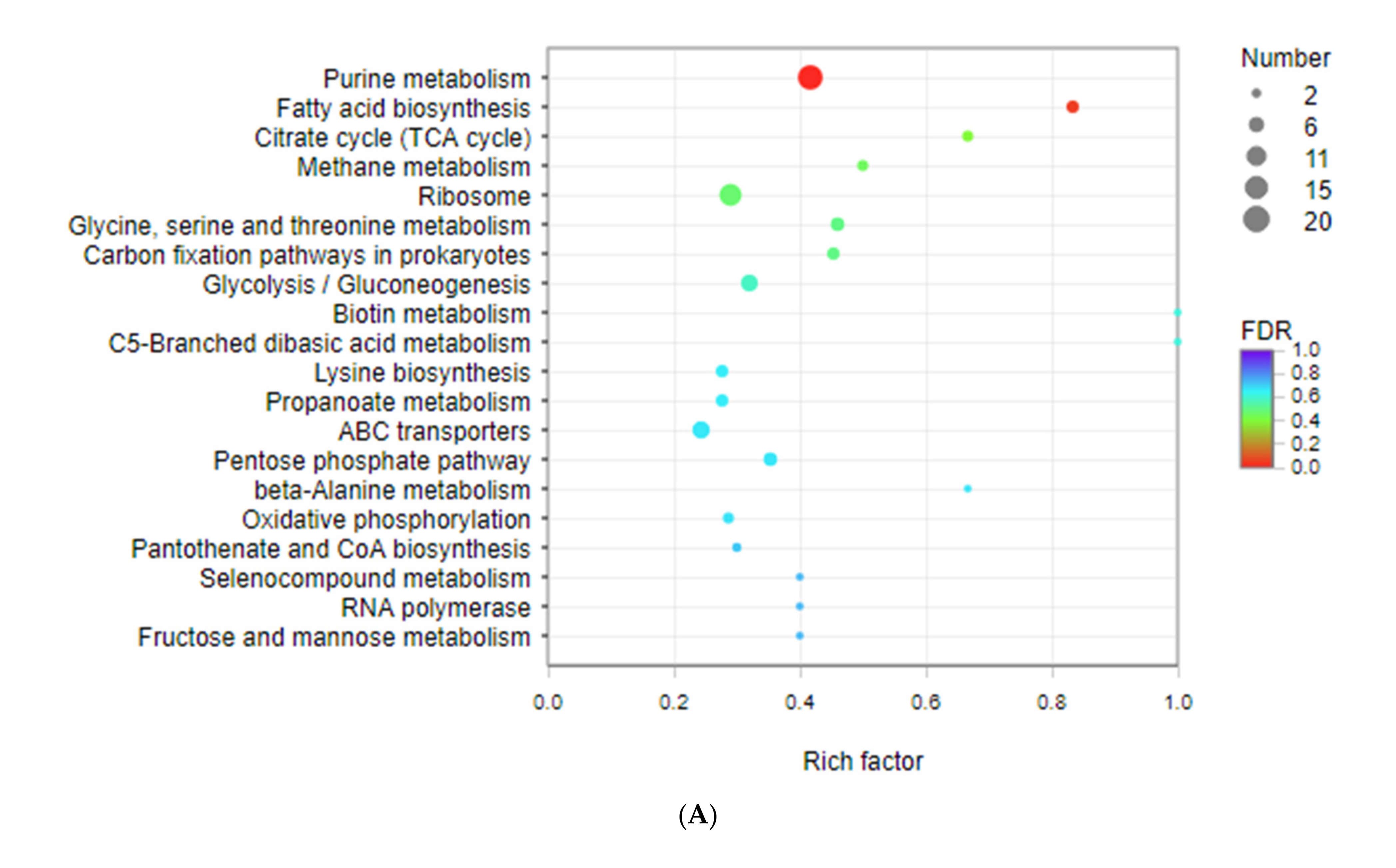
3.3. Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
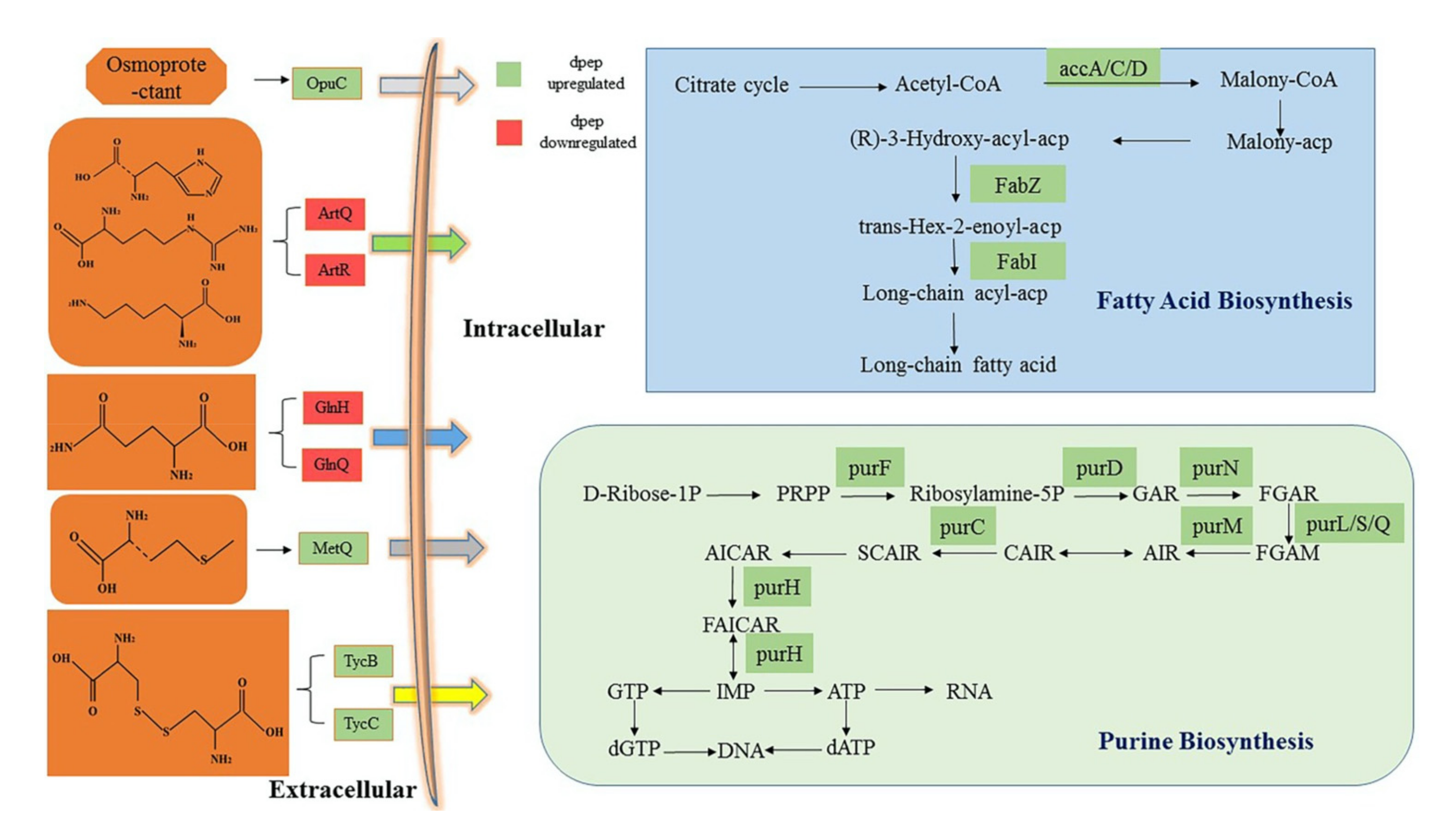
3.4. Analysis of Critical DEGs from GO and KEGG under dpep and dpro Treatment
3.4.1. DEGs Enriched in Fatty Acid Biosynthesis Pathway Were Oppositely Modulated by dpep and dpro
3.4.2. Both dpep and dpro Improve Gene Expression Enriched in Purine Biosynthesis
3.4.3. Dpep Induced Genes Enriched in ABC Transporters Pathway
3.5. Real-Time Fluorescence Quantitative PCR (RT-qPCR) Verification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hekmat, S.; Soltani, H.; Reid, G. Growth and survival of Lactobacillus reuteri RC-14 and Lactobacillus rhamnosus GR-1 in yogurt for use as a functional food. Innov. Food Sci. Emerg. Technol. 2009, 10, 293–296. [Google Scholar] [CrossRef]
- Singh, T.P.; Kaur, G.; Malik, R.K.; Schillinger, U.; Guigas, C.; Kapila, S. Characterization of Intestinal Lactobacillus reuteri Strains as Potential Probiotics. Probiotics Antimicrob. Proteins 2012, 4, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Urbanska, M.; Szajewska, H. The efficacy of Lactobacillus reuteri DSM 17938 in infants and children: A review of the current evidence. Eur. J. Pediatr. 2014, 173, 1327–1337. [Google Scholar] [CrossRef] [PubMed]
- Spinler, J.K.; Taweechotipatr, M.; Rognerud, C.L.; Ou, C.N.; Tumwasorn, S.; Versalovic, J. Human-Derived Probiotic Lactobacillus reuteri Demonstrate Antimicrobial Activities Targeting Diverse Enteric Bacterial Pathogens. Anaerobe 2008, 14, 166–171. [Google Scholar] [CrossRef]
- Chlebowska-Smigiel, A.; Gniewosz, M.; Kieliszek, M.; Bzducha-Wrobel, A. The effect of pullulan on the growth and acidifying activity of selected stool microflora of human. Curr. Pharm. Biotechnol. 2017, 18, 121–126. [Google Scholar] [CrossRef]
- Chlebowska-Smigiel, A.; Kycia, K.; Neffe-Skocinska, K.; Kieliszek, M.; Gniewosz, M.; Kolozyn-Krajewska, D. Effect of Pullulan on Physicochemical, Microbiological, and Sensory Quality of Yogurts. Curr. Pharm. Biotechnol. 2019, 20, 489–496. [Google Scholar] [CrossRef]
- Peighambardoust, S.H.; Karami, Z.; Pateiro, M.; Lorenzo, J.M. A Review on Health-Promoting, Biological, and Functional Aspects of Bioactive Peptides in Food Applications. Biomolecules 2021, 11, 631. [Google Scholar] [CrossRef]
