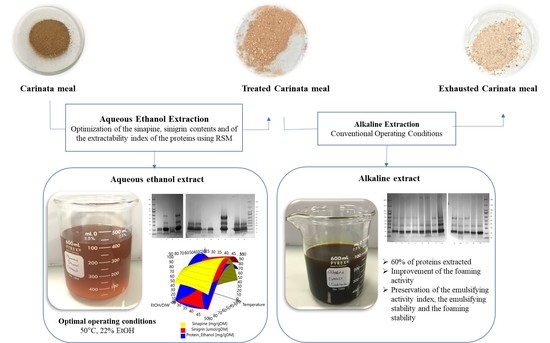
Optimization of the Recovery of Secondary Metabolites from Defatted Brassica carinata Meal and Its Effects on the Extractability and Functional Properties of Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Carinata Meal Samples and Their Characterization
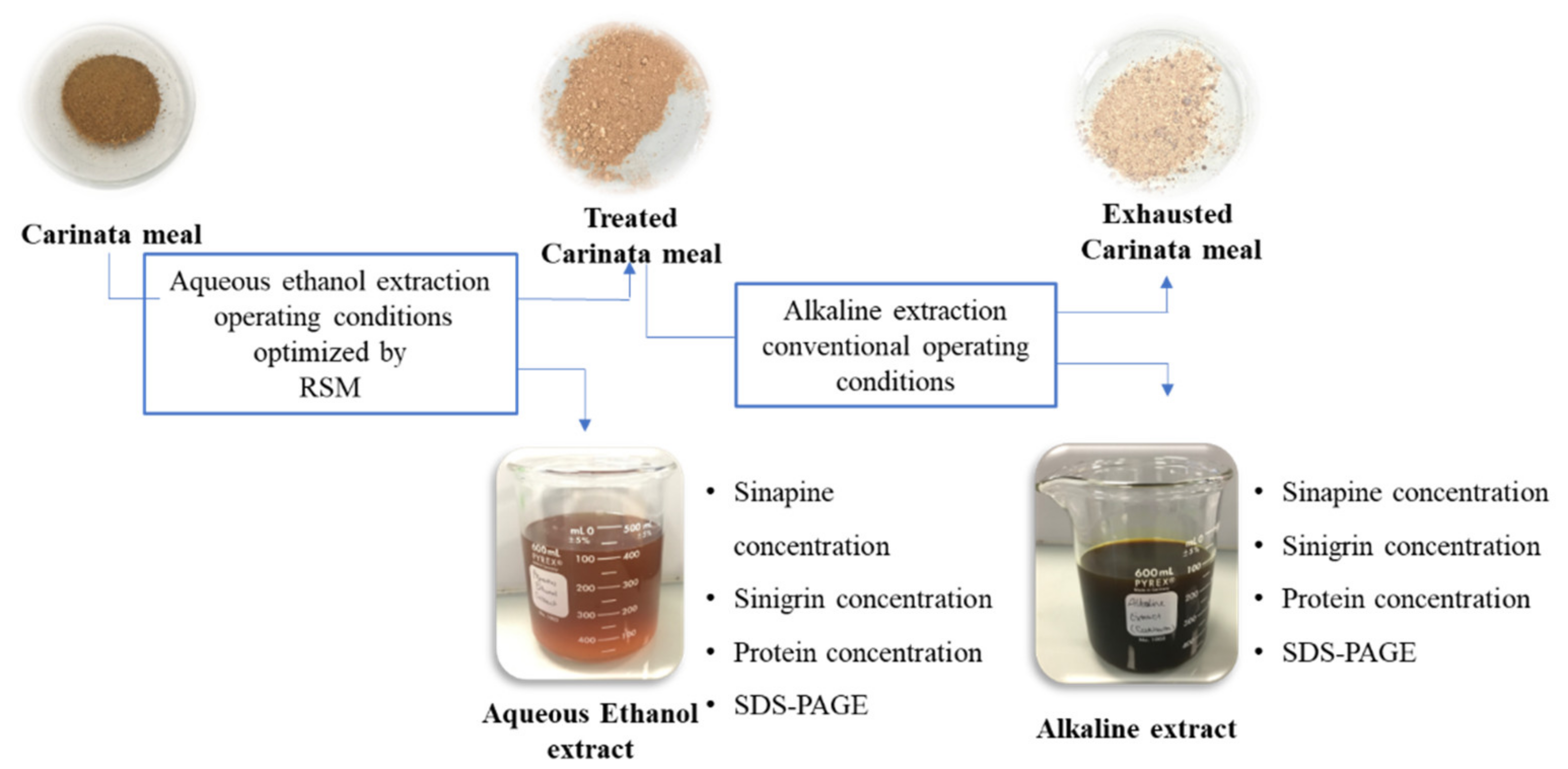
2.3. Fractionation Process of Carinata Meal
2.4. Determination of Phenolic Compound and Glucosinolate Contents
2.5. Determination of Protein Content of AE Extracts
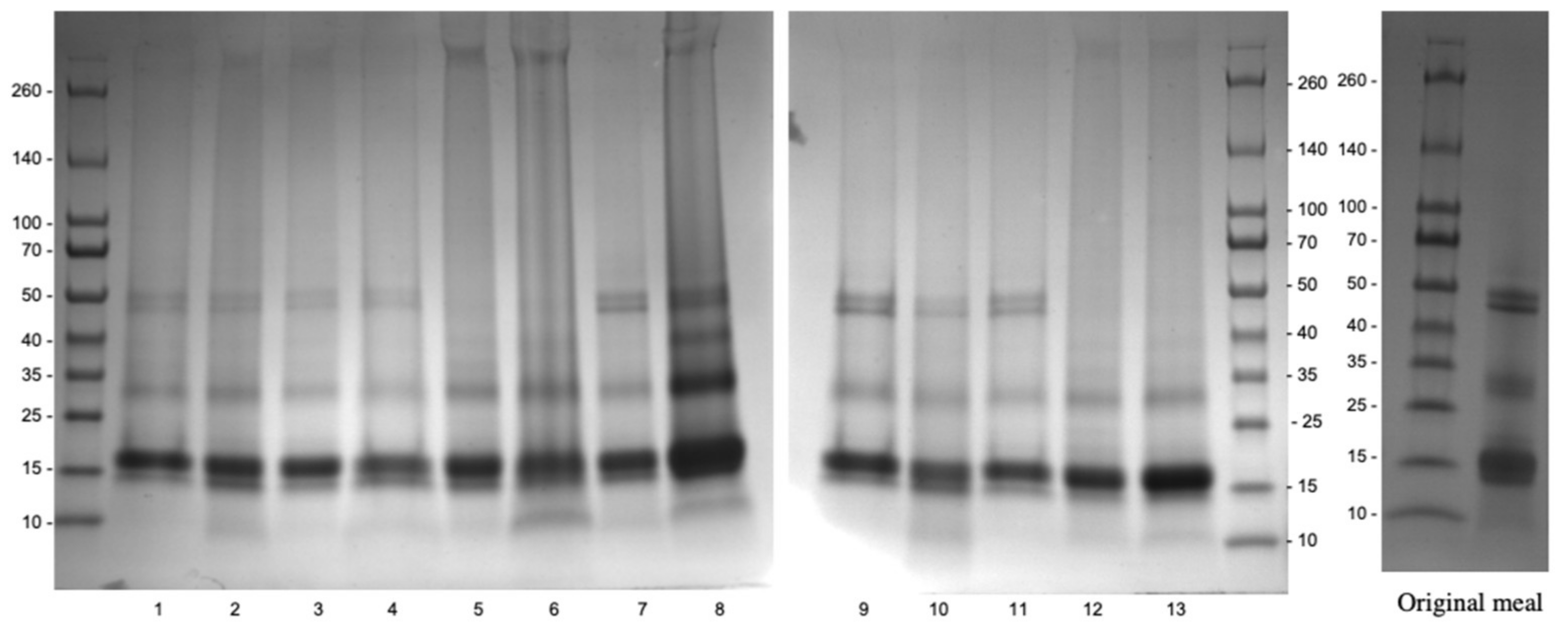
2.6. Analysis of Polypeptide Profile
2.7. Optimization of the AE Extraction of Secondary Metabolites
2.8. Determination of Optimal Operating Conditions
2.9. Functional Properties
2.9.1. Preparation of Acid-Precipitated Isoelectric Protein Isolates
2.9.2. Emulsifying Activity Index
2.9.3. Foam Expansion (FE) and Foam Stability (FS)
2.10. Statistical Analysis
3. Results
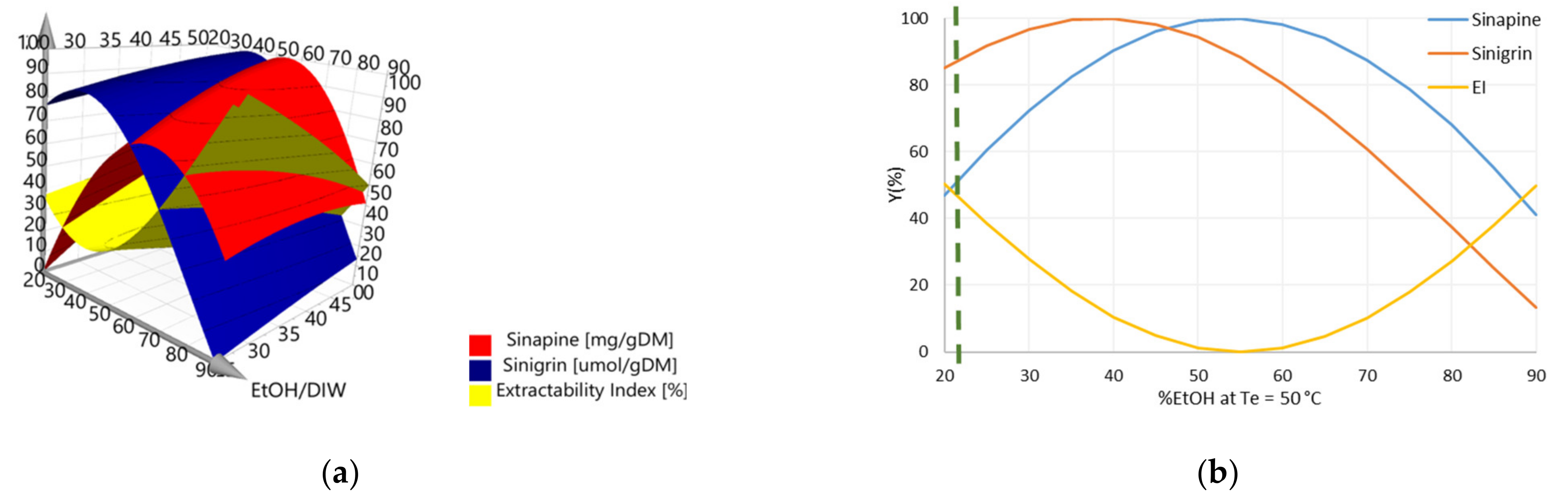
3.1. Optimization of Aqueous Ethanol Extraction using Response Surface Methodology
3.1.1. Model Adequacy
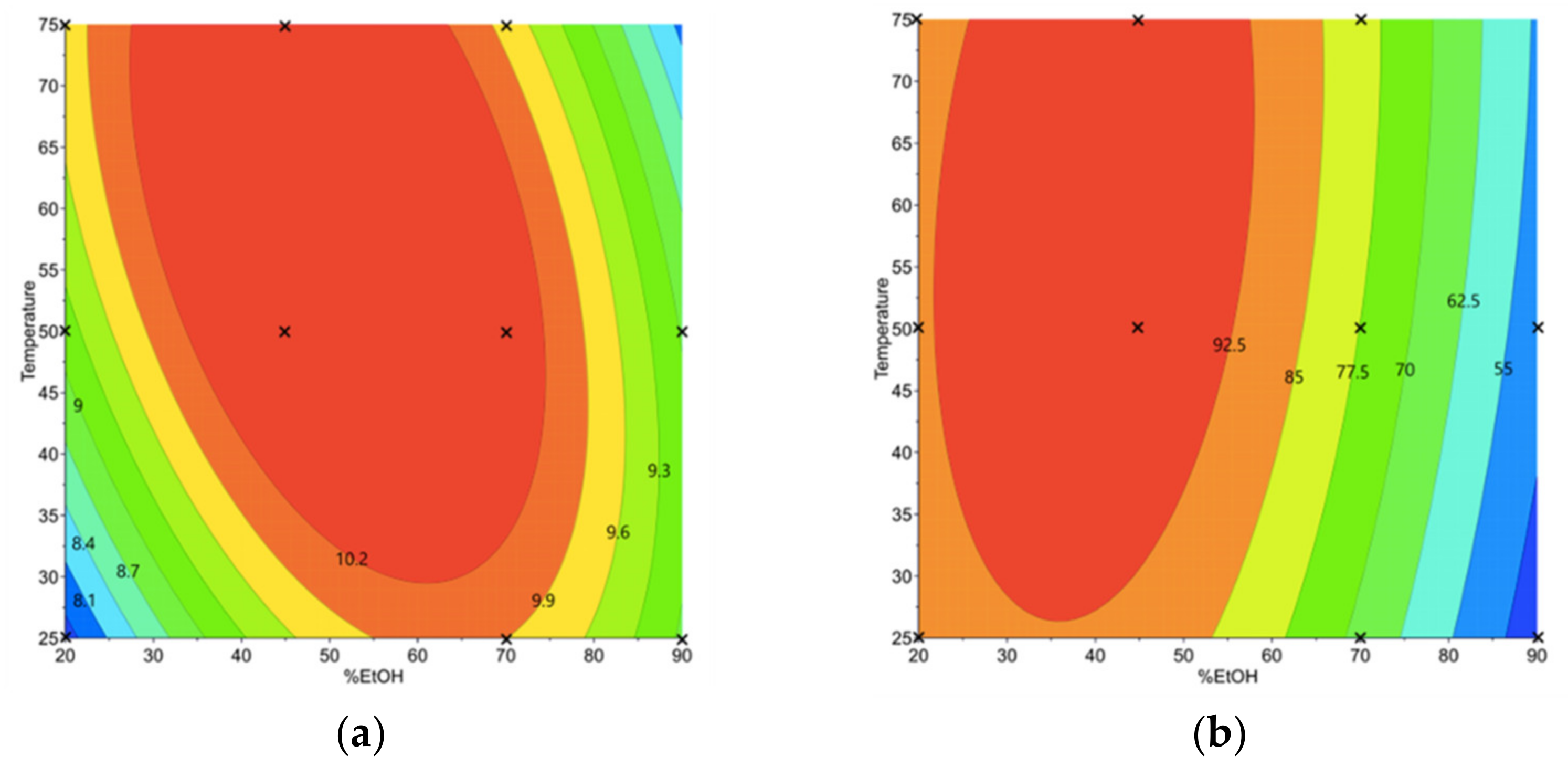
3.1.2. Phenolic Compound and Sinigrin Contents in Aqueous Ethanol Extracts (Y1 and Y2)
3.1.3. Determination of the Optimal Conditions of Aqueous Ethanol Extraction and Validation of the Prediction Models
3.2. Effects of Aqueous Ethanol Extraction on Carinata Proteins
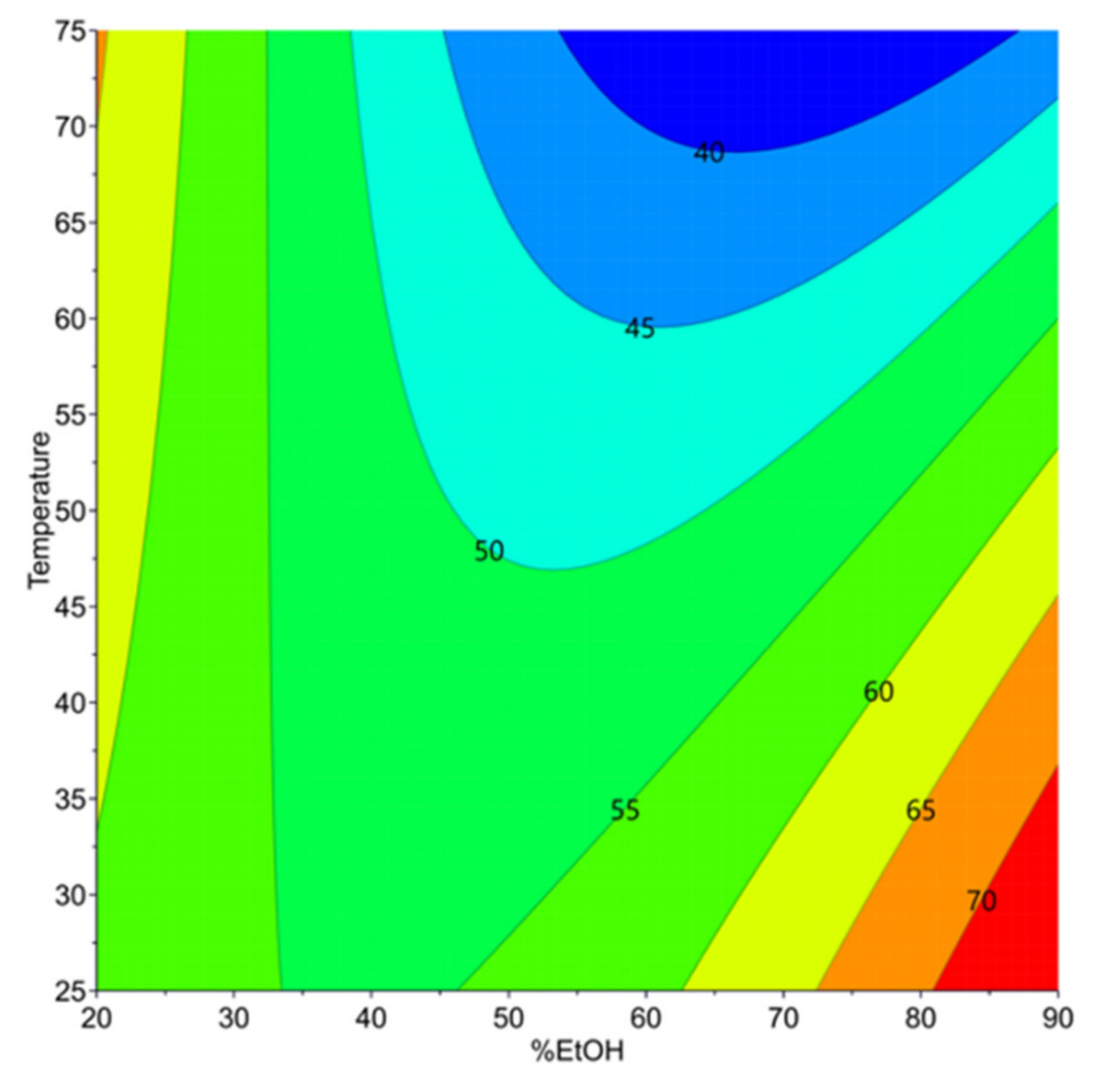
3.2.1. Extractability of Proteins Resided in Carinata Treated Meal (Y3)
3.2.2. Effects of Optimal Aqueous Ethanol Extraction on Carinata Protein Profile
3.3. Determination of Operating Conditions for the Optimization of Y1, Y2, and Y3
3.4. Effect of the New Operating Conditions on Functional Properties of Carinata Proteins upon Aqueous Ethanol Extraction
