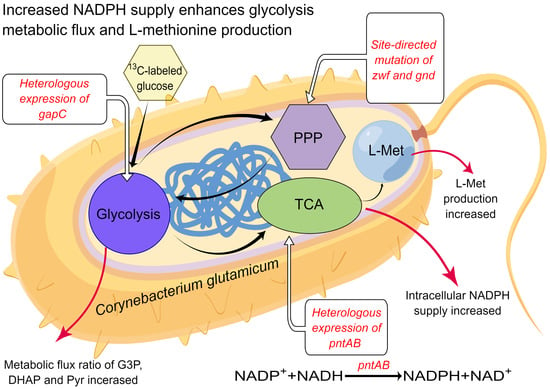
Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Primers and Plasmids
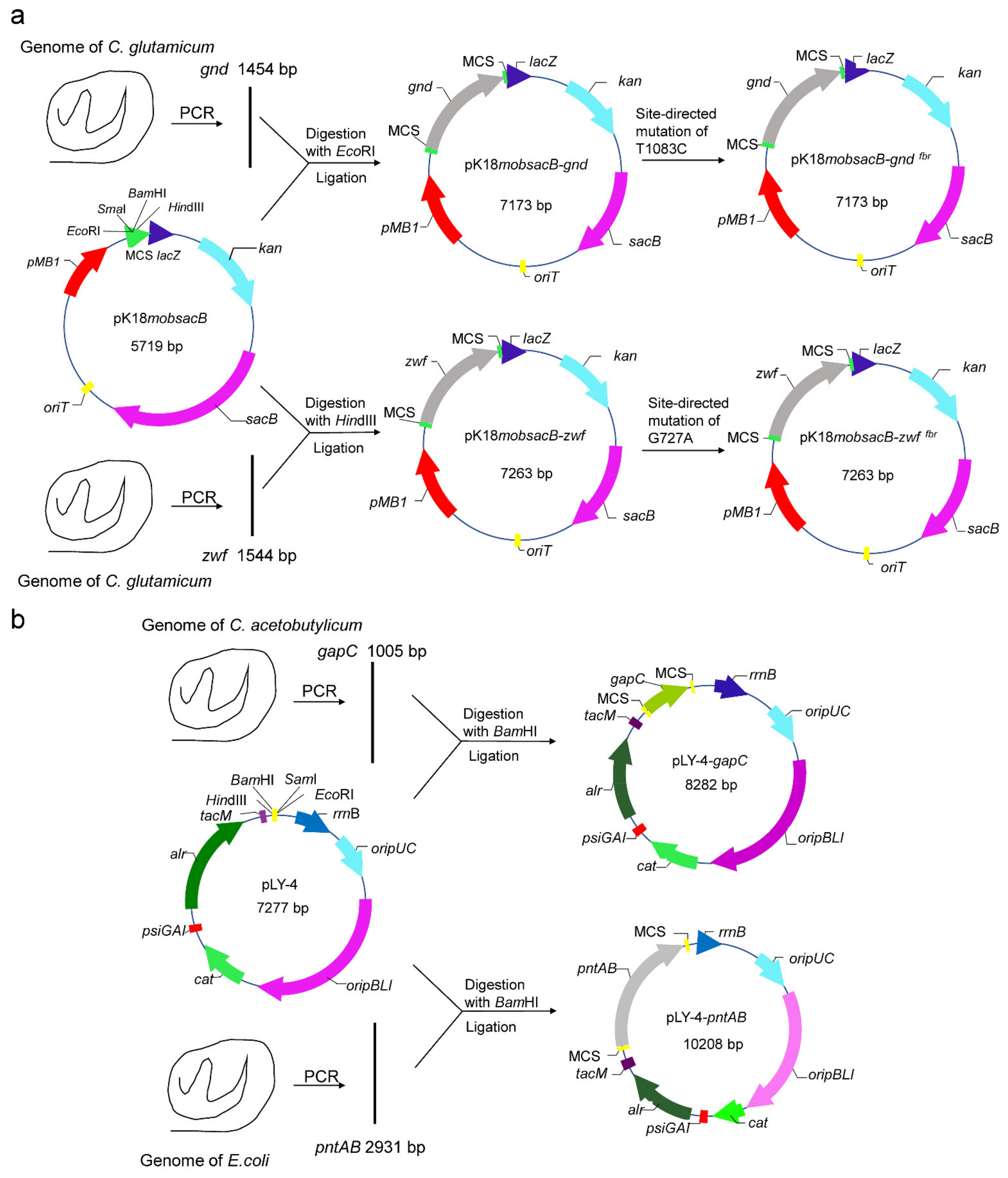
2.3. Site-Directed Mutation of zwf and gnd Gene
2.4. Construction of Expression Vectors pLY-4-gapC and pLY-4-pntAB
2.5. Transformation Method
2.6. Determination of NADPH
2.7. Determination of L-methionine
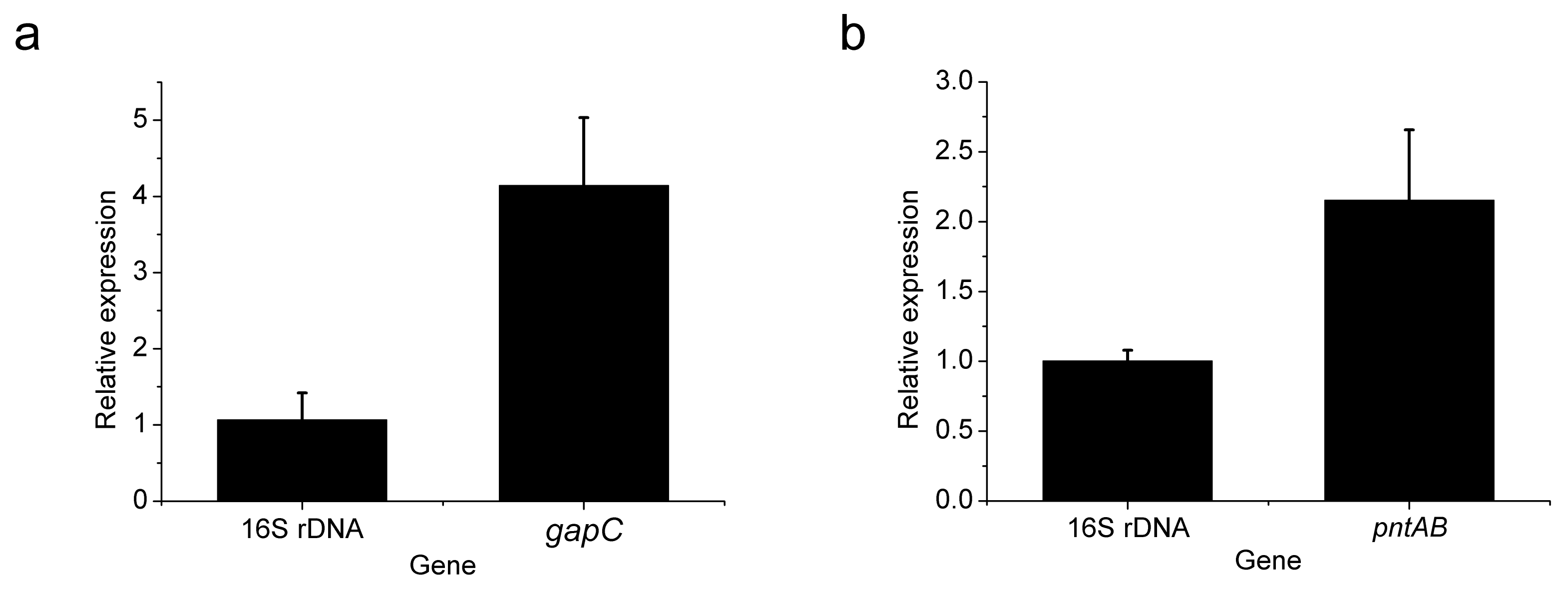
2.8. Gene Expression Analysis
2.9. 13C-Metabolic Flux Ratio of Metabolites from the Labeled Glucose
2.10. Data Processing and Statistical Analysis
3. Results
3.1. Modification of Pentose Phosphate Pathway to Increase NADPH Supplement
3.2. Heterologous Expression of gapC and pntAB
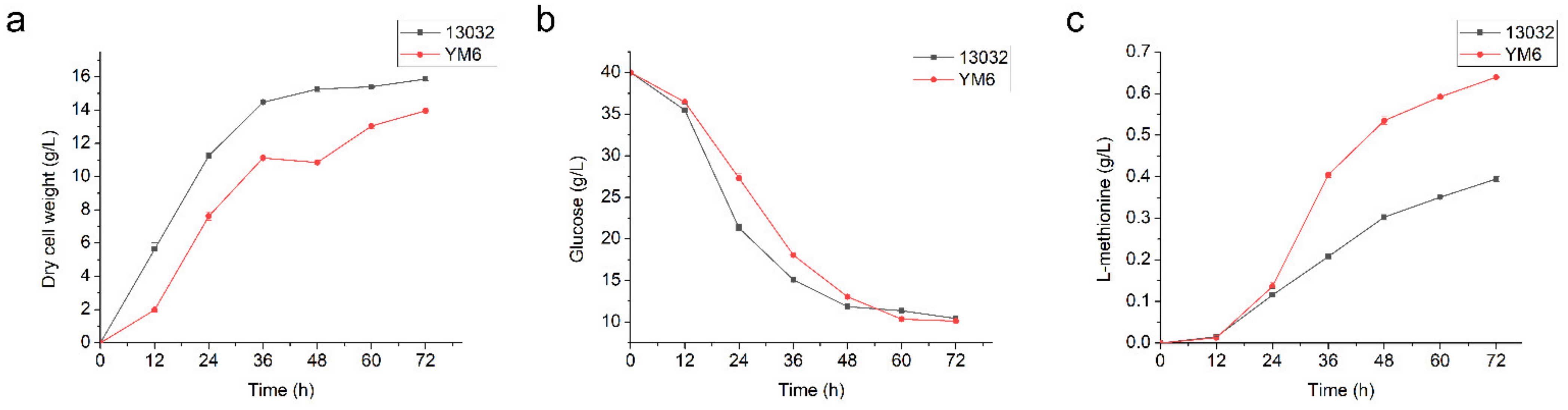
3.3. Construction and Fermentation Characteristics of C. glutamicum YM6
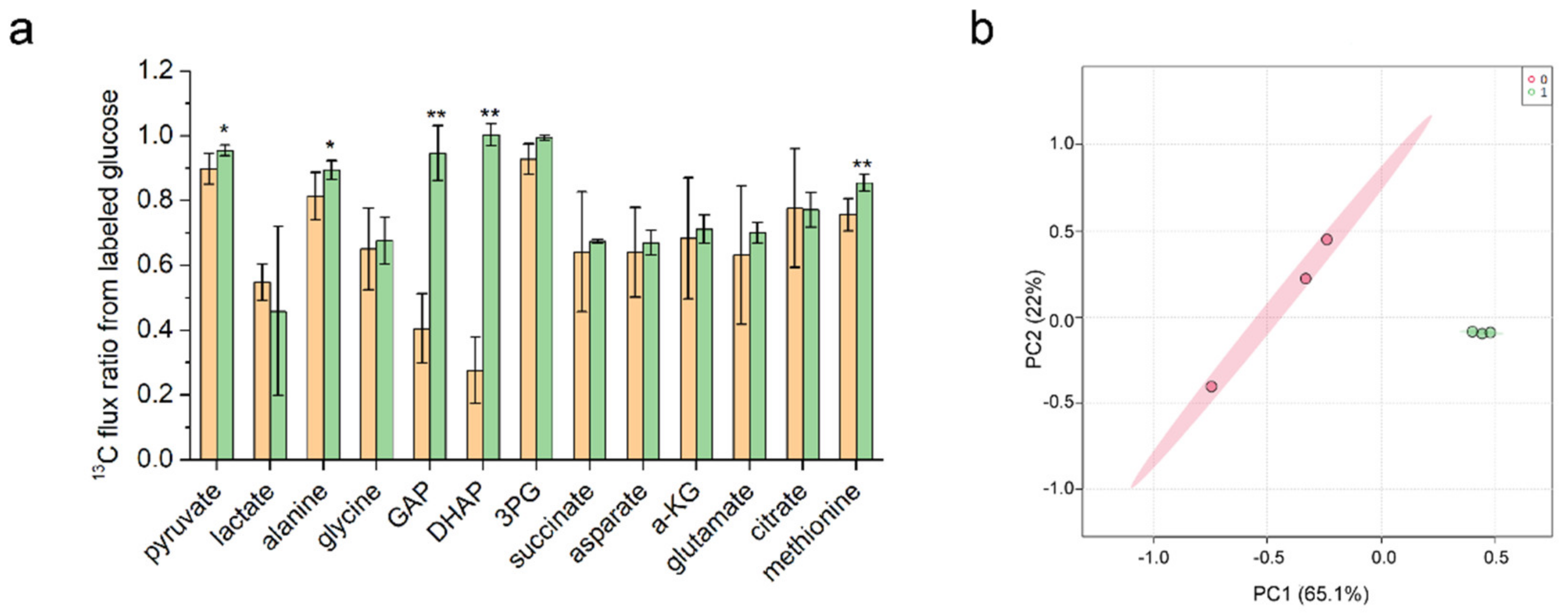
3.4. Metabolic Flux Ratio Analysis of C. glutamicum YM6
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Xue, J.J.; Xie, M.; Tang, J.; Huang, W.; Zhang, Q.; Hou, S.S. Effects of excess DL-and L-methionine on growth performance of starter Pekin ducks. Poult. Sci. 2018, 97, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, S.M.; Gao, X.; Dai, Z.; Locasale, J.W. Methionine metabolism in health and cancer: A nexus of diet and precision medicine. Nat. Rev. Cancer 2019, 19, 625–637. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qian, J.; Li, B.; Lu, M.; Le, G.; Xie, Y. Metabolomics based on 1H-NMR reveal the regulatory mechanisms of dietary methionine restriction on splenic metabolic dysfunction in obese mice. Foods 2021, 10, 2439. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.; Shin, Y.; Lee, I.; Kim, S.Y. L-Methionine Production. Adv. Biochem. Eng. Biotechnol. 2017, 159, 153–177. [Google Scholar] [CrossRef]
