Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Meat Quality Evaluation
2.3. Library Preparation and RNA Sequencing
2.4. Lncrnas Identification
2.5. Gene Ontology and Pathway Analyses
2.6. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
2.7. Statistical Analysis
3. Results
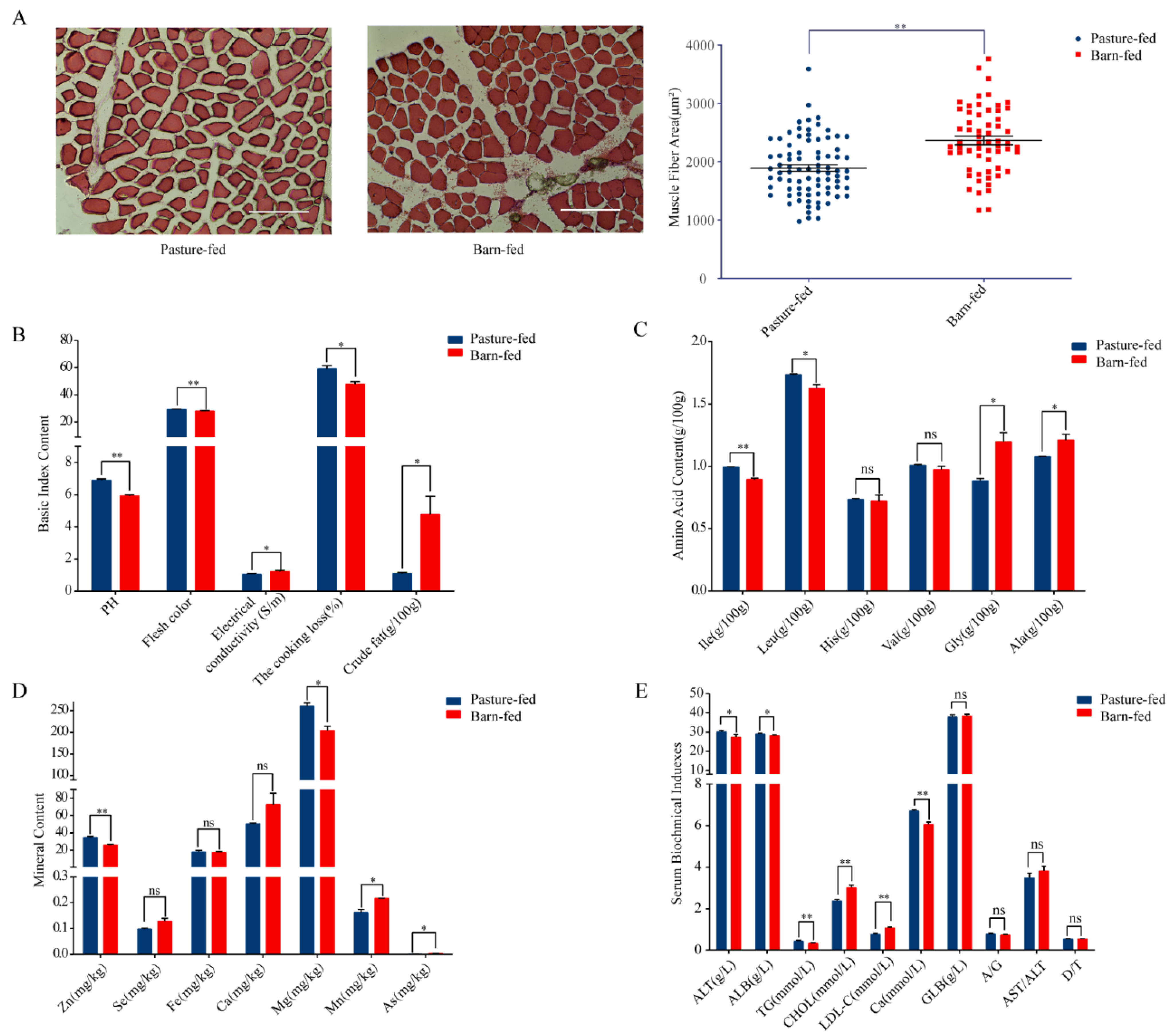
3.1. Differences of Physiological and Biochemical Characters between Pasture-Fed and Barn-Fed Goat Meat
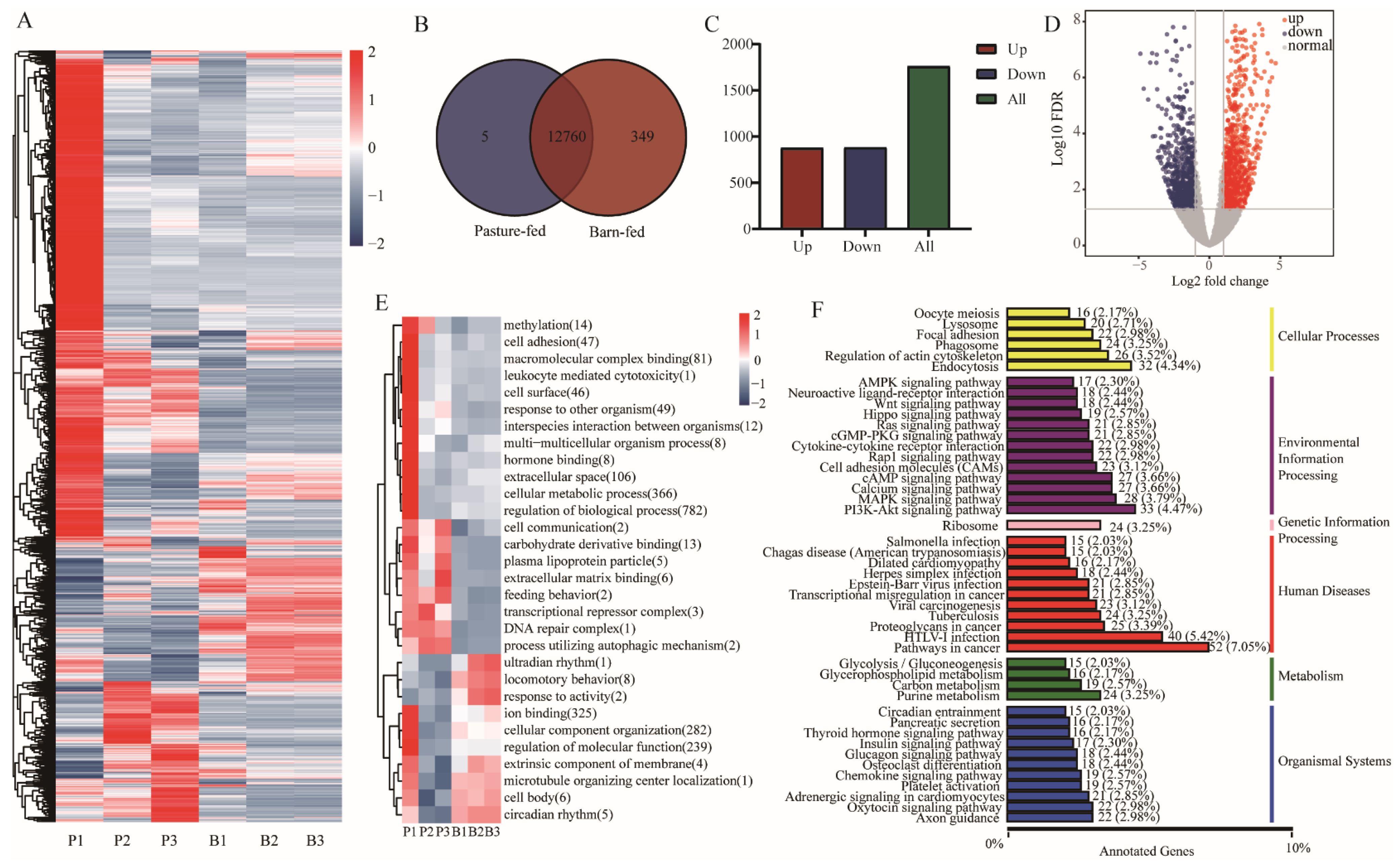
3.2. Transcriptome Differences between Muscles of Pasture-Fed and Barn-Fed Black Goats
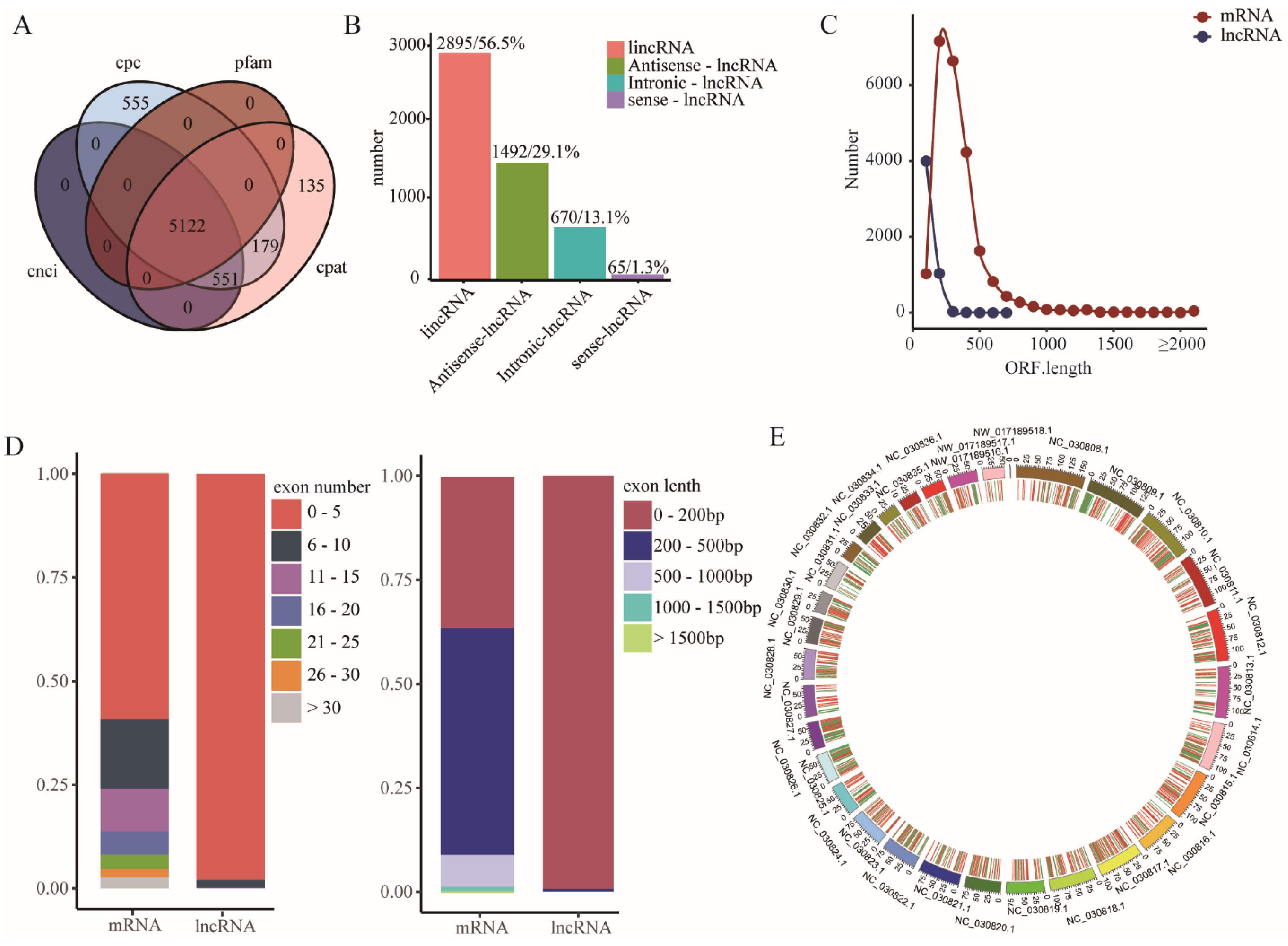
3.3. Identification of lncRNAs in Goat Muscles
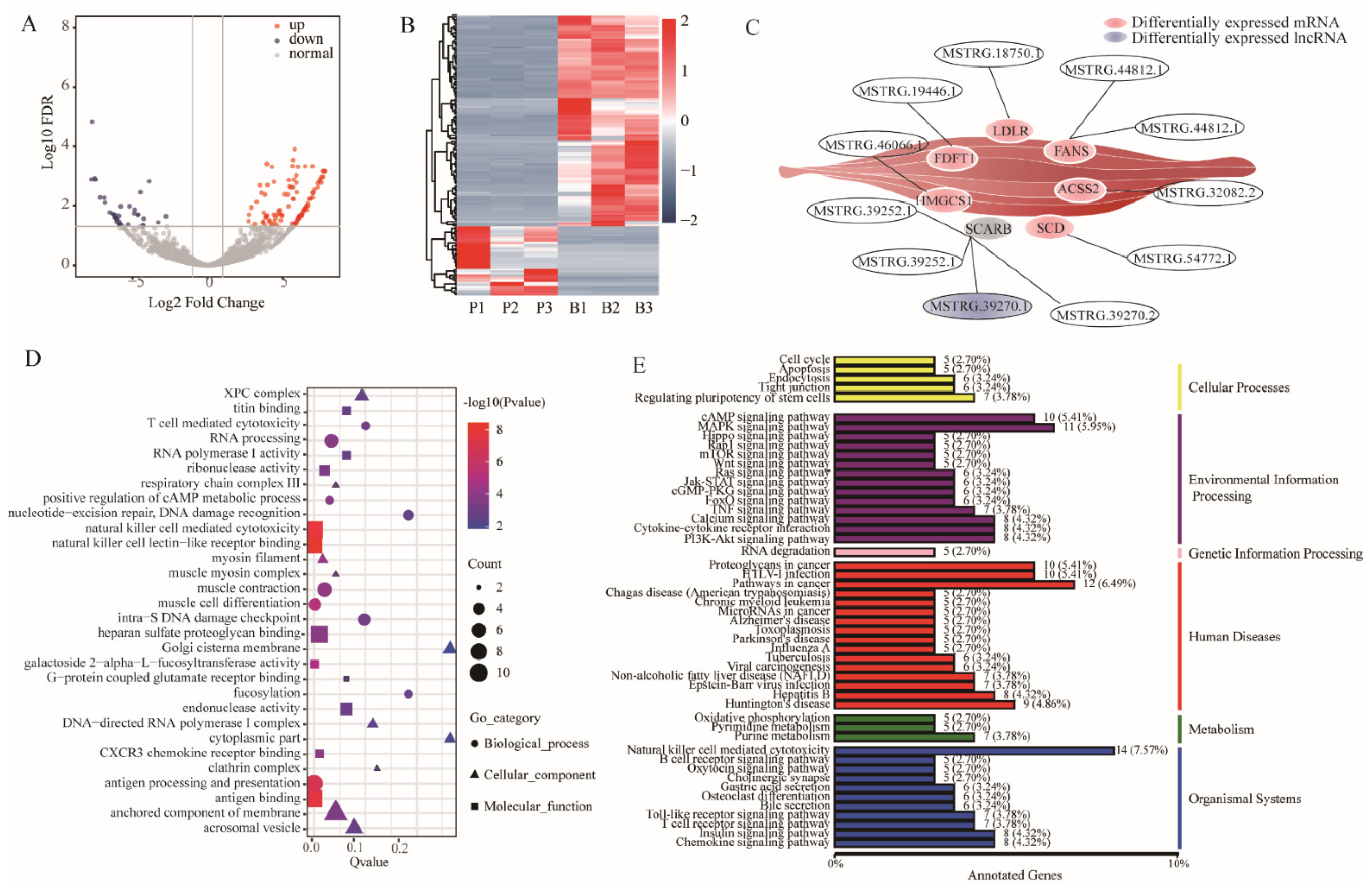
3.4. Differentially Expression and Enrichment Analysis of lncRNAs
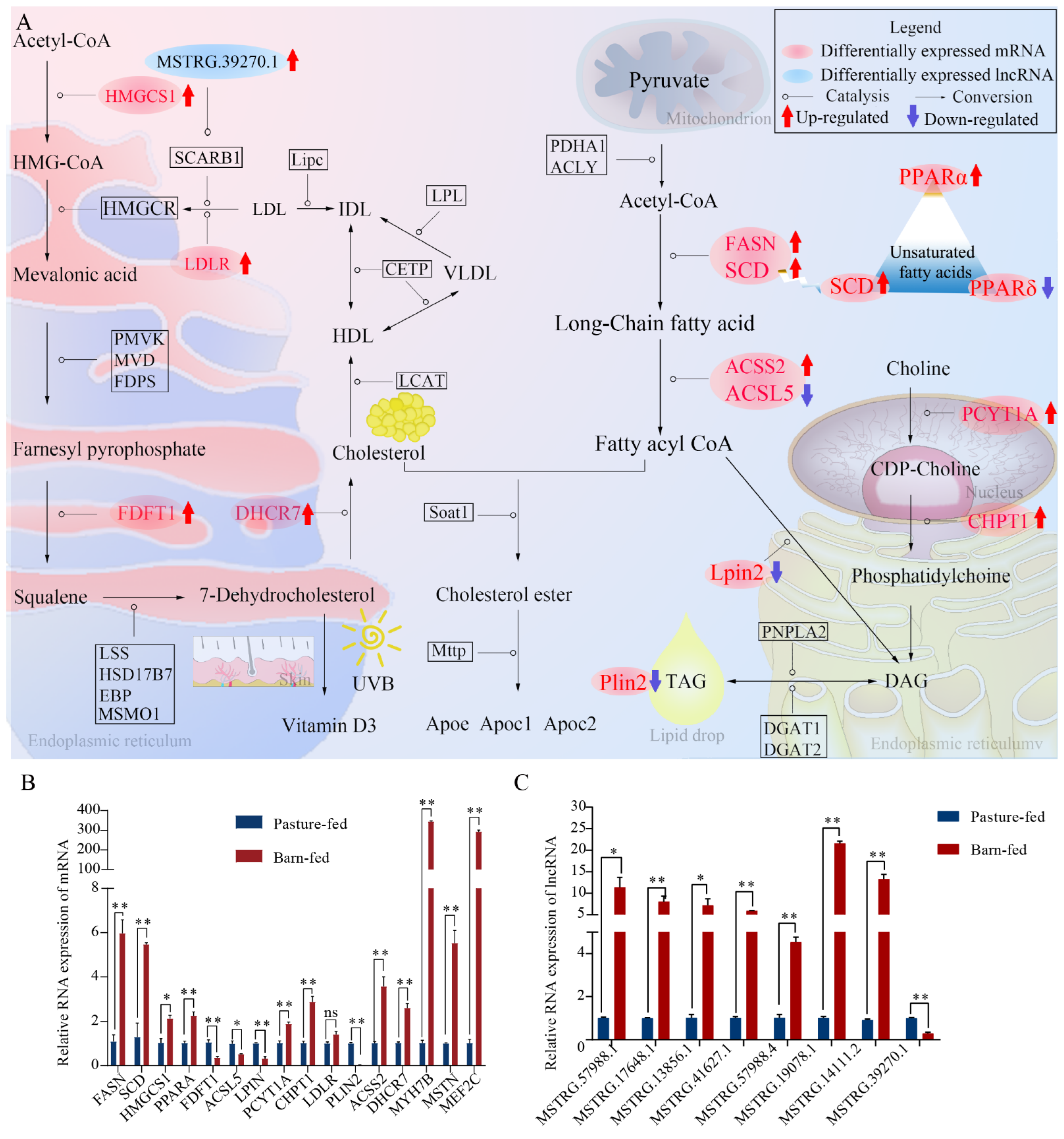
3.5. Analysis of the Underlying Molecular Mechanism and Validation of Differentially Expressed Genes and lncRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Teixeira, A.; Silva, S.; Guedes, C.; Rodrigues, S. Sheep and Goat Meat Processed Products Quality: A Review. Foods 2020, 9, 960. [Google Scholar] [CrossRef] [PubMed]
- Daley, C.A.; Abbott, A.; Doyle, P.S.; Nader, G.A.; Larson, S. A review of fatty acid profiles and antioxidant content in grass-fed and grain-fed beef. Nutr. J. 2010, 9, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spriet, L.L. New Insights into the Interaction of Carbohydrate and Fat Metabolism during Exercise. Sports Med. 2014, 44, 87–96. [Google Scholar] [CrossRef] [Green Version]