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christ, B.; Bartels, W.L.; Broughton, D.; Seepaul, R.; Geller, D. In Pursuit of a Homegrown Biofuel: Navigating Systems of Partnership, Stakeholder Knowledge, and Adoption of Brassica Carinata in the Southeast United States. Energy Res. Soc. Sci. 2020, 70, 101665. [Google Scholar] [CrossRef]
- SPARC. Southeast Partnership for Advanced Renewables from Carinata. Available online: https://Sparc-Cap.Org (accessed on 22 June 2021).
- Pedroche, J.; Yust, M.M.; Lqari, H.; Girón-Calle, J.; Alaiz, M.; Vioque, J.; Millán, F. Brassica Carinata Protein Isolates: Chemical Composition, Protein Characterization and Improvement of Functional Properties by Protein Hydrolysis. Food Chem. 2004, 88, 337–346. [Google Scholar] [CrossRef]
- Seepaul, R.; Small, I.M.; Mulvaney, M.J.; George, S.; Leon, R.G.; Geller, D.; Wright, D.L. Carinata, the Sustainable Crop for a Bio-Based Economy: 2018–2019 Production Recommendations for the Southeastern United States; University of Florida Institute of Food and Agricultural Sciences: Gainesville, FL, USA, 2019; pp. 1–12. [Google Scholar]
- Aider, M.; Barbana, C. Canola Proteins: Composition, Extraction, Functional Properties, Bioactivity, Applications as a Food Ingredient and Allergenicity—A Practical and Critical Review. Trends Food Sci. Technol. 2011, 22, 21–39. [Google Scholar] [CrossRef]
- Tan, S.H.; Mailer, R.J.; Blanchard, C.L.; Agboola, S.O. Extraction and Characterization of Protein Fractions from Australian Canola Meals. Food Res. Int. 2011, 44, 1075–1082. [Google Scholar] [CrossRef]
- Wanasundara, J.P.D. Proteins of Brassicaceae Oilseeds and Their Potential as a Plant Protein Source. Crit. Rev. Food Sci. Nutr. 2011, 51, 635–677. [Google Scholar] [CrossRef]