- Nasri, M. Protein Hydrolysates and Biopeptides: Production, Biological Activities, and Applications in Foods and Health Benefits. A Review. Adv. Food Nutr. Res. 2017, 81, 109–159. [Google Scholar] [CrossRef]
- Hartmann, R.; Meisel, H. Food-derived peptides with biological activity: From research to food applications. Curr. Opin. Biotechnol. 2007, 18, 163–169. [Google Scholar] [CrossRef]
- Lu, Y.Y.; Zhao, L.S.; Liu, L.B.; Li, C. Correlation between Moderately Hydrolyzed Whey Protein and Its Allergenicity and Probiotics Growth. J. Dairy Sci. Technol. 2018, 41, 1–5. [Google Scholar] [CrossRef]
- Pan, F.; Yang, M.; Liu, M.; Hu, W.; Wang, Y. Study on the activity of pea proteolytic products to promote the growth of probiotics. Chin. Food Sci. 2019, 19, 32–41. [Google Scholar] [CrossRef]
- Niu, J.Y.; Yin, I.X.; Mei, M.L.; Wu, W.K.K.; Li, Q.L.; Chu, C.H. The multifaceted roles of antimicrobial peptides in oral diseases. Mol. Oral Microbiol. 2021, 36, 159–171. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Liu, G.; Li, W.; Xia, S.; Li, H.; Liu, X. Effects of soybean protein isolates and peptides on the growth and metabolism of Lactobacillus rhamnosus. J. Funct. Foods 2021, 77, 104335. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Y.; Li, H.; Liu, X. The potential of proteins, hydrolysates and peptides as growth factors for Lactobacillus and Bifidobacterium: Current research and future perspectives. Food Funct. 2020, 11, 1946–1957. [Google Scholar] [CrossRef]
- Dong, Z.; Chen, Y. Transcriptomics: Advances and approaches. Sci. China Life Sci. 2013, 56, 960–967. [Google Scholar] [CrossRef]
- Zhao, F.; Liu, W.; Yu, Y.; Liu, X. Effect of small molecular weight soybean proteinderived peptide supplementation on attenuating burn injury-induced inflammation and accelerating wound healing in a rat model. RSC Adv. 2019, 9, 1247. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Zhang, Y.; He, L.; Zhang, C.; Liu, X. Different effects of soybean protein and its derived peptides on the growth and metabolism of Bifidobacterium animalis subsp. animalis JCM 1190. Food Funct. 2021, 12, 5731–5744. [Google Scholar] [CrossRef]
- Fan, L.; Yu, C.Q.; Yao, D. Selection of internal reference genes in Lp gene expression of Lactobacillus plantarum. J. Heilongjiang Bayi Agric. Univ. 2016, 28, 6. [Google Scholar] [CrossRef]
