Comparative Analysis of Fruit Metabolome Using Widely Targeted Metabolomics Reveals Nutritional Characteristics of Different Rosa roxburghii Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Experimental Methods for Fruit Basic Quality Performance
2.3. Sample Preparation and Extraction
2.4. UPLC Conditions
2.5. ESI-Q TRAP-MS/MS
2.6. Metabolite Identification and Quantification
2.7. Identification of the Key Active Ingredients
2.8. Multivariate Statistical Analysis
2.9. Other Experimental Methods
3. Results
3.1. Fruit Basic Quality Performance of Six R. roxburghii Genotypes
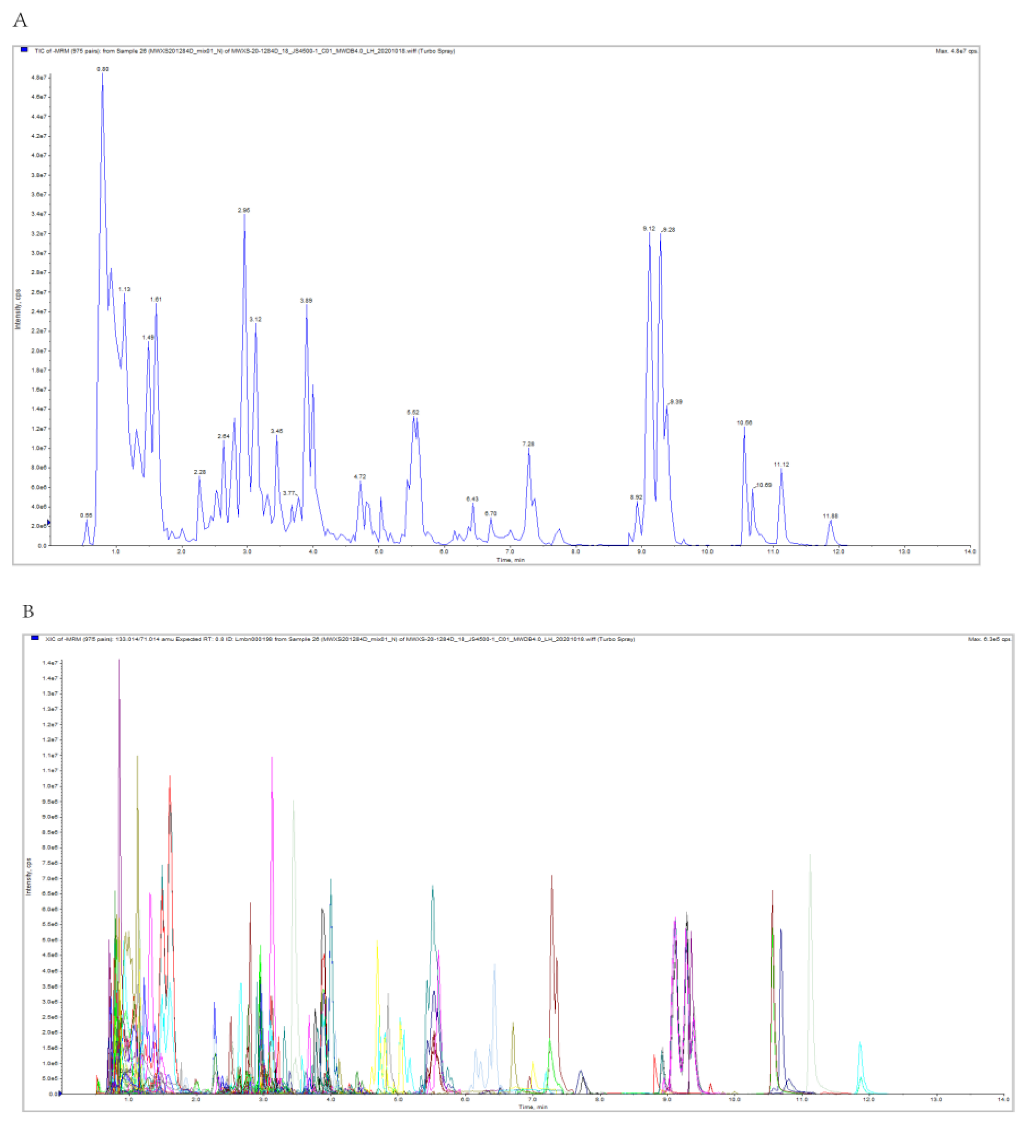
3.2. Overview of the Metabolic Profiles of Six R. roxburghii Genotypes Fruits
3.3. Identification of the Key Active Ingredients Belonging to Traditional Chinese Medicines in R. roxburghii
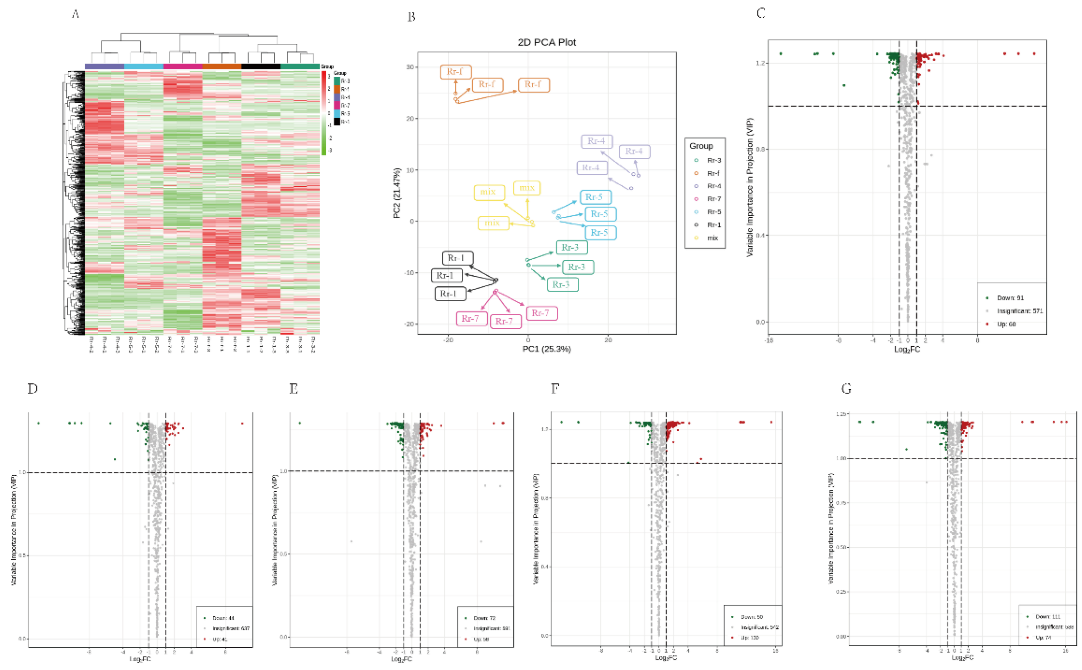
3.4. Differentially Accumulated Metabolites in Fruits of Various Genotypes
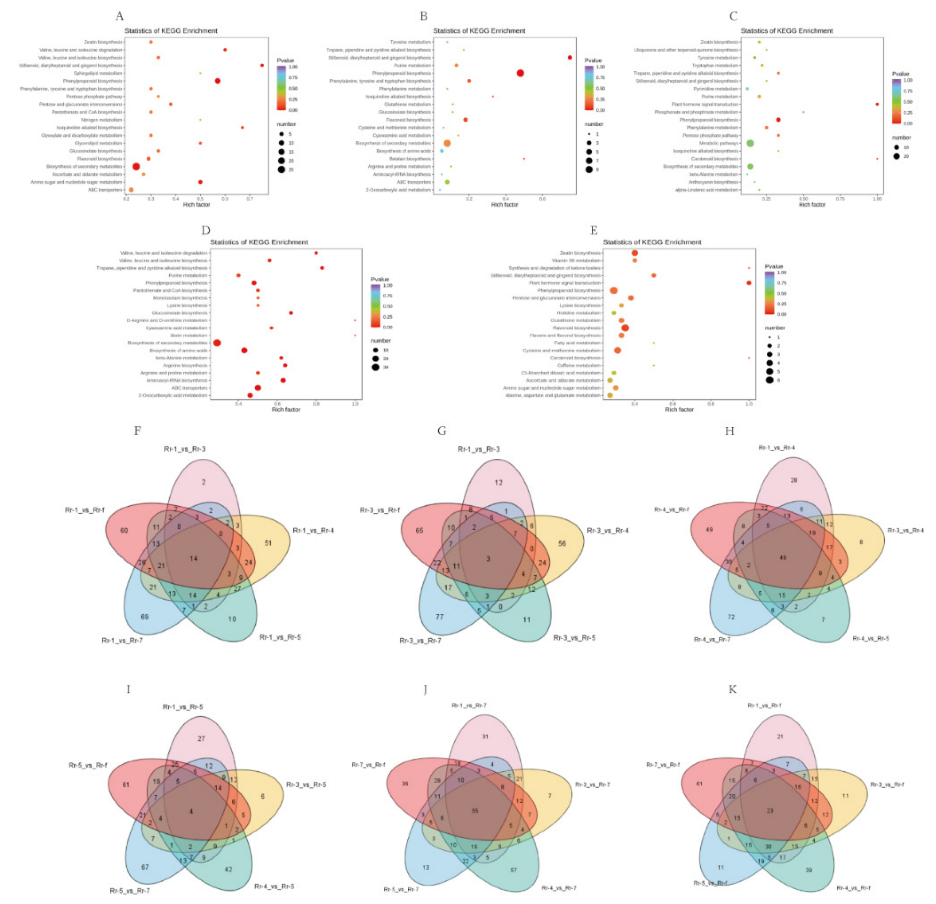
3.5. Metabolic Pathway Analysis of DAMs between Rr-5 and the Other Five Genotypes
3.6. Identification of Characteristic Metabolites of Each Genotype
3.7. Bioactive Substance Content and Antioxidant Capacity of Six R. roxburghii Genotypes
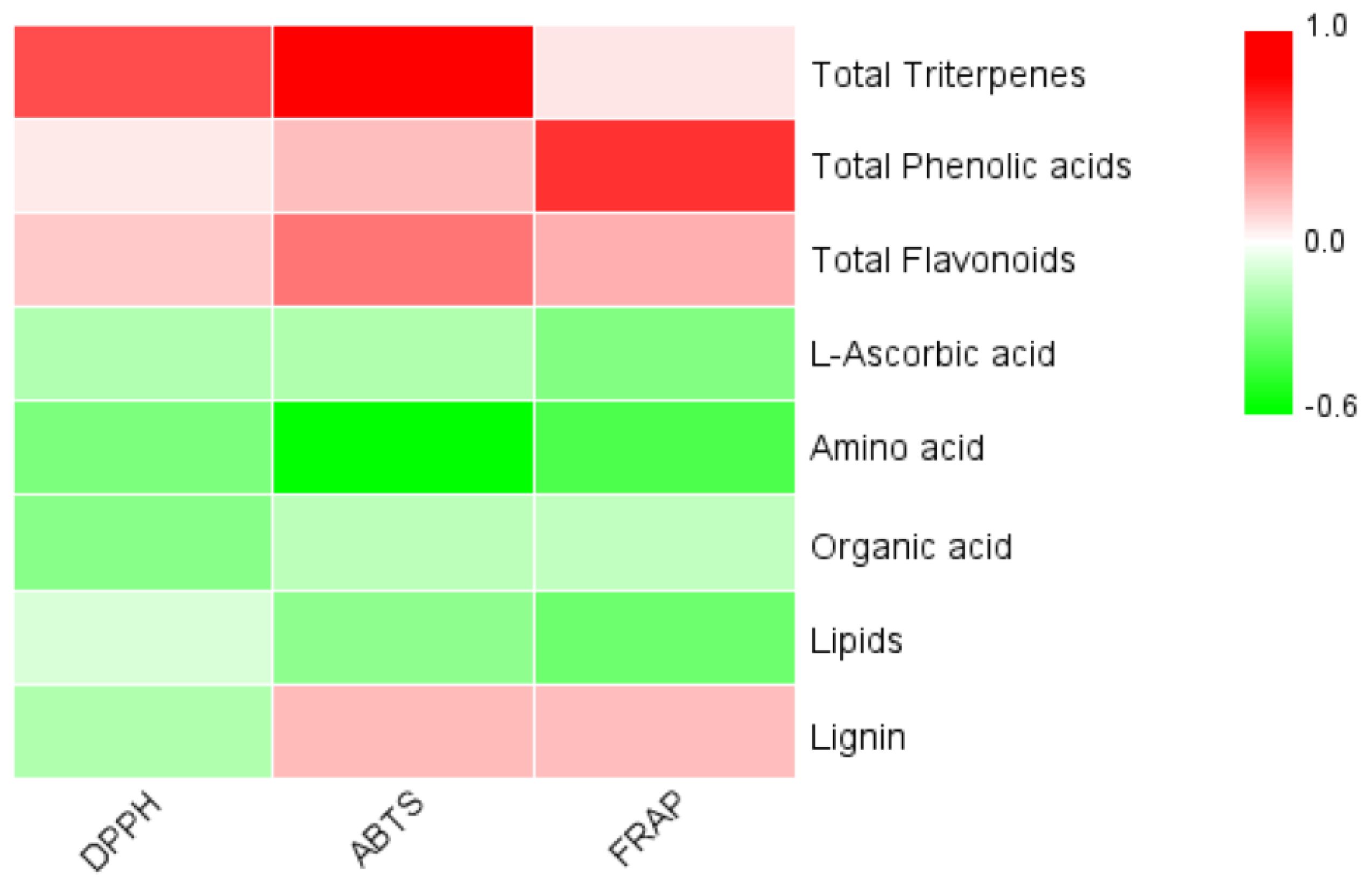
3.8. Correlation between Active Substance Content and Antioxidant Capacity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- An, H.M.; Liu, M.; Yang, M.; Fan, W.G. Analysis of Main Organic Acid Compositions in Rosa roxburghii Tratt. Sci. Agric. Sin. 2011, 44, 2094–2100. [Google Scholar] [CrossRef]
- Tao, L.; Li, J.; Fu, Y.; Yang, W. Determination of amino acids in Rosa Sterilis and wild Rosa roxburghii by HPLC. Shandong Chem. Ind. 2017, 46, 76–79. [Google Scholar] [CrossRef]