- Faroon, O.; Roney, N.; Taylor, J.; Ashizawa, A.; Lumpkin, M.H.; Plewak, D.J. Acrolein health effects. Toxicol. Ind. Health 2008, 24, 447–490. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, S.; Song, Z.; Gao, R. Immobilization of an L-aminoacylase-producing strain of Aspergillus oryzae into gelatin pellets and its application in the resolution of D, L-methionine. Biotechnol. Appl. Biochem. 2002, 35, 107–113. [Google Scholar] [CrossRef]
- Becker, J.; Gießelmann, G.; Hoffmann, S.L.; Wittmann, C. Corynebacterium glutamicum for Sustainable Bioproduction: From Metabolic Physiology to Systems Metabolic Engineering. Adv. Biochem. Eng. Biotechnol. 2018, 162, 217–263. [Google Scholar] [CrossRef]
- Qin, T.; Hu, X.; Hu, J.; Wang, X. Metabolic engineering of Corynebacterium glutamicum strain ATCC13032 to produce L-methionine. Biotechnol. Appl. Biochem. 2015, 62, 563–573. [Google Scholar] [CrossRef]
- Li, Y.; Cong, H.; Liu, B.; Song, J.; Sun, X.; Zhang, J.; Yang, Q. Metabolic engineering of Corynebacterium glutamicum for methionine production by removing feedback inhibition and increasing NADPH level. Antonie Van Leeuwenhoek 2016, 109, 1185–1197. [Google Scholar] [CrossRef]
- Krömer, J.O.; Wittmann, C.; Schröder, H.; Heinzle, E. Metabolic pathway analysis for rational design of L-methionine production by Escherichia coli and Corynebacterium glutamicum. Metab. Eng. 2006, 8, 353–369. [Google Scholar] [CrossRef]