- Akbari, A.; Wu, J. An Integrated Method of Isolating Napin and Cruciferin from Defatted Canola Meal. LWT Food Sci. Technol. 2015, 64, 308–315. [Google Scholar] [CrossRef]
- Wu, J.; Muir, A.D. Comparative Structural, Emulsifying, and Biological Properties of 2 Major Canola Proteins, Cruciferin and Napin. J. Food Sci. 2008, 73, M1–M8. [Google Scholar] [CrossRef]
- Akbari, A.; Lavasanifar, A.; Wu, J. Interaction of Cruciferin-Based Nanoparticles with Caco-2 Cells and Caco-2/HT29-MTX Co-Cultures. Acta Biomater. 2017, 64, 249–258. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Y.; Ju, X.; Udenigwe, C.C.; He, R. Polyelectrolyte Complex Nanoparticles from Chitosan and Acylated Rapeseed Cruciferin Protein for Curcumin Delivery. J. Agric. Food Chem. 2018, 66, 2685–2693. [Google Scholar] [CrossRef]
- Chen, R.; Wang, X.-J.; Zhang, Y.-Y.; Xing, Y.; Yang, L.; Ni, H.; Li, H.-H. Simultaneous Extraction and Separation of Oil, Proteins, and Glucosinolates from Moringa Oleifera Seeds. Food Chem. 2019, 300, 125162. [Google Scholar] [CrossRef]
- Nguyen, V.P.T.; Stewart, J.D.; Ioannou, I.; Allais, F. Sinapic Acid and Sinapate Esters in Brassica: Innate Accumulation, Biosynthesis, Accessibility via Chemical Synthesis or Recovery from Biomass, and Biological Activities. Front. Chem. 2021, 9, 664602. [Google Scholar] [CrossRef] [PubMed]
- Mazumder, A.; Dwivedi, A.; du Plessis, J. Sinigrin and Its Therapeutic Benefits. Molecules 2016, 21, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nićiforović, N.; Abramovič, H. Sinapic Acid and Its Derivatives: Natural Sources and Bioactivity. Compr. Rev. Food Sci. Food Saf. 2014, 13, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.R.; Erdman, J.W. Phytic Acid in Health and Disease. Crit. Rev. Food Sci. Nutr. 1995, 35, 495–508. [Google Scholar] [CrossRef]