- Li, H.; Fang, W.; Wang, Z.; Chen, Y. Physicochemical, biological properties, and flavour profile of Rosa roxburghii tratt, pyracantha fortuneana, and Rosa laevigata michx fruits: A comprehensive review. Food Chem. 2021, 366, 130509. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.H.; Zhang, Q.; Zhang, Y.H.; Lu, X.Y.; Fu, W.M.; He, J.Y. Chemical analysis of dietary constituents in Rosa roxburghii and Rosa sterilis fruits. Molecules 2016, 21, 1204. [Google Scholar] [CrossRef]
- Zhou, G.Z.; Lu, M.; An, H.M. Analysis of Bioactive Substance Contents and Antioxidant Activities in Rosa roxburghii Fruits during Development. Food Sci. 2018, 39, 20–25. [Google Scholar] [CrossRef]
- Xu, J.; Vidyarthi, S.K.; Bai, W.; Pan, Z. Nutritional constituents, health benefits and processing of Rosa roxburghii: A review. J. Funct. Foods 2019, 60, 103456. [Google Scholar] [CrossRef]
- Wang, L.T.; Lv, M.J.; An, J.Y.; Fan, X.H.; Dong, M.Z.; Zhang, S.D.; Wang, J.D.; Wang, Y.Q.; Cai, Z.H.; Fu, Y.J. Botanical characteristics, phytochemistry and related biological activities of Rosa roxburghii tratt fruit, and its potential use in functional foods: A review. Food Funct. 2021, 12, 1432–1451. [Google Scholar] [CrossRef]
- Lu, M.; An, H.M.; Zhao, X.H. Analysis of Amino Acids in Rosa sterilis and Rosa roxburghii Fruits. Food Sci. 2015, 36, 118–121. [Google Scholar] [CrossRef]
- Zou, T.; Cheng, Z.J.; Ji, Y.; Cha, Q.; Zhou, D.; Ruan, P.J.; Ge, F.H. Study on the process of supercritical CO2 extraction of volatile substances of Rosa roxburghii Tratt flesh. J. Chin. Med. Mater. 2020, 43, 942–945. [Google Scholar] [CrossRef]
- Yang, Q.Q.; Zhang, D.; Ge, Y.Y.; Arakkaveettil, K.F.; Harold, C.; Gan, R.Y. Study on the phytochemical composition and biological activity of Rosa roxburghii Tratt fruit. In Summary of Papers from the 16th Annual Meeting of the China Food Science and Technology Association and the 10th China-U.S. Food Industry High-Level Forum; Chinese Institute of Food Science and Technology: Beijing, China, 2019. [Google Scholar]