- Becker, J.; Klopprogge, C.; Zelder, O.; Heinzle, E.; Wittmann, C. Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl. Environ. Microbiol. 2005, 71, 8587–8596. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, H.; Xu, J.; Ruan, H.; Zhang, W. Deciphering the crucial roles of AraC-type transcriptional regulator Cgl2680 on NADPH metabolism and L-lysine production in Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 2020, 36, 82. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.Y.; Zhang, Y.Y.; Li, Z.; Liu, J.Z. Metabolic engineering of Corynebacterium glutamicum for increasing the production of L-ornithine by increasing NADPH availability. J. Ind. Microbiol. Biotechnol. 2013, 40, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Kan, B.; Dong, J.; Xu, G.; Han, R.; Ni, Y. Metabolic engineering of Corynebacterium glutamicum for improved L-arginine synthesis by enhancing NADPH supply. J. Ind. Microbiol. Biotechnol. 2019, 46, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Keilhauer, C.; Eggeling, L.; Sahm, H. Isoleucine synthesis in Corynebacterium glutamicum: Molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993, 175, 5595–5603. [Google Scholar] [CrossRef]
- Li, Y.; Ai, Y.; Zhang, J.; Fei, J.; Liu, B.; Wang, J.; Li, M.; Zhao, Q.; Song, J. A novel expression vector for Corynebacterium glutamicum with an auxotrophy complementation system. Plasmid 2020, 107, 102476. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J. Cloning and transformation with plasmid vectors. In Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Van der Rest, M.E.; Lange, C.; Molenaar, D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biotechnol. 1999, 52, 541–545. [Google Scholar] [CrossRef]