- Fritzen, A.M.; Lundsgaard, A.M.; Kiens, B. Tuning fatty acid oxidation in skeletal muscle with dietary fat and exercise. Nat. Rev. Endocrinol. 2020, 16, 683–696. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Chen, M.; Zhong, D.; Luo, X.; Feng, T.; Song, M.; Chen, Y.; Wei, X.; Shi, D.; Liu, Q.; et al. Circular RNA Profiling Reveals an Abundant circEch1 That Promotes Myogenesis and Differentiation of Bovine Skeletal Muscle. J. Agric. Food Chem. 2021, 69, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Hennebry, A.; Berry, C.; Siriett, V.; O’Callaghan, P.; Chau, L.; Watson, T.; Sharma, M.; Kambadur, R. Myostatin regulates fiber-type composition of skeletal muscle by regulating MEF2 and MyoD gene expression. Am. J. Physiol.-Cell Physiol. 2009, 296, C525–C534. [Google Scholar] [CrossRef]
- Prache, S.; Schreurs, N.; Guillier, L. Review: Factors affecting sheep carcass and meat quality attributes. Animal 2021, 100330. [Google Scholar] [CrossRef]
- Morales, P.E.; Bucarey, J.L.; Espinosa, A. Muscle Lipid Metabolism: Role of Lipid Droplets and Perilipins. J. Diabetes Res. 2017, 2017, 1789395. [Google Scholar] [CrossRef]
- Muscella, A.; Stefàno, E.; Lunetti, P.; Capobianco, L.; Marsigliante, S. The Regulation of Fat Metabolism during Aerobic Exercise. Biomolecules 2020, 10, 1699. [Google Scholar] [CrossRef]
- Frank, D.; Joo, S.; Warner, R. Consumer Acceptability of Intramuscular Fat. Korean J. Food Sci. Anim. Resour. 2016, 36, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, X.; Yang, G.; Du, M. Review: Enhancing intramuscular fat development via targeting fibro-adipogenic progenitor cells in meat animals. Animal 2020, 14, 312–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knobloch, M.; Braun, S.M.G.; Zurkirchen, L.; von Schoultz, C.; Zamboni, N.; Araúzo-Bravo, M.J.; Kovacs, W.J.; Karalay, Ö.; Suter, U.; Machado, R.A.C.; et al. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature 2013, 493, 226–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, M.; Shen, L.; Fan, Y.; Tan, Y.; Liu, L.; Chen, L.; Zhao, Y.; Niu, L.; Tang, G.; Li, Q.; et al. ssc-miR-451 Regulates Porcine Primary Adipocyte Differentiation by Targeting ACACA. Animals 2020, 10, 1891. [Google Scholar] [CrossRef]
- Kojta, I.; Zabielski, P.; Roszczyc-Owsiejczuk, K.; Imierska, M.; Sokołowska, E.; Błachnio-Zabielska, A. GPAT Gene Silencing in Muscle Reduces Diacylglycerols Content and Improves Insulin Action in Diet-Induced Insulin Resistance. Int. J. Mol. Sci. 2020, 21, 7369. [Google Scholar] [CrossRef]
- Ackerman, D.; Tumanov, S.; Qiu, B.; Michalopoulou, E.; Spata, M.; Azzam, A.; Xie, H.; Simon, M.C.; Kamphorst, J.J. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep. 2018, 24, 2596–2605.e5. [Google Scholar] [CrossRef] [Green Version]
- Landgraf, K.; Klöting, N.; Gericke, M.; Maixner, N.; Guiu-Jurado, E.; Scholz, M.; Witte, A.V.; Beyer, F.; Schwartze, J.T.; Lacher, M.; et al. The Obesity-Susceptibility Gene TMEM18 Promotes Adipogenesis through Activation of PPARG. Cell Rep. 2020, 33, 108295. [Google Scholar] [CrossRef]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Bio. 2013, 14, 699–712. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.; Bai, Y.; Liu, J.; Cai, W.; Liu, L.; He, Y.; Song, J. Metabolic Regulations by lncRNA, miRNA, and ceRNA Under Grass-Fed and Grain-Fed Regimens in Angus Beef Cattle. Front. Genet. 2021, 12, 220. [Google Scholar] [CrossRef]
- Ling, Y.; Zheng, Q.; Sui, M.; Zhu, L.; Xu, L.; Zhang, Y.; Liu, Y.; Fang, F.; Chu, M.; Ma, Y.; et al. Comprehensive Analysis of LncRNA Reveals the Temporal-Specific Module of Goat Skeletal Muscle Development. Int. J. Mol. Sci. 2019, 20, 3950. [Google Scholar] [CrossRef] [Green Version]