- Ren, B.; Chen, C.; Li, C.; Fu, X.; You, L.; Liu, R.H. Optimization of microwave-assisted extraction of Sargassum thunbergii polysaccharides and its antioxidant and hypoglycemic activities. Carbohydr. Polym. 2017, 173, 192–201. [Google Scholar] [CrossRef]
- He, J.Y.; Zhang, Y.H.; Ma, N.; Zhang, X.L.; Liu, M.H.; Fu, W.M. Comparative analysis of multiple ingredients in Rosa roxburghii and R. sterilis fruits and their antioxidant activities. J. Funct. Foods 2016, 27, 29–41. [Google Scholar] [CrossRef]
- Wu, P.H.; Han, C.H.; Wu, M.H. Beneficial effects of hydroalcoholic extract from Rosa Roxburghii tratt fruit on hyperlipidemia in high-fat-fed rats. Acta Cardiol. Sin. 2020, 36, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, H.S.; Bai, J.; An, H.M. Genetic Diversity Analysis and Core Collection Construction in Rosa roxburghii Based on Fruits Quality and EST-SSR Mar. Mol. Plant Breed. 2020, 18, 3098–3106. [Google Scholar] [CrossRef]
- Fan, W.G.; Xiang, X.; An, H.M.; Liu, J.P. A new Rosa roxburghii cultivar ‘Guinong 5’. Acta Hortic Sin. 2011, 38, 1609–1610. [Google Scholar] [CrossRef]
- Yan, N.; Du, Y.M.; Liu, X.M.; Chu, M.J.; Shi, J.; Zhang, H.B.; Liu, Y.H.; Zhang, Z.F. A comparative UHPLC-QqQ-MS-based metabolomics approach for evaluating Chinese and North American wild rice. Food Chem. 2019, 275, 618–627. [Google Scholar] [CrossRef]
- Li, H.Y.; Lv, Q.Y.; Liu, A.K.; Wang, J.R.; Sun, X.Q.; Deng, J.; Chen, Q.F.; Wu, Q. Comparative metabolomics study of Tartary (Fagopyrum tataricum (L.) Gaertn) and common (Fagopyrum esculentum Moench) buckwheat seeds. Food Chem. 2022, 371, 131125. [Google Scholar] [CrossRef]