- Yu, S.; Feng, S.; Sun, Y.; Tang, T.; Han, J.; Wang, F.; Li, T. Determination of sulfur amino acids in feedstuffs by high performance liquid chromatography coupled with pre-column derivatization. Chin. J. Chromatogr. 2011, 29, 239–243. [Google Scholar] [CrossRef]
- Bolten, C.J.; Schröder, H.; Dickschat, J.; Wittmann, C. Towards methionine overproduction in Corynebacterium glutamicum--methanethiol and dimethyldisulfide as reduced sulfur sources. J. Microbiol. Biotechnol. 2010, 20, 1196–1203. [Google Scholar] [CrossRef]
- Khosla, C.; Keasling, J.D. Metabolic engineering for drug discovery and development. Nat. Rev. Drug Discov. 2003, 2, 1019–1025. [Google Scholar] [CrossRef]
- Xu, J.Z.; Yang, H.K.; Zhang, W.G. NADPH metabolism: A survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit. Rev. Biotechnol. 2018, 38, 1061–1076. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhang, H.; Hung, H.C.; Kuo, C.C.; Tsai, L.C.; Yuan, H.S.; Chou, W.Y.; Chang, G.G.; Tong, L. Structural studies of the pigeon cytosolic NADP (+)-dependent malic enzyme. Protein Sci. 2002, 11, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Yao, J.; Liu, L.; Ma, X.; Li, W.; Sun, X.; Wang, Y. Improving squalene production by enhancing the NADPH/NADP (+) ratio, modifying the isoprenoid-feeding module and blocking the menaquinone pathway in Escherichia coli. Biotechnol. Biofuels 2019, 12, 68. [Google Scholar] [CrossRef]
- Becker, J.; Klopprogge, C.; Herold, A.; Zelder, O.; Bolten, C.J.; Wittmann, C. Metabolic flux engineering of L-lysine production in Corynebacterium glutamicum—Over expression and modification of G6P dehydrogenase. J. Biotechnol. 2007, 132, 99–109. [Google Scholar] [CrossRef]