- Griffiths, D.W.; Birch, A.N.E.; Hillman, J.R. Antinutritional Compounds in the Brassicaceae: Analysis, Biosynthesis, Chemistry and Dietary Effects. J. Hortic. Sci. Biotechnol. 1998, 73, 1–18. [Google Scholar] [CrossRef]
- Chuo, S.C.; Nasir, H.M.; Mohd-Setapar, S.H.; Mohamed, S.F.; Ahmad, A.; Wani, W.A.; Muddassir, M.; Alarifi, A. A Glimpse into the Extraction Methods of Active Compounds from Plants. Crit. Rev. Anal. Chem. 2020, 20, 1–30. [Google Scholar] [CrossRef]
- Fang, J.; Reichelt, M.; Kai, M.; Schneider, B. Metabolic Profiling of Lignans and Other Secondary Metabolites from Rapeseed (Brassica napus L.). J. Agric. Food Chem. 2012, 60, 10523–10529. [Google Scholar] [CrossRef]
- Flourat, A.L.; Willig, G.; Teixeira, A.R.S.; Allais, F. Eco-Friendly Extraction of Sinapine from Residues of Mustard Production. Front. Sustain. Food Syst. 2019, 3, 12. [Google Scholar] [CrossRef]
- Li, J.; Guo, Z. Complete Utilization of Rapeseed Meal to Produce Lipophilic Antioxidants, Protein, and Monosugars in a Concordant Manner. ACS Sustain. Chem. Eng. 2017, 5, 6218–6226. [Google Scholar] [CrossRef]
- Reungoat, V.; Gaudin, M.; Flourat, A.L.; Isidore, E.; Mouterde, L.M.M.; Allais, F.; Ducatel, H.; Ioannou, I. Optimization of an Ethanol/Water-Based Sinapine Extraction from Mustard Bran Using Response Surface Methodology. Food Bioprod. Process. 2020, 122, 322–331. [Google Scholar] [CrossRef]
- Laguna, O.; Odinot, E.; Bisotto, A.; Baréa, B.; Villeneuve, P.; Sigoillot, J.C.; Record, E.; Faulds, C.B.; Fine, F.; Lesage-Meessen, L.; et al. Release of Phenolic Acids from Sunflower and Rapeseed Meals Using Different Carboxylic Esters Hydrolases from Aspergillus Niger. Ind. Crop. Prod. 2019, 139, 111579. [Google Scholar] [CrossRef]