- Carlos, G.F.; Brian, H.C.; Ronald, J.M.; Erika, M.Z. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, xcms, and chemometrics. Anal Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Ru, J.; Li, P.; Wang, J.; Zhou, W.; Li, B.; Huang, C. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014, 6, 13. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acid Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Meda, A.; Lamien, C.E.; Romito, M.; Millogo, J.; Nacoulma, O.G. Determination of the total phenolic, flavonoid and proline contents in Burkina Fasan honey, as well as their radical scavenging activity. Food Chem. 2005, 91, 571–577. [Google Scholar] [CrossRef]
- Huang, R.S.; Yu, Y.X.; Yan, H.; Sheng, X.B. Determination of total flavonoids in abrus cantoniensis and its dynamic changes. China J. Chin. Mater. Med. 2006, 31, 1428–1431. [Google Scholar]
- Zhou, Q.X.; Zhang, J.S.; Gu, M.; Gu, Z.L. Determination of total triterpenoid acids in Hawthorn and its extract. Chin. Wild Plant Resour. 2004, 5, 43–44. [Google Scholar] [CrossRef]
- Wang, X.K.; Huang, J.L. Principles and Techniques of Plant Physiological Biochemical Experiment; Higher Education Press: Beijing, China, 2015. [Google Scholar]
- Moure, A.; Franco, D.; Sineiro, J.; Domínguez, H.; Núñez, M.J.; Lema, J.M. Antioxidant activity of extracts from Gevuina avellana and Rosa rubiginosa defatted seeds. Food Res. Int. 2001, 34, 103–109. [Google Scholar] [CrossRef]
- Martinez-Villaluenga, C.; Peñas, E.; Ciska, E.; Piskuła, M.K.; Kozlowska, H.; Vidal-Valverde, C. Time dependence of bioactive compounds and antioxidant capacity during germination of different cultivars of broccoli and radish seeds. Food Chem. 2010, 120, 710–716. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of ‘‘antioxidant power’’: The FRAP assay. Anal Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.T.; Lu, M.; Wang, D.J.; An, H.M. Morphological structure of trichomes on Rosa sterilis fruit and expression of associated genes during development. Plant Physiol. J. 2021, 57, 362–372. [Google Scholar] [CrossRef]