- Zhan, S.; Qin, C.; Li, D.; Zhao, W.; Nie, L.; Cao, J.; Guo, J.; Zhong, T.; Wang, L.; Li, L.; et al. A Novel Long Noncoding RNA, lncR-125b, Promotes the Differentiation of Goat Skeletal Muscle Satellite Cells by Sponging miR-125b. Front. Genet. 2019, 10, 1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Cai, R.; Wang, Y.; Zhao, R.; Qin, J.; Pang, W. A Newly Identified LncRNA LncIMF4 Controls Adipogenesis of Porcine Intramuscular Preadipocyte through Attenuating Autophagy to Inhibit Lipolysis. Animals 2020, 10, 926. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.; Feng, X.; Wei, Y.; Guo, D.; Li, J.; Liu, Q.; Jiang, J.; Shi, D.; Huang, J. lncSAMM50 Enhances Adipogenic Differentiation of Buffalo Adipocytes with No Effect on Its Host Gene. Front. Genet. 2021, 12, 333. [Google Scholar] [CrossRef] [PubMed]
- Wereńska, M.; Haraf, G.; Wołoszyn, J.; Goluch, Z.; Okruszek, A.; Teleszko, M. Fatty acid profile and health lipid indicies of goose meat in relation to various types of heat treatment. Poult. Sci. 2021, 100, 101237. [Google Scholar] [CrossRef]
- Li, H.; Huang, K.; Wang, P.; Feng, T.; Shi, D.; Cui, K.; Luo, C.; Shafique, L.; Qian, Q.; Ruan, J.; et al. Comparison of Long Non-Coding RNA Expression Profiles of Cattle and Buffalo Differing in Muscle Characteristics. Front. Genet. 2020, 11, 98. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−△△CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- D’Alessandro, A.G.; Maiorano, G.; Kowaliszyn, B.; Loiudice, P.; Martemucci, G. How the nutritional value and consumer acceptability of suckling lambs meat is affected by the maternal feeding system. Small Ruminant. Res. 2012, 106, 83–91. [Google Scholar] [CrossRef]