- Lin, L.-Z.; Harnly, J.M. Phenolic Component Profiles of Mustard Greens, Yu Choy, and 15 Other Brassica Vegetables. J. Agric. Food Chem. 2010, 58, 6850–6857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalaydzhiev, H.; Georgiev, R.; Ivanova, P.; Stoyanova, M.; Silva, C.L.M.; Chalova, V.I. Enhanced Solubility of Rapeseed Meal Protein Isolates Prepared by Sequential Isoelectric Precipitation. Foods 2020, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Wanasundara, J.P.D.; McIntosh, T.C.; Perera, S.P.; Withana-Gamage, T.S.; Mitra, P. Canola/Rapeseed Protein-Functionality and Nutrition. OCL 2016, 23, D407. [Google Scholar] [CrossRef] [Green Version]
- Kalaydzhiev, H.; Ivanova, P.; Stoyanova, M.; Pavlov, A.; Rustad, T.; Silva, C.L.M.; Chalova, V.I. Valorization of Rapeseed Meal: Influence of Ethanol Antinutrients Removal on Protein Extractability, Amino Acid Composition and Fractional Profile. Waste Biomass Valorization 2020, 11, 2709–2719. [Google Scholar] [CrossRef]
- Mouterde, L.M.M.; Peru, A.A.M.; Mention, M.M.; Brunissen, F.; Allais, F. Sustainable Straightforward Synthesis and Evaluation of the Antioxidant and Antimicrobial Activity of Sinapine and Analogues. J. Agric. Food Chem. 2020, 68, 6998–7004. [Google Scholar] [CrossRef]
- AOAC International. AOAC Official Methods for Analysis Association of Official Analytical Chemists; AOAC International: Rockville, MD, USA, 2000. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Reichwald, K.; Hatzack, F. Application of a Modified Haug and Lantzsch Method for the Rapid and Accurate Photometrical Phytate Determination in Soybean, Wheat, and Maize Meals. J. Agric. Food Chem. 2008, 56, 2888–2891. [Google Scholar] [CrossRef]