- Yan, H.Q.; Wu, Z.M.; Liu, Y.J.; Weng, Q.B.; Yi, Y.; Huang, X.L. Functional divergence of RrGL3 and RrEGL3 from Rosa roxburghii in mediating trichome development. Plant Cell Tissue Organ Cult. (PCTOC) 2021, 147, 313–324. [Google Scholar] [CrossRef]
- Ma, W.T.; Lu, M.; Ludlow, A.R.; Wang, D.J.; Zeng, J.W.; An, H.M. Contrastive analysis of trichome distribution, morphology, structure, and associated gene expression reveals the formation factors of different trichome types in two commercial rosa species. Sci Hortic. 2021, 285, 110131. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Lu, M.; Ludlow, R.A.; Ma, W.T.; An, H.M. Transcriptome analysis reveals candidate genes for dietary fiber metabolism in rosa roxburghii fruit grown under different light intensities. Hortic. Environ. Biotechnol. 2021, 62, 751–764. [Google Scholar] [CrossRef]
- Wang, D.J.; Lu, M.; Ludlow, R.A.; Zeng, J.W.; Ma, W.T.; An, H.M. Comparative ultrastructure of trichomes on various organs of Rosa roxburghii. Microsc. Res. Tech. 2021, 84, 2095–2103. [Google Scholar] [CrossRef]
- Li, J.H.; Yang, T.G.; Zhang, N.; Sun, Y.J.; Qu, Y.X.; Cao, Y.J.; Lin, H.Y.; Tao, O. Rapid Identification of Chemical Constituents in the Seed of Rosa roxburghii by UPLC-Q-TOF-MSE Combined with UNIFI Informatics Platform. J. Chin. Mass Spectrom. Soc. 2020, 41, 76–86. [Google Scholar] [CrossRef]
- Zhang, D.; Wei, G.X.; Wang, W.; Feng, F.; Zeng, F.K. Comparative research on basic ingredients and volatile aroma compounds of Rosa roxburghii Tratt and Rosa sterilis D. shi. Sci. Technol. Food Ind. 2016, 37, 149–154, 177. [Google Scholar]
- Werker, E. Trichome diversity and development. Adva. Bot. Res. 2000, 31, 1–35. [Google Scholar]