- Moritz, B.; Striegel, K.; de Graaf, A.A.; Sahm, H. Kinetic properties of the glucose-6-phosphate and 6 phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 2000, 267, 3442–3452. [Google Scholar] [CrossRef]
- Ohnishi, J.; Katahira, R.; Mitsuhashi, S.; Kakita, S.; Ikeda, M. A novel gnd mutation leading to increased L-lysine production in Corynebacterium glutamicum. FEMS Microbiol. Lett. 2005, 242, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Yukawa, T.; Bamba, T.; Guirimand, G.; Matsuda, M.; Hasunuma, T.; Kondo, A. Optimization of 1,2,4-butanetriol production from xylose in Saccharomyces cerevisiae by metabolic engineering of NADH/NADPH balance. Biotechnol. Bioeng. 2021, 118, 175–185. [Google Scholar] [CrossRef]
- Ma, W.; Wang, J.; Li, Y.; Hu, X.; Shi, F.; Wang, X. Enhancing pentose phosphate pathway in Corynebacterium glutamicum to improve l-isoleucine production. Biotechnol. Appl. Biochem. 2016, 63, 877–885. [Google Scholar] [CrossRef]
- Richard, J.P. Restoring a metabolic pathway. ACS Chem. Biol. 2008, 3, 605–607. [Google Scholar] [CrossRef][Green Version]

| Strains or Plasmids | Relevant Characteristics | Source |
|---|---|---|
| E. coli Top10 | Wild type | Takara |
| Clostrdium acetobutylicum ATCC824 | Wild type | ATCC |
| C. glutamicun ATCC13032 | Wild type | ATCC |
| C. glutamicun YM1 | C. glutamicum ATCC13032 chromosome with zwffbr gene | This study |
| C. glutamicun YM2 | C. glutamicum ATCC13032 chromosome with gndfbr gene | This study |
| C. glutamicun YM3 | C. glutamicum ATCC13032 chromosome with zwffbr and gndfbr gene | This study |