- Grosser, K.; van Dam, N.M. A Straightforward Method for Glucosinolate Extraction and Analysis with High-Pressure Liquid Chromatography (HPLC). J. Vis. Exp. 2017, 121, 55425. [Google Scholar] [CrossRef] [Green Version]
- Perera, S.P.; McIntosh, T.C.; Wanasundara, J.P.D. Structural Properties of Cruciferin and Napin of Brassica Napus (Canola) Show Distinct Responses to Changes in PH and Temperature. Plants 2016, 5, 64–74. [Google Scholar] [CrossRef] [Green Version]
- Aluko, R.E.; McIntosh, T. Polypeptide Profile and Functional Properties of Defatted Meals and Protein Isolates of Canola Seeds. J. Sci. Food Agric. 2001, 81, 391–396. [Google Scholar] [CrossRef]
- Pearce, K.N.; Kinsella, J.E. Emulsifying Properties of Proteins: Evaluation of a Turbidimetric Technique. J. Agric. Food Chem. 1978, 26, 716–723. [Google Scholar] [CrossRef]
- Xin, H.; Yu, P. Chemical Profile, Energy Values, and Protein Molecular Structure Characteristics of Biofuel/Bio-Oil Co-Products (Carinata Meal) in Comparison with Canola Meal. J. Agric. Food Chem. 2013, 61, 3926–3933. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, F.; Valentão, P.; Sousa, C.; Pereira, J.A.; Seabra, R.M.; Andrade, P.B. Chemical and Antioxidative Assessment of Dietary Turnip (Brassica rapa var. Rapa L.). Food Chem. 2007, 105, 1003–1010. [Google Scholar] [CrossRef]
- Laguna, O.; Barakat, A.; Alhamada, H.; Durand, E.; Baréa, B.; Fine, F.; Villeneuve, P.; Citeau, M.; Dauguet, S.; Lecomte, J. Production of Proteins and Phenolic Compounds Enriched Fractions from Rapeseed and Sunflower Meals by Dry Fractionation Processes. Ind. Crop. Prod. 2018, 118, 160–172. [Google Scholar] [CrossRef]