- Alves, K.C.M.; Gobbo-Neto, L.; Lopes, N.P. Sesquiterpene lactones and flavonoids from Lychnophora reticulata Gardn. (Asteraceae). Biochem. Syst. Ecol. 2008, 36, 434–436. [Google Scholar] [CrossRef]
- Andreucci, A.C.; Ciccarelli, D.; Desideri, I.; Pagni, A.M. Glandular hairs and secretory ducts of Matricaria chamomilla (Asteraceae):Morphology and histochemistry. Ann. Bot. Fenn. 2008, 45, 11–18. [Google Scholar] [CrossRef]
- Chen, O.; Deng, L.; Ruan, C.; Yi, L.; Zeng, L. Pichia galeiformis Induces Resistance in Postharvest Citrus by Activating the Phenylpropanoid Biosynthesis Pathway. J. Agric. Food Chem. 2021, 69, 2619–2631. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Kobayashi, T.; Kamata, K. Role of Lysophosphatidylcholine (LPC) in Atherosclerosis. Curr. Med. Chem. 2007, 14, 3209–3220. [Google Scholar] [CrossRef] [Green Version]
- Mayne, S.T. Antioxidant nutrients and chronic disease: Use of biomarkers of exposure and oxidative stress status in epidemiologic research. J. Nutr. 2003, 133 (Suppl. 3), 933S–940S. [Google Scholar] [CrossRef] [Green Version]
- Kawaguchi, K.; Matsumoto, T.; Kumazawa, Y. Effects of antioxidant polyphenols on tnf-alpha-related diseases. Curr. Top. Med. Chem. 2011, 11, 1767–1779. [Google Scholar] [CrossRef] [Green Version]
- Rensburg, C.; Erasmus, E.; Loots, D.T.; Oosthuizen, W.; Jerling, J.C.; Kruger, H.S.; Louw, R.; Brits, M.; Westhuizen, F.H. Rosa roxburghii supplementation in a controlled feeding study increases plasma antioxidant capacity and glutathione redox state. Eur. J. Nutr. 2005, 44, 452–457. [Google Scholar] [CrossRef]
- Chen, F.J.; Li, L.Q.; Peng, M.; Yan, Y.F.; Wang, L.; Li, L.L.; Yang, L.S.; Wang, Y.; Yang, J.; Yan, Y.; et al. Identification of triterpenoids and hepatoprotective property of fru ctus Rosa roxburghii against alcohol-induced liver injury by regulating keap1-Nrf2 signaling. Phytomedicine 2021, 1, 100102. [Google Scholar] [CrossRef]