- O’Reilly, R.A.; Pannier, L.; Gardner, G.E.; Garmyn, A.J.; Luo, H.; Meng, Q.; Miller, M.F.; Pethick, D.W. Minor differences in perceived sheepmeat eating quality scores of Australian, Chinese and American consumers. Meat Sci. 2020, 164, 108060. [Google Scholar] [CrossRef]
- Ricardo, H.A.; Fernandes, A.R.M.; Mendes, L.C.N.; Oliveira, M.A.G.; Protes, V.M.; Scatena, E.M.; Roça, R.O.; Athayde, N.B.; Girão, L.V.C.; Alves, L.G.C. Carcass traits and meat quality differences between a traditional and an intensive production model of market lambs in Brazil: Preliminary investigation. Small Ruminant. Res. 2015, 130, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Ardö, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef]
- Wang, J.; Xu, Z.; Zhang, H.; Wang, Y.; Liu, X.; Wang, Q.; Xue, J.; Zhao, Y.; Yang, S. Meat differentiation between pasture-fed and concentrate-fed sheep/goats by liquid chromatography quadrupole time-of-flight mass spectrometry combined with metabolomic and lipidomic profiling. Meat Sci. 2021, 173, 108374. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, R.; Kiyimba, F.; Gonzalez, J.; Mafi, G.; DeSilva, U. Impact of Up- and Downregulation of Metabolites and Mitochondrial Content on pH and Color of the Longissimus Muscle from Normal-pH and Dark-Cutting Beef. J. Agric. Food Chem. 2020, 68, 7194–7203. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 2015, 44, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, D.; Ono, Y.; Hirano, S.; Kan-no, N.; Watabe, S. Lampreys have a single gene cluster for the fast skeletal myosin heavy chain gene family. PLoS ONE 2013, 8, e85500. [Google Scholar] [CrossRef]
