Sensomics-Assisted Aroma Decoding of Pea Protein Isolates (Pisum sativum L.)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Pea Protein Isolates
2.3. Protein Content (PC)
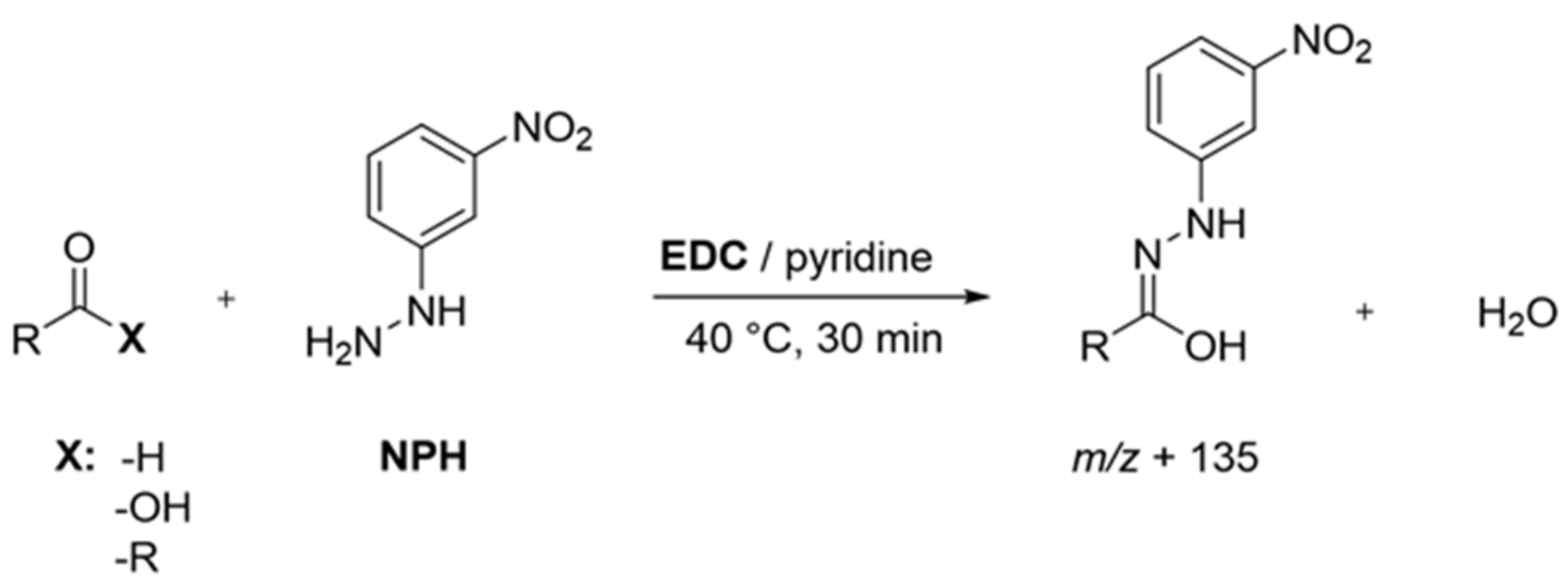
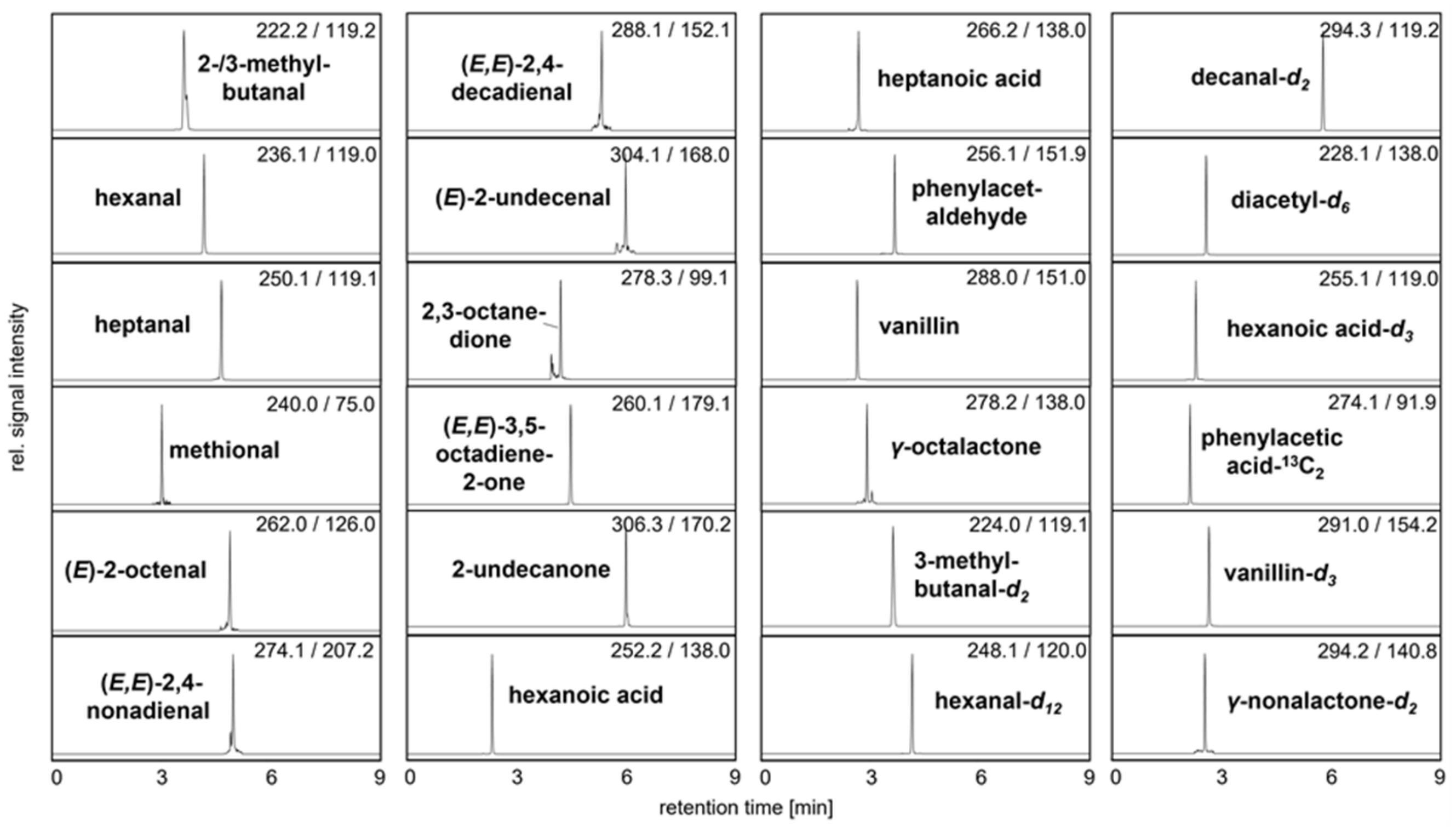
2.4. Identification and Quantitation of Odor-Active Acids, Aldehydes, Ketones, and Pyrazines