| Single Fruit Weight (g) | Longitudinal Diameter (cm) | Transverse Diameter (cm) | Shape Index | Titratable Acidity (%) | Soluble Solids (%) | Soluble Solids to Acidity Ratio | |
|---|---|---|---|---|---|---|---|
| Rr-1 | 17.22 ± 1.03 c | 3.69 ± 0.11 a | 3.38 ± 0.13 b | 1.11 ± 0.05 a | 1.81 ± 0.13 a | 12.30 ± 0.41 b | 7.08 ± 0.45 a |
| Rr-3 | 25.50 ± 1.78 ab | 3.39 ± 0.11 ab | 4.37 ± 0.07 a | 0.78 ± 0.04 b | 1.58 ± 0.19 a | 11.73 ± 0.44 b | 7.66 ± 0.94 a |
| Rr-4 | 23.65 ± 2.17 b | 3.15 ± 0.13 bc | 4.12 ± 0.19 a | 0.77 ± 0.02 b | 1.52 ± 0.10 a | 11.30 ± 0.65 b | 7.45 ± 0.28 a |
| Rr-5 | 29.48 ± 1.12 a | 3.16 ± 0.03 bc | 4.64 ± 0.09 a | 0.68 ± 0.01 b | 1.81 ± 0.23 a | 10.30 ± 0.25 b | 5.99 ± 0.66 a |
| Rr-7 | 13.94 ± 0.90 c | 2.78 ± 0.13 cd | 3.27 ± 0.06 b | 0.84 ± 0.03 b | 2.02 ± 0.23 a | 14.88 ± 0.55 a | 7.68 ± 0.98 a |
| Rr-f | 13.89 ± 1.09 c | 2.58 ± 0.07 d | 3.21 ± 0.09 b | 0.80 ± 0.01 b | 2.17 ± 0.22 a | 11.48 ± 0.85 b | 5.44 ± 0.56 a |
| L-Ascorbic Acid (mg/g Protein) | Total Flavonoids (mg/100 g) | Total Phenolic Acids (mg/100 g) | Total Triterpenes (mg/100 g) | Amino Acid (umol/g) | Organic Acid (%) | Lignin (mg/100 g) | Lipids (%) | DPPH (mmol/L) | FRAP (mmol/L) | ABTS (mmol/L) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Rr-1 | 851.96 ± 112.27 ab | 958.92 ± 78.85 a | 1190.15 ± 109.68 bc | 3062.30 ± 195.02 bc | 89.64 ± 2.63 ab | 3.28 ± 0.14 ab | 2063.67 ± 153.34 bc | 1.50 ± 0.27 a | 8.84 ± 0.06 a | 2.39 ± 0.30 b | 4.45 ± 0.30 c |
| Rr-3 | 924.91 ± 117.64 ab | 1025.16 ± 32.72 a | 1558.38 ± 169.39 ab | 3306.17 ± 111.23 ab | 108.85 ± 6.56 a | 3.49 ± 0.32 ab | 1900.04 ± 253.65 c | 1.45 ± 0.32 a | 9.26 ± 0.27 a | 4.45 ± 0.45 ab | 5.25 ± 0.20 abc |
| Rr-4 | 1044.99 ± 123.88 ab | 509.82 ± 58.35 b | 857.48 ± 118.01 c | 2281.02 ± 262.28 cd | 88.08 ± 4.77 b | 2.79 ± 0.23 b | 2213.40 ± 270.76 abc | 1.56 ± 0.60 a | 8.75 ± 0.16 a | 4.62 ± 0.63 ab | 4.46 ± 0.08 c |