- Huang, J.; Jiao, J.; Tan, Z.; He, Z.; Beauchemin, K.A.; Forster, R.; Han, X.; Tang, S.; Kang, J.; Zhou, C. Inferring the Skeletal Muscle Developmental Changes of Grazing and Barn-Fed Goats from Gene Expression Data. J. Agric. Food Chem. 2016, 64, 6791–6800. [Google Scholar] [CrossRef]
- Yokoyama, S.; Ohno, Y.; Egawa, T.; Yasuhara, K.; Nakai, A.; Sugiura, T.; Ohira, Y.; Yoshioka, T.; Okita, M.; Origuchi, T.; et al. Heat shock transcription factor 1-associated expression of slow myosin heavy chain in mouse soleus muscle in response to unloading with or without reloading. Acta Physiol. 2016, 217, 325–337. [Google Scholar] [CrossRef]
- Alapat, D.; Chaudhry, T.; Ardakanytaghavi, R.; Kohtz, D. Fiber-types of sarcomeric proteins expressed in cultured myogenic cells are modulated by the dose of myogenin activity. Cell Signal. 2009, 21, 128–135. [Google Scholar] [CrossRef]
- Hargreaves, M.S.; Priet, L.L. Exercise Metabolism: Fuels for the Fire. CSH Perspect. Med. 2018, 8, a029744. [Google Scholar] [CrossRef]
- Hondares, E.; Iglesias, R.; Giralt, A.; Gonzalez, F.J.; Giralt, M.; Mampel, T.; Villarroya, F. Thermogenic Activation Induces FGF21 Expression and Release in Brown Adipose Tissue. J. Biol. Chem. 2011, 286, 12983–12990. [Google Scholar] [CrossRef] [Green Version]
- Yao, W.; Jiao, Y.; Zhou, Y.; Luo, X. KLF13 suppresses the proliferation and growth of colorectal cancer cells through transcriptionally inhibiting HMGCS1-mediated cholesterol biosynthesis. Cell Biosci. 2020, 10, 76. [Google Scholar] [CrossRef]
- Kohlhaas, J.; Jäger, M.A.; Lust, L.; De La Torre, C.; Hecker, M.; Korff, T. Endothelial cells control vascular smooth muscle cell cholesterol levels by regulating 24-dehydrocholesterol reductase expression. Exp. Cell Res. 2021, 399, 112446. [Google Scholar] [CrossRef] [PubMed]