2.5. Sensory Analysis
2.6. Determination of Odor Thresholds
2.7. Estimation of Aroma Contribution
2.8. Statistical Analysis
3. Results
3.1. Method Development and Validation Experiments
3.2. Sensomics-Assisted Aroma Decoding
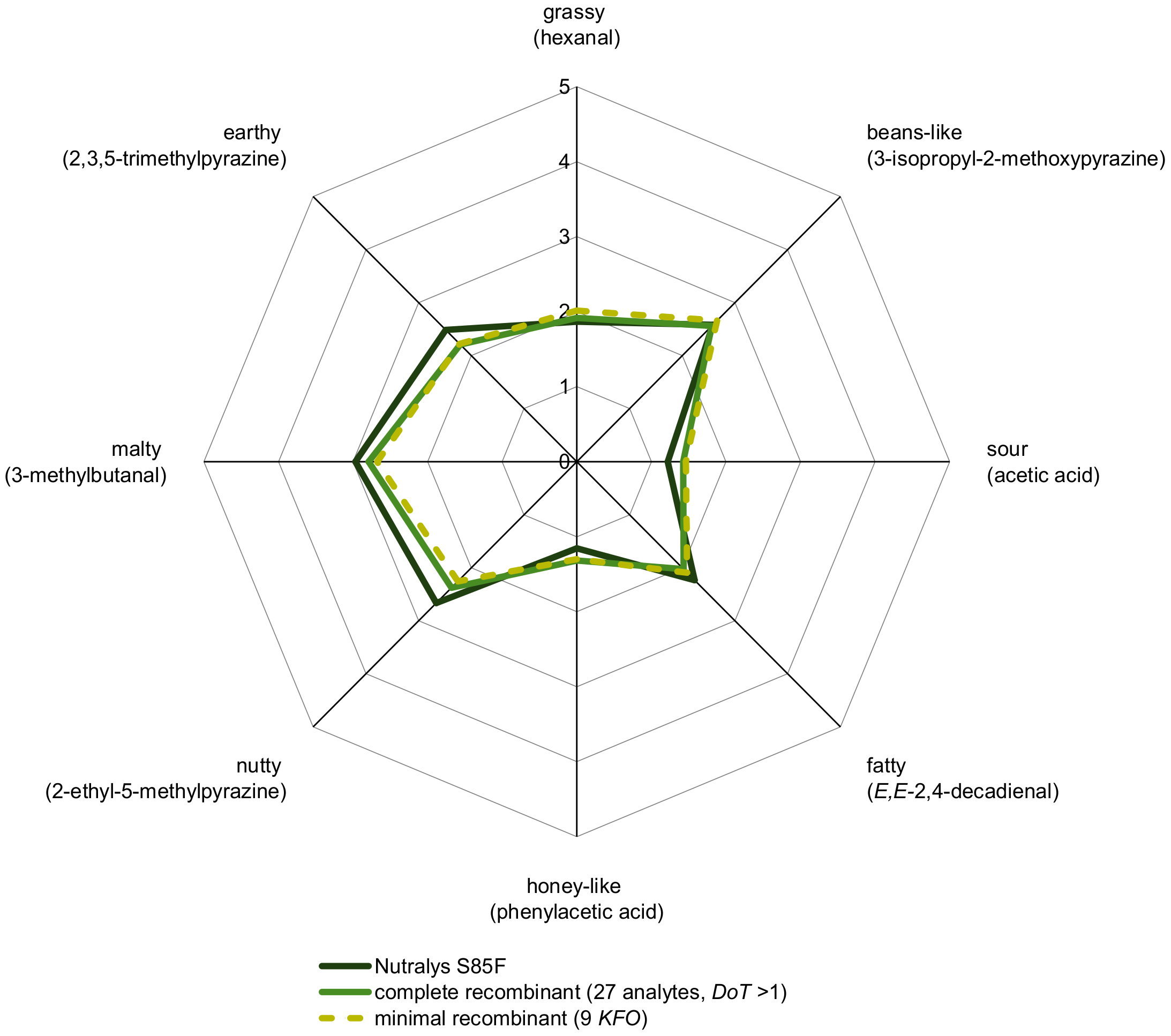
3.3. Pea Protein Aroma Simulation and Omission Experiments
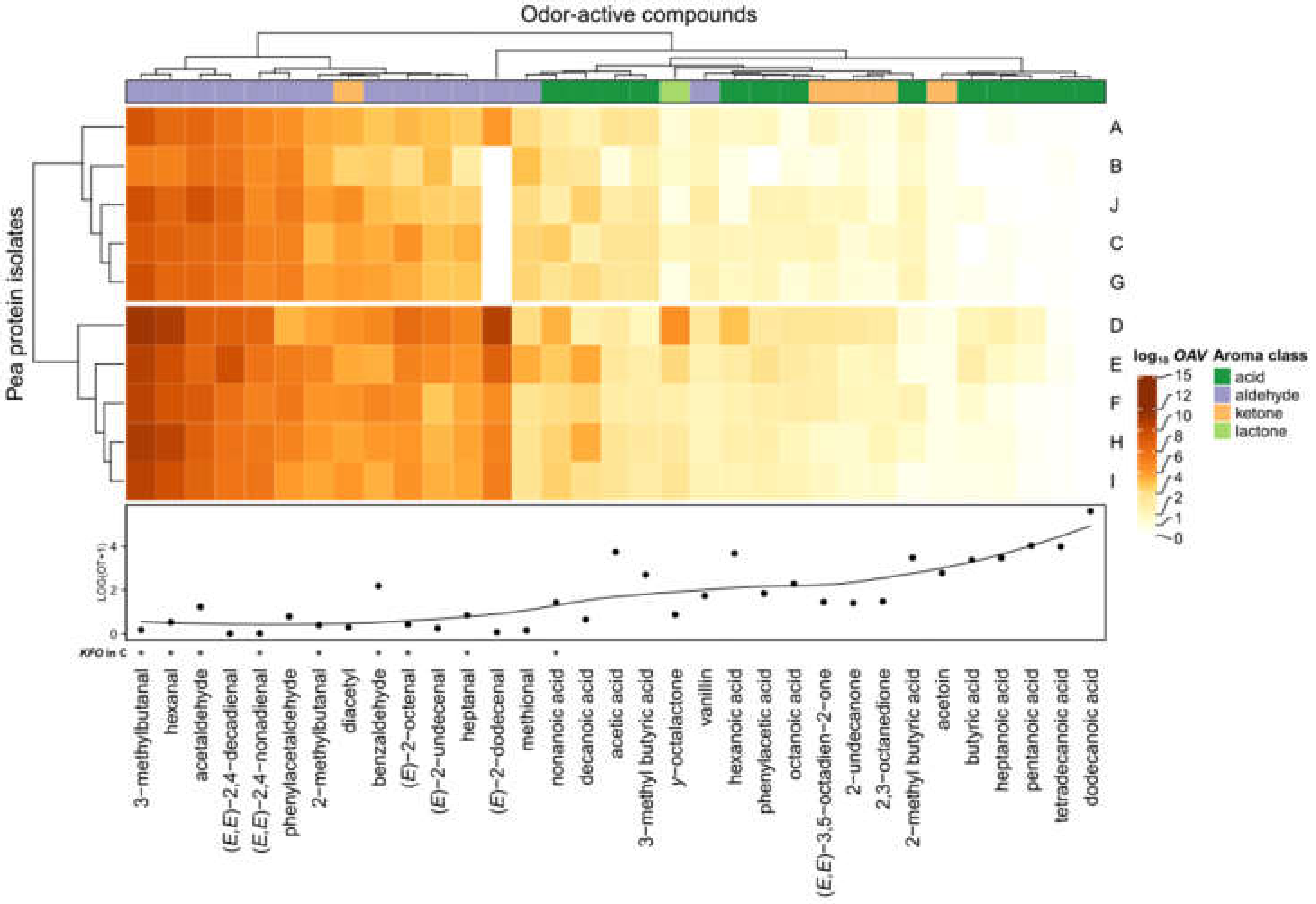
3.4. OAV Mapping of Commercially Available Pea Proteins
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nielsen Company. Umsatz mit Pflanzlichen Milchersatzprodukten in Deutschland Bis 2020. Available online: https://de.statista.com/statistik/daten/studie/1185175/umfrage/umsatz-mit-pflanzlichen-milchersatzprodukten-in-deutschland/ (accessed on 14 April 2021).