| C. glutamicun YM4 | C. glutamicum ATCC13032 chromosome with gapC gene | This study |
| C. glutamicun YM5 | C. glutamicum ATCC13032 chromosome with pntAB gene | This study |
| C. glutamicun YM6 | C. glutamicum YM3 chromosome with gapC gene | This study |
| pK18mobsacB | pK18mobsacB non-replicating suicide plasmid | [9] |
| pLY-4 | E. coli -C. glutamicun shuttle expression vector | [16] |
| pK18mobsacB-zwffbr | pK18mobsacB carrying mutant zwf (G727A) | This study |
| pK18mobsacB-gndfbr | pK18mobsacB carrying mutant gnd(T1083C) | This study |
| pLY-4-gapC | pLY-4 carrying gapC gene | This study |
| pLY-4-pntAB | pLY-4-pntAB carrying pntAB gene | This study |
| Name | Sequence (5′—3′) | Intention |
|---|---|---|
| zwf-F | CACAAGCTTATGGTGATCTTCGGTGTCACTGG | amplification of zwf gene |
| zwf-R | TATAAGCTTTTATGGCCTGCGCCAGGTGT | |
| gnd-F | CACGAATTCTTAAGCTTCAACCTCGGAGC′ | amplification of gnd gene |
| gnd-R | GTGGAATTCATGCCGTCAAGTACGATCAA | |
| pntAB-F | CGAGGATCCATGCGAATTGGCATACCAAG | amplification of pntAB gene |
| pntAB-R | TCGGGATCCTTACAGAGCTTTCAGGATTG | |
| gapC-F | CGAGGATCCATGGCAAAGATAGCTATTAATG | amplification of gapC gene |
| gapC-R | TCGGGATCCCTATTTTGCTATTTTTGCA | |
| zwf-TBF | ACGTCCAGATCACCATGACTGAAGATATTGG | mutation of zwf gene |
| zwf-TBR | TCATGGTGATCTGGACGTGGTCAACGTA | |
| gnd-TBF | CGAGATCAAGGCTGGCCCCGACGAGAA | mutation of gnd gene |
| gnd-TBR | GGCCAGCCTTGATCTCGTCGAAGCCCT | |
| gndYZ-F | ATCGAAATCACCGCAGAGGTTC | validation of mutant zwf gene |
| gndYZ-R | GGAAGGAGCCATCCTTGTCGAT | |
| zwfYZ-F | ATGATGCAGCTTTCGACAACCT | validation of mutant gnd gene |
| zwfYZ-R | GATCTCTAAGGTACAAGCCGC | |
| zwfRT-F | TTGACCACGTCCAGATCACCATG | qRT-PCR of zwf gene |