- Rijk, J.C.; Peijnenburg, A.A.; Hendriksen, P.J.; Van Hende, J.M.; Groot, M.J.; Nielen, M.W. Feasibility of a liver transcriptomics approach to assess bovine treatment with the prohormone dehydroepiandrosterone (DHEA). BMC Vet. Res. 2010, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Caroca, R.; Howell, K.A.; Malinova, I.; Burgos, A.; Tiller, N.; Pellizzer, T.; Annunziata, M.G.; Hasse, C.; Ruf, S.; Karcher, D.; et al. Knockdown of the plastid-encoded acetyl-CoA carboxylase gene uncovers functions in metabolism and development. Plant Physiol. 2021, 185, 1091–1110. [Google Scholar] [CrossRef] [PubMed]
- Kozaeva, E.; Volkova, S.; Matos, M.R.A.; Mezzina, M.P.; Wulff, T.; Volke, D.C.; Nielsen, L.K.; Nikel, P.I. Model-guided dynamic control of essential metabolic nodes boosts acetyl-coenzyme A-dependent bioproduction in rewired Pseudomonas putida. Metab. Eng. 2021, 67, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Kiatpapan, P.; Kobayashi, H.; Sakaguchi, M.; Ono, H.; Yamashita, M.; Kaneko, Y.; Murooka, Y. Molecular characterization of Lactobacillus plantarum genes for beta-ketoacyl-acyl carrier protein synthase III (fabH) and acetyl coenzyme A carboxylase (accBCDA), which are essential for fatty acid biosynthesis. Appl. Environ. Microbiol. 2001, 67, 426–433. [Google Scholar] [CrossRef] [PubMed]
- Rana, P.; Ghouse, S.M.; Akunuri, R.; Madhavi, Y.V.; Chopra, S.; Nanduri, S. FabI (enoyl acyl carrier protein reductase)-A potential broad spectrum therapeutic target and its inhibitors. Eur. J. Med. Chem. 2020, 208, 112757. [Google Scholar] [CrossRef]
- Sharma, S.K.; Kapoor, M.; Ramya, T.N.; Kumar, S.; Kumar, G.; Modak, R.; Sharma, S.; Surolia, N.; Surolia, A. Identification, characterization, and inhibition of Plasmodium falciparum beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J. Biol. Chem. 2003, 278, 45661–45671. [Google Scholar] [CrossRef] [PubMed]
- Guerzoni, M.E.; Lanciotti, R.; Cocconcelli, P.S. Alteration in cellular fatty acid composition as a response to salt, acid, oxidative and thermal stresses in Lactobacillus helveticus. Microbiology 2001, 147, 2255–2264. [Google Scholar] [CrossRef]
- Zhang, C.; Xia, S.; Zhang, Y.; Zhu, S.; Li, H.; Liu, X. Identification of soybean peptides and their effect on the growth and metabolism of Limosilactobacillus reuteri LR08. Food Chem. 2022, 369, 130923. [Google Scholar] [CrossRef]