- Statistisches Bundesamt. Vegetarische und Vegane Lebensmittel: Produktion steigt im 1. Quartal 2020 um 37%. Available online: https://www.destatis.de/DE/Presse/Pressemitteilungen/Zahl-der-Woche/2020/PD20_30_p002.html (accessed on 14 April 2021).
- KPMG. Nachhaltigkeit ist mehr als nur ein Kurzzeitiger Trend: Consumer Barometer 01/20. Available online: https://home.kpmg/de/de/home/media/press-releases/2020/03/nachhaltigkeit-ist-mehr-als-nur-ein-kurzzeitiger-trend-consumer-barometer.html (accessed on 14 April 2021).
- Steinfeld, H.; Gerber, P.; Wassenaar, T.; Castel, T.; Rosales, M.; de Haan, C. Livestock’s Long Shadow: Environmental Issues and Options; FAO: Rome, Italy, 2006. [Google Scholar]
- Tanger, C.; Schmidt, F.; Utz, F.; Kreissl, J.; Dawid, C.; Kulozik, U. Pea protein microparticulation using extrusion cooking: Influence of extrusion parameters and drying on microparticle characteristics and sensory by application in a model milk dessert. Innov. Food Sci. Emerg. Technol. 2021, 74, 102851. [Google Scholar] [CrossRef]
- Mordor Intelligence. Europe Pea Protein Market—Growth, Trends and Forecasts (2017–2022). Available online: https://www.researchandmarkets.com/reports/4472989/global-pea-protein-market-growth-trends-and (accessed on 21 April 2021).
- Global Market Insights. Pea Protein Market Size by Product (Isolate, Concentrate, Textured), by Application (Meat Substitutes, Nutraceuticals, Sports Supplements), Industry Analysis Report, Regional Outlook, Application Potential, Price Trends, Competitive Market Share & Forecast, 2020–2026. Available online: https://www.gminsights.com/industry-analysis/pea-protein-market-report (accessed on 21 April 2021).
- Gläser, P.; Dawid, C.; Meister, S.; Bader-Mittermaier, S.; Schott, M.; Eisner, P.; Hofmann, T. Molecularization of Bitter Off-Taste Compounds in Pea-Protein Isolates (Pisum sativum L.). J. Agric. Food Chem. 2020, 68, 10374–10387. [Google Scholar] [CrossRef]
- Bi, S.; Xu, X.; Luo, D.; Lao, F.; Pang, X.; Shen, Q.; Hu, X.; Wu, J. Characterization of Key Aroma Compounds in Raw and Roasted Peas (Pisum sativum L.) by Application of Instrumental and Sensory Techniques. J. Agric. Food Chem. 2020, 68, 2718–2727. [Google Scholar] [CrossRef] [PubMed]
- Murat, C.; Bard, M.-H.; Dhalleine, C.; Cayot, N. Characterisation of odour active compounds along extraction process from pea flour to pea protein extract. Food Res. Int. 2013, 53, 31–41. [Google Scholar] [CrossRef]
- Trikusuma, M.; Paravisini, L.; Peterson, D.G. Identification of aroma compounds in pea protein UHT beverages. Food Chem. 2020, 312, 126082. [Google Scholar] [CrossRef]
- Flaig, M.; Qi, S.; Wei, G.; Yang, X.; Schieberle, P. Characterization of the Key Odorants in a High-Grade Chinese Green Tea Beverage (Camellia sinensis; Jingshan cha) by Means of the Sensomics Approach and Elucidation of Odorant Changes in Tea Leaves Caused by the Tea Manufacturing Process. J. Agric. Food Chem. 2020, 68, 5168–5179. [Google Scholar] [CrossRef]
- Poehlmann, S.; Schieberle, P. Characterization of the aroma signature of Styrian pumpkin seed oil (Cucurbita pepo subsp. pepo var. Styriaca) by molecular sensory science. J. Agric. Food Chem. 2013, 61, 2933–2942. [Google Scholar] [CrossRef]
- Wagner, J.; Granvogl, M.; Schieberle, P. Characterization of the Key Aroma Compounds in Raw Licorice (Glycyrrhiza glabra L.) by Means of Molecular Sensory Science. J. Agric. Food Chem. 2016, 64, 8388–8396. [Google Scholar] [CrossRef]