| zwfRT-R | AGAGAGCACCTTGATCTTTTCTGC | |
| gndRT-F | GTTCTCTCCCAGGTGGATGCTG | qRT-PCR of gnd gene |
| gndRT-R | AAGTGCTTCCAGATCGGTGAGG | |
| pntABRT-F | CACCAACGCGATTTCAGGGAT | qRT-PCR of pntAB gene |
| pntABRT-R | ACCCCTTAATTTTTGCGGAAC | |
| gapCRT-F | GCATCATGCACAACTAACTGCTTAG | qRT-PCR of gapC gene |
| gapCRT-R | TGGCTTATAGCTTTAGCAGCAC |
| Strains | NADPH (nmol/106 cell) | L-methionine (g/L) |
|---|---|---|
| C. glutamicum 13032 | 0.56 ± 0.01 | 0.39 ± 0.02 |
| C. glutamicum YM1 | 0.94 ± 0.02 | 0.45 ± 0.03 |
| C. glutamicum YM2 | 0.88 ± 0.02 | 0.46 ± 0.03 |
| C. glutamicum YM3 | 1.41 ± 0.03 | 0.50 ± 0.04 |
| C. glutamicum YM4 | 0.98 ± 0.02 | 0.58 ± 0.03 |
| C. glutamicum YM5 | 1.06 ± 0.02 | 0.53 ± 0.01 |
| C. glutamicum YM6 | 2.51 ± 0.02 | 0.64 ± 0.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Sun, X.; Liu, Y.; Yang, M.; Wang, L.; Li, Y.; Wang, J. Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum. Foods 2022, 11, 1031. https://doi.org/10.3390/foods11071031
Liu B, Sun X, Liu Y, Yang M, Wang L, Li Y, Wang J. Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum. Foods. 2022; 11(7):1031. https://doi.org/10.3390/foods11071031
Chicago/Turabian StyleLiu, Bingnan, Xinyu Sun, Yue Liu, Mengmeng Yang, Liang Wang, Ying Li, and Jihui Wang. 2022. "Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum" Foods 11, no. 7: 1031. https://doi.org/10.3390/foods11071031
APA StyleLiu, B., Sun, X., Liu, Y., Yang, M., Wang, L., Li, Y., & Wang, J. (2022). Increased NADPH Supply Enhances Glycolysis Metabolic Flux and L-methionine Production in Corynebacterium glutamicum. Foods, 11(7), 1031. https://doi.org/10.3390/foods11071031