- Morar, M.; Hoskins, A.A.; Stubbe, J.; Ealick, S.E. Formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima: Structural insights into complex formation. Biochemistry 2008, 47, 7816–7830. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Morar, M.; Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 2008, 65, 3699–3724. [Google Scholar] [CrossRef]
- Pedley, A.M.; Benkovic, S.J. A New View into the Regulation of Purine Metabolism: The Purinosome. Trends Biochem. Sci. 2017, 42, 141–154. [Google Scholar] [CrossRef]
- Liu, Z.-S.; Lin, C.-F.; Chen, P.-W. Transcriptome analysis of Lactobacillus rhamnosus GG strain treated with prebiotic-bovine lactoferrin under a cold environment. J. Food Drug Anal. 2021, 29, 402–418. [Google Scholar] [CrossRef]
- Laakso, K.; Koskenniemi, K.; Koponen, J.; Kankainen, M.; Surakka, A.; Salusjarvi, T.; Auvinen, P.; Savijoki, K.; Nyman, T.A.; Kalkkinen, N.; et al. Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microb. Biotechnol. 2011, 4, 746–766. [Google Scholar] [CrossRef]
- Thomas, C.; Tampe, R. Structural and Mechanistic Principles of ABC Transporters. Annu. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef]
- Ribic, U.; Jakse, J.; Toplak, N.; Koren, S.; Kovac, M.; Klancnik, A.; Jersek, B. Transporters and Efflux Pumps Are the Main Mechanisms Involved in Staphylococcus epidermidis Adaptation and Tolerance to Didecyldimethylammonium Chloride. Microorganisms 2020, 8, 344. [Google Scholar] [CrossRef]
- Nguyen, P.T.; Lai, J.Y.; Kaiser, J.T.; Rees, D.C. Structures of the Neisseria meningitides methionine-binding protein MetQ in substrate-free form and bound to l- and d-methionine isomers. Protein Sci. 2019, 28, 1750–1757. [Google Scholar] [CrossRef]
- Hartel, T.; Klein, M.; Koedel, U.; Rohde, M.; Petruschka, L.; Hammerschmidt, S. Impact of glutamine transporters on pneumococcal fitness under infection-related conditions. Infect. Immun. 2011, 79, 44–58. [Google Scholar] [CrossRef]
- Heuveling, J.; Landmesser, H.; Schneider, E. Evidence from Mutational Analysis for a Single Transmembrane Substrate Binding Site in the Histidine ATP-Binding Cassette Transporter of Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2019, 201, e00521-18. [Google Scholar] [CrossRef]
- Teichmann, L.; Chen, C.; Hoffmann, T.; Smits, S.H.J.; Schmitt, L.; Bremer, E. From substrate specificity to promiscuity: Hybrid ABC transporters for osmoprotectants. Mol. Microbiol. 2017, 104, 761–780. [Google Scholar] [CrossRef]