- Hofstetter, C.K.; Dunkel, A.; Hofmann, T. Unified Flavor Quantitation: Toward High-Throughput Analysis of Key Food Odorants and Tastants by Means of Ultra-High-Performance Liquid Chromatography Tandem Mass Spectrometry. J. Agric. Food Chem. 2019, 67, 8599–8608. [Google Scholar] [CrossRef]
- Utz, F.; Kreissl, J.; Stark, T.D.; Schmid, C.; Tanger, C.; Kulozik, U.; Hofmann, T.; Dawid, C. Sensomics-Assisted Flavor Decoding of Dairy Model Systems and Flavor Reconstitution Experiments. J. Agric. Food Chem. 2021, 69, 6588–6600. [Google Scholar] [CrossRef]
- Jakobsen, H.B.; Hansen, M.; Christensen, M.R.; Brockhoff, P.B.; Olsen, C.E. Aroma Volatiles of Blanched Green Peas (Pisum sativum L.). J. Agric. Food Chem. 1998, 46, 3727–3734. [Google Scholar] [CrossRef]
- Ebert, S.; Michel, W.; Nedele, A.-K.; Baune, M.-C.; Terjung, N.; Zhang, Y.; Gibis, M.; Weiss, J. Influence of protein extraction and texturization on odor-active compounds of pea proteins. J. Sci. Food Agric. 2021, 102, 1021–1029. [Google Scholar] [CrossRef]
- Bader, S.; Czerny, M.; Eisner, P.; Buettner, A. Characterisation of odour-active compounds in lupin flour. J. Sci. Food Agric. 2009, 89, 2421–2427. [Google Scholar] [CrossRef]
- Buettner, A.; Schieberle, P. Evaluation of aroma differences between hand-squeezed juices from Valencia late and Navel oranges by quantitation of key odorants and flavor reconstitution experiments. J. Agric. Food Chem. 2001, 49, 2387–2394. [Google Scholar] [CrossRef]
- Semmelroch, P.; Laskawy, G.; Blank, I.; Grosch, W. Determination of potent odourants in roasted coffee by stable isotope dilution assays. Flavour Fragr. J. 1995, 10, 1–7. [Google Scholar] [CrossRef]
- Poisson, L.; Schieberle, P. Characterization of the key aroma compounds in an american bourbon whisky by quantitative measurements, aroma recombination, and omission studies. J. Agric. Food Chem. 2008, 56, 5820–5826. [Google Scholar] [CrossRef]
- Mariotti, F.; Tomé, D.; Mirand, P.P. Converting nitrogen into protein—Beyond 6.25 and Jones’ factors. Crit. Rev. Food Sci. Nutr. 2008, 48, 177–184. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Sequeira, C.; Borchers, C.H. An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography–tandem mass spectrometry. Anal. Chim. Acta 2015, 854, 86–94. [Google Scholar] [CrossRef]
- Dunkel, A.; Steinhaus, M.; Kotthoff, M.; Nowak, B.; Krautwurst, D.; Schieberle, P.; Hofmann, T. Nature’s chemical signatures in human olfaction: A foodborne perspective for future biotechnology. Angew. Chem. Int. Ed. 2014, 53, 7124–7143. [Google Scholar] [CrossRef]
- Stone, H.; Sidel, J.L. Sensory Evaluation Practices, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Quadt, A.; Schönberger, S.; Schwarz, M. Statistische Auswertungen in der Sensorik: Leitfaden für die Praxis; Behr’s Verlag: Hamburg, Germany, 2009; ISBN 978-3-89947-531-9. [Google Scholar]
- Czerny, M.; Christlbauer, M.; Christlbauer, M.; Fischer, A.; Granvogl, M.; Hammer, M.; Hartl, C.; Moran Hernandez, N.; Schieberle, P. Re-investigation on odor thresholds of key food aroma compounds and development of an aroma language based on odor qualities of defined aqueous odorant solutions. Food Chem. 2008, 228, 265–273. [Google Scholar] [CrossRef]
- Kreissl, J.; Mall, V.; Steinhaus, P.; Steinhaus, M. Leibniz-LSB@TUM Odorant Database. Available online: https://www.leibniz-lsb.de/en/databases/leibniz-lsbtum-odorant-database (accessed on 29 July 2021).