- Nguyen, V.P.T.; Stewart, J.; Lopez, M.; Ioannou, I.; Allais, F. Glucosinolates: Natural Occurrence, Biosynthesis, Accessibility, Isolation, Structures, and Biological Activities. Molecules 2020, 25, 4537. [Google Scholar] [CrossRef] [PubMed]
- Fetzer, A.; Herfellner, T.; Stäbler, A.; Menner, M.; Eisner, P. Influence of Process Conditions during Aqueous Protein Extraction upon Yield from Pre-Pressed and Cold-Pressed Rapeseed Press Cake. Ind. Crop. Prod. 2018, 112, 236–246. [Google Scholar] [CrossRef]
| Experiments | %EtOH (%) X1 | Te (°C) X2 |
|---|---|---|
| 0 | 0 | 25 |
| 1 | 70 | 50 |
| 2 | 20 | 25 |
| 3 | 70 | 50 |
| 4 | 20 | 50 |
| 5 | 45 | 50 |
| 6 | 20 | 75 |
| 7 | 70 | 25 |
| 8 | 70 | 50 |
| 9 | 90 | 25 |
| 10 | 20 | 25 |
| 11 | 90 | 50 |
| 12 | 45 | 75 |
| 13 | 70 | 75 |
| Factors | Coefficient Values | ||
|---|---|---|---|
| Y1 | Y2 | Y3 | |
| Constant | 1.0333 | 1.9744 | −1.7016 |
| %EtOH | 0.0028 NS | −0.0864 | −0.0080 NS |
| Te | 0.0144 NS | 0.0221 NS | −0.0529 |
| %EtOH×%EtOH | −0.0457 | −0.0847 | 0.0818 |
| Te × Te | −0.0145 NS | −0.0126 NS | −0.0108 NS |
| %EtOH× Te | −0.0220 | 0.0143 NS | −0.0717 |
| R2 | 0.869 | 0.929 | 0.849 |
| R2 adjusted | 0.775 | 0.870 | 0.763 |
| Regression (p-value) | 5.4 × 10−3 | 2.2 × 10−3 | 5.3 × 10−3 |
| Lack of fit | 0.324 | 0.708 | 0.273 |
| Reproducibility | 0.847 | 0.820 | 0.900 |
| Condition number | 4.654 | 4.683 | 3.605 |
| Te (°C) | Optimal %EtOH | Y1 (mg/gDM) | Y2 (µmol/gDM) | Y3 (%) |
|---|---|---|---|---|
| 25 | 67% | 9.94 a ± 0.08 | 71.47 b ± 0.90 | 62.15 a ± 0.56 |
| 30 | 70% | 10.10 a ± 0.08 | 70.31 b,c ± 0.92 | 61.56 a ± 0.57 |
| 35 | 73% | 10.13 a ± 0.09 | 68.63 c,d ± 0.92 | 60.85 a,b ± 0.58 |