- Nau-Wagner, G.; Opper, D.; Rolbetzki, A.; Boch, J.; Kempf, B.; Hoffmann, T.; Bremer, E. Genetic Control of Osmoadaptive Glycine Betaine Synthesis in Bacillus subtilis through the Choline-Sensing and Glycine Betaine-Responsive GbsR Repressor. J. Bacteriol. 2012, 194, 2703–2714. [Google Scholar] [CrossRef]
- Romeo, Y.; Bouvier, J.; Gutierrez, C. Osmotic stress response of lactic acid bacteria Lactococcus lactis and Lactobacillus plantarum. Lait 2001, 81, 49–55. [Google Scholar] [CrossRef]
- Li, E.; Zhu, Q.; Pang, D.; Liu, F.; Liao, S.; Zou, Y. Analysis of Lactobacillus rhamnosus GG in Mulberry Galacto-Oligosaccharide Medium by Comparative Transcriptomics and Metabolomics. Front. Nutr. 2022, 9, 853271. [Google Scholar] [CrossRef] [PubMed]


| Composition | MRS (g/L) | dpep (L) | dpro (g/L) |
|---|---|---|---|
| Peptone | 10 | 5 | 5 |
| Beef powder | 5 | 2.5 | 2.5 |
| Yeast extract powder | 4 | 2 | 2 |
| Dipotassium hydrogen phosphate | 2 | 2 | 2 |
| Anhydrous sodium acetate | 5 | 5 | 5 |
| Magnesium sulfate | 0.2 | 0.2 | 0.2 |
| Triamine citrate | 2 | 2 | 2 |
| Manganese sulfate | 0.05 | 0.05 | 0.05 |
| Twain 80 | 1 | 1 | 1 |
| Glucose | 20 | 20 | 20 |
| Digested soybean peptides | - | 13.214 | - |
| Digested soybean protein | - | - | 12.259 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Zhang, Y.; Wang, J.; Zhang, C.; Liu, X. Investigating Differential Expressed Genes of Limosilactobacillus reuteri LR08 Regulated by Soybean Protein and Peptides. Foods 2022, 11, 1251. https://doi.org/10.3390/foods11091251
Zhu S, Zhang Y, Wang J, Zhang C, Liu X. Investigating Differential Expressed Genes of Limosilactobacillus reuteri LR08 Regulated by Soybean Protein and Peptides. Foods. 2022; 11(9):1251. https://doi.org/10.3390/foods11091251
Chicago/Turabian StyleZhu, Shuya, Yinxiao Zhang, Jingyi Wang, Chi Zhang, and Xinqi Liu. 2022. "Investigating Differential Expressed Genes of Limosilactobacillus reuteri LR08 Regulated by Soybean Protein and Peptides" Foods 11, no. 9: 1251. https://doi.org/10.3390/foods11091251
APA StyleZhu, S., Zhang, Y., Wang, J., Zhang, C., & Liu, X. (2022). Investigating Differential Expressed Genes of Limosilactobacillus reuteri LR08 Regulated by Soybean Protein and Peptides. Foods, 11(9), 1251. https://doi.org/10.3390/foods11091251