- van den Boomen, D.J.H.; Sienkiewicz, A.; Berlin, I.; Jongsma, M.L.M.; van Elsland, D.M.; Luzio, J.P.; Neefjes, J.J.C.; Lehner, P.J. A trimeric Rab7 GEF controls NPC1-dependent lysosomal cholesterol export. Nat. Commun. 2020, 11, 5559. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Zhu, Y.; Wang, S.; Luo, Y.; Lu, S.; Nan, F.; Sun, G.; Sun, X. Bavachinin inhibits cholesterol synthesis enzyme FDFT1 expression via AKT/mTOR/SREBP-2 pathway. Int. Immunopharmacol. 2020, 88, 106865. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Cheng, X.; Shi, G.; Zou, C.; Chen, L.; Li, J.; Li, M.; Fang, C.; Li, C. Transcriptome Analysis Reveals the Effect of Long Intergenic Noncoding RNAs on Pig Muscle Growth and Fat Deposition. Biomed. Res. Int. 2019, 2019, 2951427. [Google Scholar] [CrossRef] [Green Version]
- Hitachi, K.; Nakatani, M.; Funasaki, S.; Hijikata, I.; Maekawa, M.; Honda, M.; Tsuchida, K. Expression Levels of Long Non-Coding RNAs Change in Models of Altered Muscle Activity and Muscle Mass. Int. J. Mol. Sci. 2020, 21, 1628. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.; Liu, J.; Xiao, J.; Yang, L.; Cai, M.; Shen, H.; Chen, X.; Ma, Y.; Hu, S.; Wang, Z.; et al. Lnc-mg is a long non-coding RNA that promotes myogenesis. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; He, W.; Huang, Y.; Xiang, H.; Guo, J.; Che, Y.; Cheng, X.; Hu, F.; Hu, M.; Ma, T.; et al. Fatty Acid Synthase–Suppressor Screening Identifies Sorting Nexin 8 as a Therapeutic Target for NAFLD. Hepatology 2021, 74, 2508–2525. [Google Scholar] [CrossRef]
- Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats. Available online: https://www.ncbi.nlm.nih.gov/sra (accessed on 30 November 2021).