| Rr-5 | 1209.30 ± 129.95 a | 1141.13 ± 84.53 a | 1152.05 ± 122.29 bc | 3209.38 ± 297.73 b | 46.81 ± 4.65 c | 3.74 ± 0.29 a | 2962.14 ± 252.28 a | 1.51 ± 0.53 a | 8.77 ± 0.23 a | 5.05 ± 0.77 ab | 5.32 ± 0.36 ab |
| Rr-7 | 672.14 ± 38.60 b | 1172.26 ± 53.10 a | 1945.55 ± 57.68 a | 4014.20 ± 199.50 a | 43.43 ± 2.81 c | 3.16 ± 0.01 ab | 2683.00 ± 180.11 ab | 1.30 ± 0.41 a | 9.32 ± 0.22 a | 8.17 ± 2.08 a | 6.00 ± 0.22 a |
| Rr-f | 886.82 ± 123.25 ab | 1000.72 ± 161.42 a | 1731.80 ± 118.41 a | 2110.92 ± 247.97 d | 104.81 ± 9.9 ab | 3.49 ± 0.18 ab | 2101.20 ± 111.44 bc | 1.16 ± 0.50 a | 8.39 ± 0.52 a | 6.75 ± 1.22 a | 4.55 ± 0.22 bc |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, L.; Lu, M.; Rao, T.; Liu, Z.; Wu, X.; An, H. Comparative Analysis of Fruit Metabolome Using Widely Targeted Metabolomics Reveals Nutritional Characteristics of Different Rosa roxburghii Genotypes. Foods 2022, 11, 850. https://doi.org/10.3390/foods11060850
Jiang L, Lu M, Rao T, Liu Z, Wu X, An H. Comparative Analysis of Fruit Metabolome Using Widely Targeted Metabolomics Reveals Nutritional Characteristics of Different Rosa roxburghii Genotypes. Foods. 2022; 11(6):850. https://doi.org/10.3390/foods11060850
Chicago/Turabian StyleJiang, Lanlan, Min Lu, Tianzhi Rao, Zeyang Liu, Xiaomao Wu, and Huaming An. 2022. "Comparative Analysis of Fruit Metabolome Using Widely Targeted Metabolomics Reveals Nutritional Characteristics of Different Rosa roxburghii Genotypes" Foods 11, no. 6: 850. https://doi.org/10.3390/foods11060850
APA StyleJiang, L., Lu, M., Rao, T., Liu, Z., Wu, X., & An, H. (2022). Comparative Analysis of Fruit Metabolome Using Widely Targeted Metabolomics Reveals Nutritional Characteristics of Different Rosa roxburghii Genotypes. Foods, 11(6), 850. https://doi.org/10.3390/foods11060850