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Harb, S.; Saint-Eve, A.; Panouillé, M.; Souchon, I.; Bonnarme, P.; Dugat-Bony, E.; Irlinger, F. Design of microbial consortia for the fermentation of pea-protein-enriched emulsions. Int. J. Food Microbiol. 2019, 293, 124–136. [Google Scholar] [CrossRef] [PubMed]

| Code | Commercial Name | PC (a) | Batch | Producer |
|---|---|---|---|---|
| A | Nutralys F85F | 67% | W084M | Roquette, Lestrem, France |
| B | Prestige | 42% | KO67X | Parrheim Foods, Saskatoon, Canada |
| C | Nutralys S85F | 68% | W317M | Roquette, Lestrem, France |
| D | Bio Erbsen Protein | 70% | 15720-30118 | Golden Peanut, Garstedt, Germany |
| E | Bio Erbsen Protein | 68% | 170180323 | Piowald, Mühbrook, Germany |
| F | 1501018 | 68% | 83526272 | Döhler, Darmstadt, Germany |
| G | Pea Pro | 68% | 06102016 | LSP Sports Nutrition, Bonn, Germany |
| H | Empro E86HV | 72% | 41266 | Emsland-Stärke, Emlichheim, Germany |
| I | Empro E86 | 71% | 41266 | Emsland-Stärke, Emlichheim, Germany |
| J | Pisane C9 | 69% | 817021 | Cosucra Group, Warcoing, Belgium |
| Validated Analyte | Add. (a) (µmol/L) | Add. Found ± SD (b) (µmol/L) | RSD (c) (%) | Recovery (%) | LOD (d) (nmol/L) | LOQ (e) (nmol/L) |
|---|---|---|---|---|---|---|
| 2-/3-methylbutanal, sum | 0.668 | 0.667 ± 0.038 | 5.7 | 99.8 | <0.1 | <0.1 |
| 2-methylbutanal | 0.346 | 0.344 ± 0.009 | 2.7 | 99.5 | <0.1 | <0.1 |
| hexanal | 0.333 | 0.277 ± 0.004 | 1.5 | 83.0 | 2.7 | 9.0 |
| heptanal | 0.301 | 0.301 ± 0.006 | 1.8 | 99.8 | <0.1 | <0.1 |
| methional | 0.370 | 0.336 ± 0.008 | 2.4 | 90.8 | 0.2 | 0.5 |
| (E)-2-octenal | 0.292 | 0.259 ± 0.003 | 1.0 | 88.8 | 0.5 | 1.7 |
| (E,E)-2,4-nonadienal | 0.274 | 0.272 ± 0.004 | 1.5 | 99.3 | 0.2 | 0.5 |
| (E,Z)-2,6-nonadienal | 0.294 | 0.294 ± 0.008 | 2.8 | 100.0 | 0.2 | 0.8 |
| (E,E)-2,4-decadienal | 0.246 | 0.241 ± 0.003 | 1.3 | 98.0 | 0.2 | 0.5 |
| (E)-2-undecenal | 0.278 | 0.267 ± 0.020 | 7.4 | 96.2 | 0.3 | 0.9 |
| (E)-2-dodecenal | 0.275 | 0.258 ± 0.015 | 5.7 | 93.7 | 1.4 | 4.7 |
| 2,3-octanedione | 0.293 | 0.290 ± 0.003 | 0.9 | 99.0 | 0.4 | 1.2 |
| (E,E)-3,5-octadien-2-one | 0.256 | 0.264 ± 0.006 | 2.1 | 103.2 | <0.1 | 0.3 |
| 2-undecanone | 0.291 | 0.288 ± 0.011 | 3.8 | 99.1 | <0.1 | <0.1 |
| hexanoic acid | 0.305 | 0.288 ± 0.012 | 4.3 | 94.4 | <0.1 | <0.1 |
| heptanoic acid | 0.280 | 0.285 ± 0.013 | 4.7 | 101.7 | 5.8 | 19.2 |