| Content in Different Feeding Model (%) | Content in Different Feeding Model (%) | ||||
|---|---|---|---|---|---|
| Fatty Acid | Pasture-Fed (n = 3) | Barn-Fed (n = 3) | Fatty Acid% | Pasture-Fed (n = 3) | Barn-Fed (n = 3) |
| (C 6:0) | 0.600 ± 0.077 A | 0.130 ± 0.054 B | (C 16:1) cis | 2.960 ± 0.173 A | 1.230 ± 0.119 B |
| (C 8:0) | 0.084 ± 0.009 | 0.100 ± 0.030 | (C 17:1) cis | 1.240 ± 0.079 | 2.900 ± 0.649 |
| (C 10:0) | 0.199 ± 0.019 | 0.176 ± 0.024 | (C 18:1n9c) trans | 32.200 ± 1.329 | 0.620 ± 0.615 |
| (C 12:0) | 0.090 ± 0.011 b | 0.160 ± 0.017 a | (C 18:1n9t) cis | 2.450 ± 0.797 | 52.680 ± 0.553 |
| (C 13:0) | 0.000 ± 0.000 b | 0.070 ± 0.011 a | (C 20:1) cis | 0.040 ± 0.042 | 0.040 ± 0.007 |
| (C 14:0) | 2.050± 0.179 B | 3.360 ± 0.182 A | (C 24:1) cis | 0.000 ± 0.000 b | 0.130 ± 0.046 a |
| (C 15:0) | 0.890 ± 0.082 b | 1.620 ± 0.145 a | (C 18:2n6t) trans | 0.300 ± 0.010 | 0.488 ± 0.223 |
| (C 16:0) | 25.060 ± 0.928 b | 54.730 ± 9.748 a | (C 18:2n6c) cis | 1.210 ± 0.311 | 0.360 ± 0.054 |
| (C 17:0) | 2.770 ± 0.128 | 3.840 ± 0.541 | (C 18:3n6) cis | 0.040 ± 0.039 | 0.250 ± 0.041 |
| (C 18:0) | 27.370 ± 1.357 | 22.350 ± 11.160 | (C 18:3n3) cis | 0.000± 0.000 | 0.180 ± 0.076 |
| (C 20:0) | 0.050 ± 0.050 | 0.026 ± 0.008 | (C 20:3n6) cis | 0.000± 0.000 | 0.090 ± 0.075 |
| (C 21:0) | 0.080 ± 0.076 | 0.160 ± 0.027 | (C 20:4n6) cis | 0.000± 0.000 | 0.050 ± 0.024 |
| (C 23:0) | 0.000 ± 0.000 B | 0.070 ± 0.006 A | (C 20:5n3) cis | 0.000± 0.000 b | 0.030 ± 0.008 a |
| (C 14:1) cis | 0.320 ± 0.027 b | 0.450 ± 0.024 a | (C 20:2) cis | 0.000± 0.000 | 0.050 ± 0.027 |
| (C 15:1) cis | 0.000 ± 0.000 | 0.070 ± 0.055 | Total FA | 100.000 ± 0.003 | 100.100 ± 15.790 |
| PUFA n-3 | 0.000 ± 0.000 | 0.100 ± 0.048 | PUFA n-6 | 0.660 ± 0.228 a | 0.270 ± 0.062 b |
| MUFA | 39.210 ± 1.843 | 42.810 ± 14.970 | PUFA | 1.550 ± 0.314 | 1.280 ± 0.245 |
| Total SFA | 59.250 ± 2.065 b | 68.790 ± 2.603 a | Total UFA | 40.760 ± 0.021 a | 31.770 ± 0.086 b |
| AI | 23.530 ± 4.907 | 41.990 ± 9.237 | NVI | 2.490 ± 0.143 A | 1.360 ± 0.183 B |
| OFA | 27.110 ± 1.106 b | 39.920 ± 2.655 a | TI | 2.700 ± 0.226 | 3.940 ± 2.691 |
| DFA | 65.680 ± 1.257 a | 18.070 ± 11.550 b | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wang, Y.; Shan, H.; Zheng, Y.; Xuan, Z.; Hu, J.; Wei, M.; Wang, Z.; Liu, Q.; Li, Z. Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats. Foods 2022, 11, 381. https://doi.org/10.3390/foods11030381
Yang Y, Wang Y, Shan H, Zheng Y, Xuan Z, Hu J, Wei M, Wang Z, Liu Q, Li Z. Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats. Foods. 2022; 11(3):381. https://doi.org/10.3390/foods11030381
Chicago/Turabian StyleYang, Yufeng, Yan Wang, Huiquan Shan, Yalin Zheng, Zeyi Xuan, Jinling Hu, Mingsong Wei, Zhiqiang Wang, Qingyou Liu, and Zhipeng Li. 2022. "Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats" Foods 11, no. 3: 381. https://doi.org/10.3390/foods11030381
APA StyleYang, Y., Wang, Y., Shan, H., Zheng, Y., Xuan, Z., Hu, J., Wei, M., Wang, Z., Liu, Q., & Li, Z. (2022). Novel Insights into the Differences in Nutrition Value, Gene Regulation and Network Organization between Muscles from Pasture-Fed and Barn-Fed Goats. Foods, 11(3), 381. https://doi.org/10.3390/foods11030381