| 40 | 77% | 10.01 a ± 0.09 | 66.89 d ± 0.93 | 60.45 b ± 0.59 |
| 45 | 23% | 9.09 b ± 0.09 | 92.19 a ± 0.93 | 59.94 b ± 0.60 |
| 50 | 22% | 9.15 b ± 0.09 | 91.02 a ± 0.93 | 61.12 a ± 0.61 |
| Type of Meal | EAI (m2/g) | ES (%) | FA (%) | FS (%) |
|---|---|---|---|---|
| Original meal | 10.7 ± 1.4 | 63.5 ± 0.2 | 123.8 ± 1.8 | 94.9 ± 1.5 |
| Treated meal | 10.0 ± 0.6 | 61.4 ± 1.1 | 147.3 ± 3.2 | 94.2 ± 1.6 |
| Student’s test (p-value) | 0.571 | 0.035 | 4.07E−12 | 0.467 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, V.P.T.; Stewart, J.D.; Allais, F.; Ioannou, I. Optimization of the Recovery of Secondary Metabolites from Defatted Brassica carinata Meal and Its Effects on the Extractability and Functional Properties of Proteins. Foods 2022, 11, 429. https://doi.org/10.3390/foods11030429
Nguyen VPT, Stewart JD, Allais F, Ioannou I. Optimization of the Recovery of Secondary Metabolites from Defatted Brassica carinata Meal and Its Effects on the Extractability and Functional Properties of Proteins. Foods. 2022; 11(3):429. https://doi.org/10.3390/foods11030429
Chicago/Turabian StyleNguyen, V. P. Thinh, Jon D. Stewart, Florent Allais, and Irina Ioannou. 2022. "Optimization of the Recovery of Secondary Metabolites from Defatted Brassica carinata Meal and Its Effects on the Extractability and Functional Properties of Proteins" Foods 11, no. 3: 429. https://doi.org/10.3390/foods11030429
APA StyleNguyen, V. P. T., Stewart, J. D., Allais, F., & Ioannou, I. (2022). Optimization of the Recovery of Secondary Metabolites from Defatted Brassica carinata Meal and Its Effects on the Extractability and Functional Properties of Proteins. Foods, 11(3), 429. https://doi.org/10.3390/foods11030429