| phenylacetaldehyde | 0.286 | 0.306 ± 0.009 | 2.9 | 106.9 | <0.1 | 0.3 |
| 4-ethyl benzaldehyde | 0.400 | 0.378 ± 0.023 | 6.1 | 94.5 | 4.7 | 15.8 |
| vanillin | 0.285 | 0.258 ± 0.012 | 4.7 | 90.6 | 0.2 | 0.6 |
| γ-octalactone | 0.302 | 0.243 ± 0.010 | 4.0 | 80.5 | 1.0 | 3.2 |
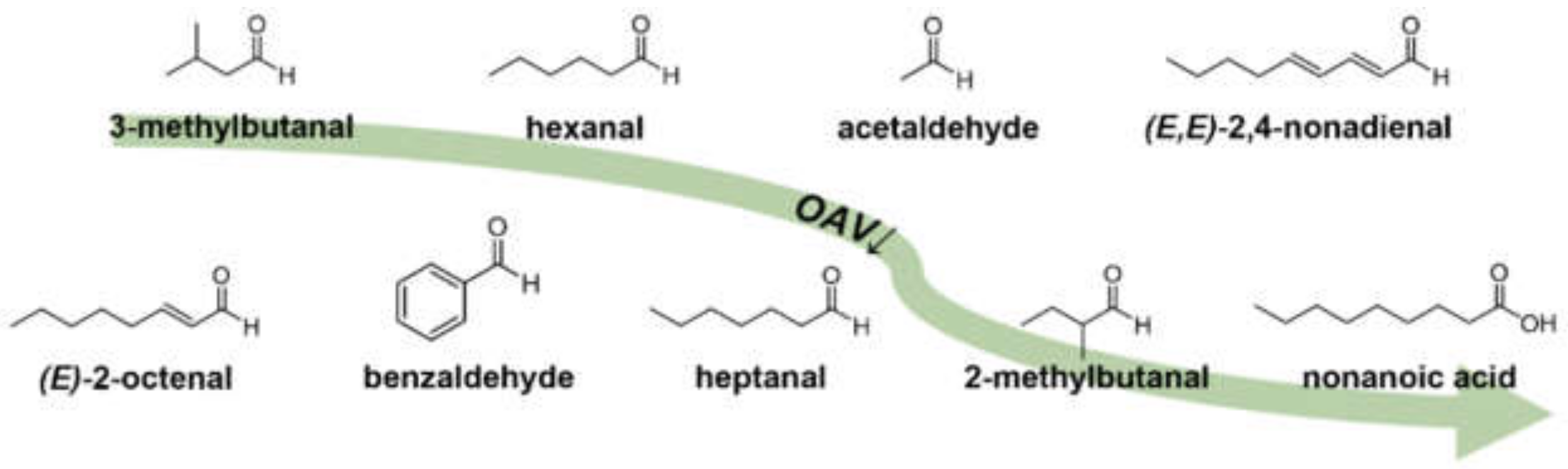
| No. | Aroma-Active Analytes | Used IS | Odor Quality | Mean ± SD (a) (µg/kg) | RSD (b) (%) | OT (c) (µg/kg) | OAV (d) |
|---|---|---|---|---|---|---|---|
| 1 | 3-methylbutanal (e) | 3-methylbutanal-d2 | malty | 5093 (5360 ± 547) (e) | (10.2) (e) | 0.5 | 10186 |
| 2 | hexanal | hexanal-d12 | green, grassy | 14,886 ± 1904 | 12.8 | 2.4 | 6202 |
| 3 | acetaldehyde (f) | acetaldehyde-d3 | fresh, green | 72,197 ± 4231 | 5.9 | 16 | 4512 |
| 4 | (E,E)-2,4-decadienal | decanal-d2 | fatty, deep-fried | 101 ± 10 | 9.8 | 0.027 | 3736 |
| 5 | phenylacetaldehyde | phenylacetic acid-13C2 | flowery, honey-like | 6097 ± 303 | 5.0 | 5.2 | 1173 |
| 6 | (E,E)-2,4-nonadienal | decanal-d2 | fatty, green | 53 ± 7 | 12.5 | 0.046 | 1156 |
| 7 | (E)-2-octenal | hexanal-d12 | fatty, nutty | 907 ± 8 | 0.8 | 1.7 | 533 |
| 8 | diacetyl (f) | diacetyl-d6 | butter-like | 316 ± 25 | 8.0 | 0.96 | 329 |
| 9 | benzaldehyde (f) | phenylacetic acid-13C2 | bitter almond-like, marzipan-like | 37,201 ± 7850 | 21.1 | 150 | 248 |
| 10 | heptanal | hexanal-d12 | citrus-like, fatty | 1326 ± 130 | 9.8 | 6.1 | 217 |
| 11 | 2-methylbutanal | 3-methylbutanal-d2 | malty | 267 ± 8 | 2.9 | 1.5 | 178 |
| 12 | (E)-2-undecenal | decanal-d2 | soapy, metallic | 123 ± 8 | 6.5 | 0.78 | 157 |
| 13 | nonanoic acid (f) | octanoic acid-d15 | moldy, pungent | 2776 ± 167 | 6.0 | 26 | 107 |
| 14 | methional | hexanal-d12 | cooked potato-like | 37 ± 5 | 4.7 | 0.43 | 85 |
| 15 | acetic acid (f) | acetic acid-13C2 | vinegar-like | 262,037 ± 10,440 | 4.0 | 5600 | 47 |
| 16 | 3-methylbutanoic acid (f) | butyric acid-13C4 | sweaty | 23,274 ± 1425 | 6.1 | 490 | 47 |
| 17 | decanoic acid (f) | octanoic acid-d15 | soapy, musty | 51 ± 3 | 5.5 | 3.5 | 15 |
| 18 | vanillin | vanillin-d3 | vanilla-like, sweet | 561 ± 18 | 3.2 | 53 | 11 |
| 19 | (E,E)-3,5-octadien-2-one | diacetyl-d6 | woody, mushroom-like, green | 231 ± 18 | 7.9 | 27 | 9 |
| 20 | hexanoic acid | hexanoic acid-d3 | sweaty | 36,802 ± 1978 | 5.4 | 4800 | 8 |
| 21 | octanoic acid (f) | octanoic acid-d15 | carrot-like, musty | 1536 ± 48 | 3.1 | 190 | 8 |
| 22 | phenylacetic acid (f) | phenylacetic acid-13C2 | honey-like, beeswax-like | 516 ± 61 | 11.8 | 68 | 8 |
| 23 | γ-octalactone | γ-nonalactone-d2 | coconut-like | 47 ± 5 | 10.5 | 6.5 | 7 |
| 24 | 2-methylbutanoic acid (f) | butyric acid-13C4 | malty, fruity, sweaty | 21,419 ± 1014 | 4.7 | 3100 | 7 |
| 25 | 2,3-octanedione | diacetyl-d6 | mushroom-like, dill-like, broccoli-like | 141 ± 13 | 8.9 | 29 | 4.8 |
| 26 | 2-undecanone | decanal-d2 | soapy, green | 68 ± 4 | 6.5 | 24 | 2.8 |
| 27 | acetoin (f) | diacetyl-d6 | butter-like, carrot-like | 978 ± 7 | 0.7 | 590 | 1.7 |
| Test | Odorant(s) Omitted (a) | OAV | p Value (%) | Significance (b) |
|---|---|---|---|---|
| O1 | 19–27 | <10 | 23.8 | NS |
| O2 | 14–18 | <100 | 19.1 | NS |
| O3 | 1, 3-methylbutanal | 10186 | <0.1 | *** |
| O4 | 2, hexanal | 6202 | 2.6 | * |
| O5 | 3, acetaldehyde | 4512 | 4.8 | * |
| O6 | 4, (E,E)-2,4-decadienal | 3736 | 9.2 | NS |
| O7 | 5, phenylacetaldehyde | 1173 | 21.4 | NS |
| O8 | 6, (E,E)-2,4-nonadienal | 1156 | 4.8 | * |
| O9 | 7, (E)-2-octenal | 533 | 1.3 | * |
| O10 | 8, diacetyl | 329 | 15.6 | NS |
| O11 | 9, benzaldehyde | 248 | 4.0 | * |
| O12 | 10, heptanal | 217 | <0.1 | *** |
| O13 | 11, 2-methylbutanal | 178 | 0.7 | ** |
| O14 | 12, (E)-2-undecenal | 157 | 23.0 | NS |
| O15 | 13, nonanoic acid | 107 | 4.0 | * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Utz, F.; Spaccasassi, A.; Kreissl, J.; Stark, T.D.; Tanger, C.; Kulozik, U.; Hofmann, T.; Dawid, C. Sensomics-Assisted Aroma Decoding of Pea Protein Isolates (Pisum sativum L.). Foods 2022, 11, 412. https://doi.org/10.3390/foods11030412
Utz F, Spaccasassi A, Kreissl J, Stark TD, Tanger C, Kulozik U, Hofmann T, Dawid C. Sensomics-Assisted Aroma Decoding of Pea Protein Isolates (Pisum sativum L.). Foods. 2022; 11(3):412. https://doi.org/10.3390/foods11030412
Chicago/Turabian StyleUtz, Florian, Andrea Spaccasassi, Johanna Kreissl, Timo D. Stark, Caren Tanger, Ulrich Kulozik, Thomas Hofmann, and Corinna Dawid. 2022. "Sensomics-Assisted Aroma Decoding of Pea Protein Isolates (Pisum sativum L.)" Foods 11, no. 3: 412. https://doi.org/10.3390/foods11030412
APA StyleUtz, F., Spaccasassi, A., Kreissl, J., Stark, T. D., Tanger, C., Kulozik, U., Hofmann, T., & Dawid, C. (2022). Sensomics-Assisted Aroma Decoding of Pea Protein Isolates (Pisum sativum L.). Foods, 11(3), 412. https://doi.org/10.3390/foods11030412







