Identification and Evolution of a Natural Tetr Protein Based on Molecular Docking and Development of a Fluorescence Polari-Zation Assay for Multi-Detection of 10 Tetracyclines in Milk
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Identification of Natural TetR Protein
2.3. Molecular Docking
2.4. Surface Plasmon Resonance (SPR)
2.5. Mutation of Natural TetR
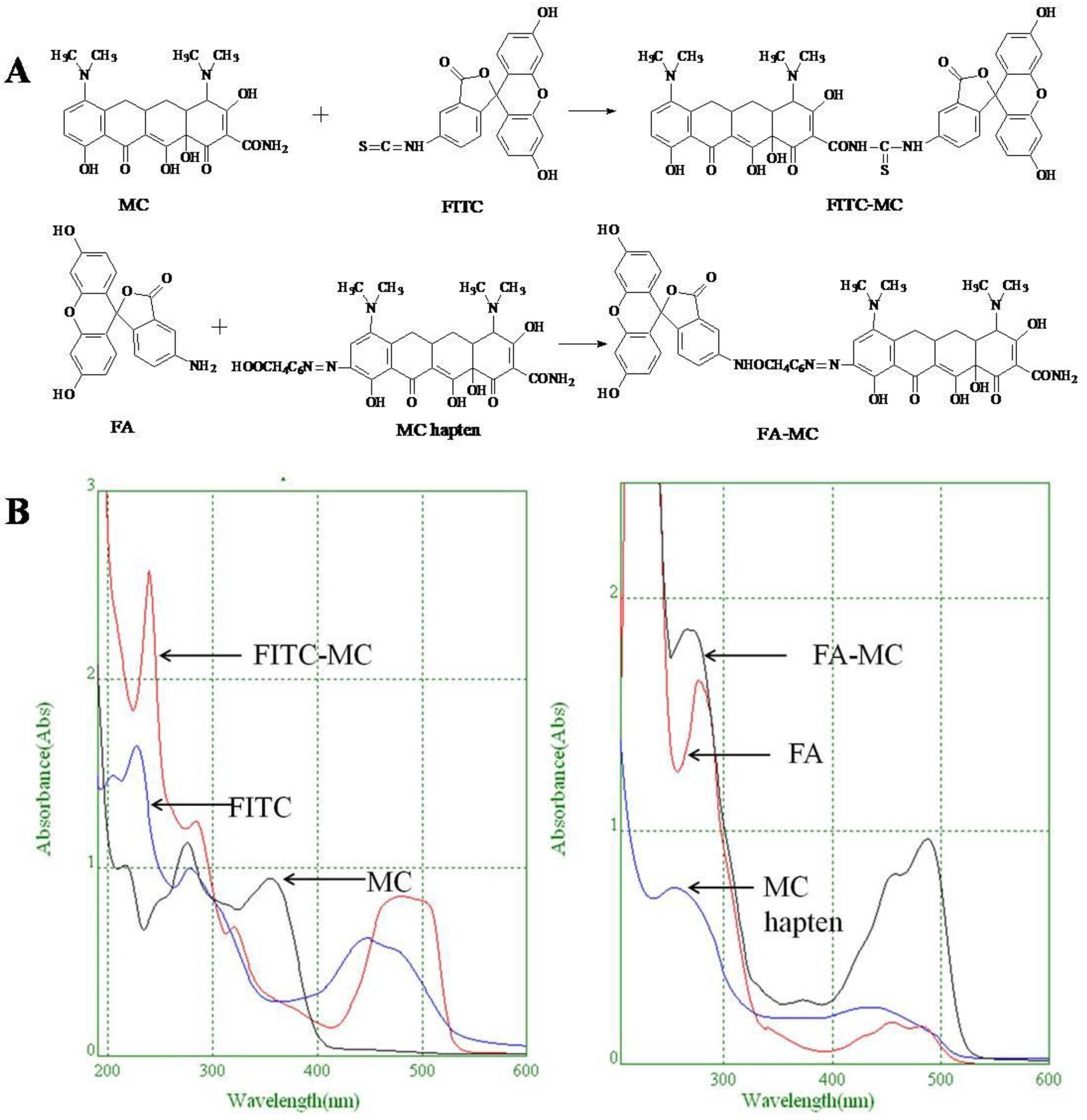
2.6. Synthesis of Fluorescent Tracers
2.7. Development of FPA Method
2.8. Method Application
3. Results and Discussions
3.1. Characterization of TetR Identity
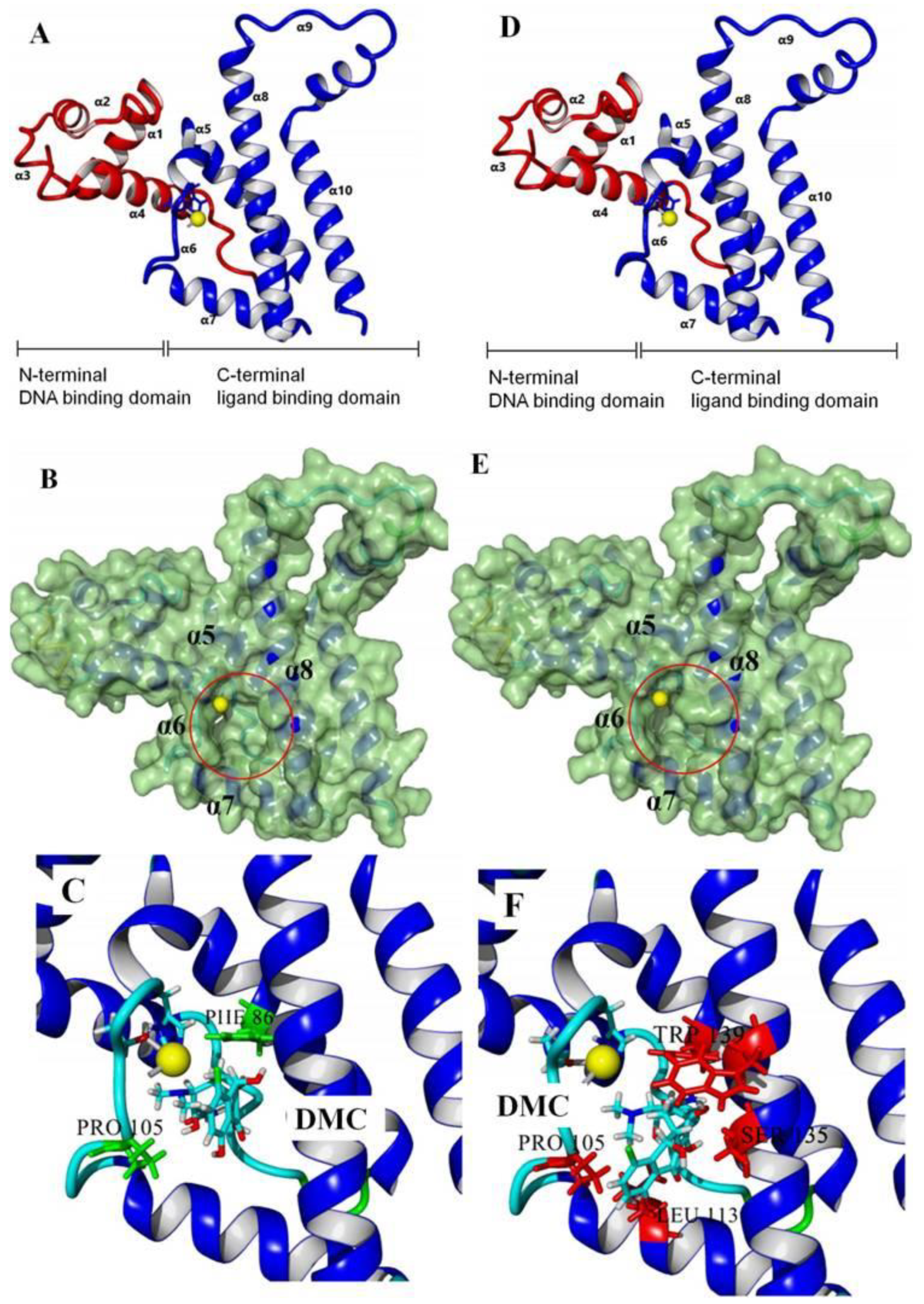
3.2. Recognition Mechanism of Natural TetR
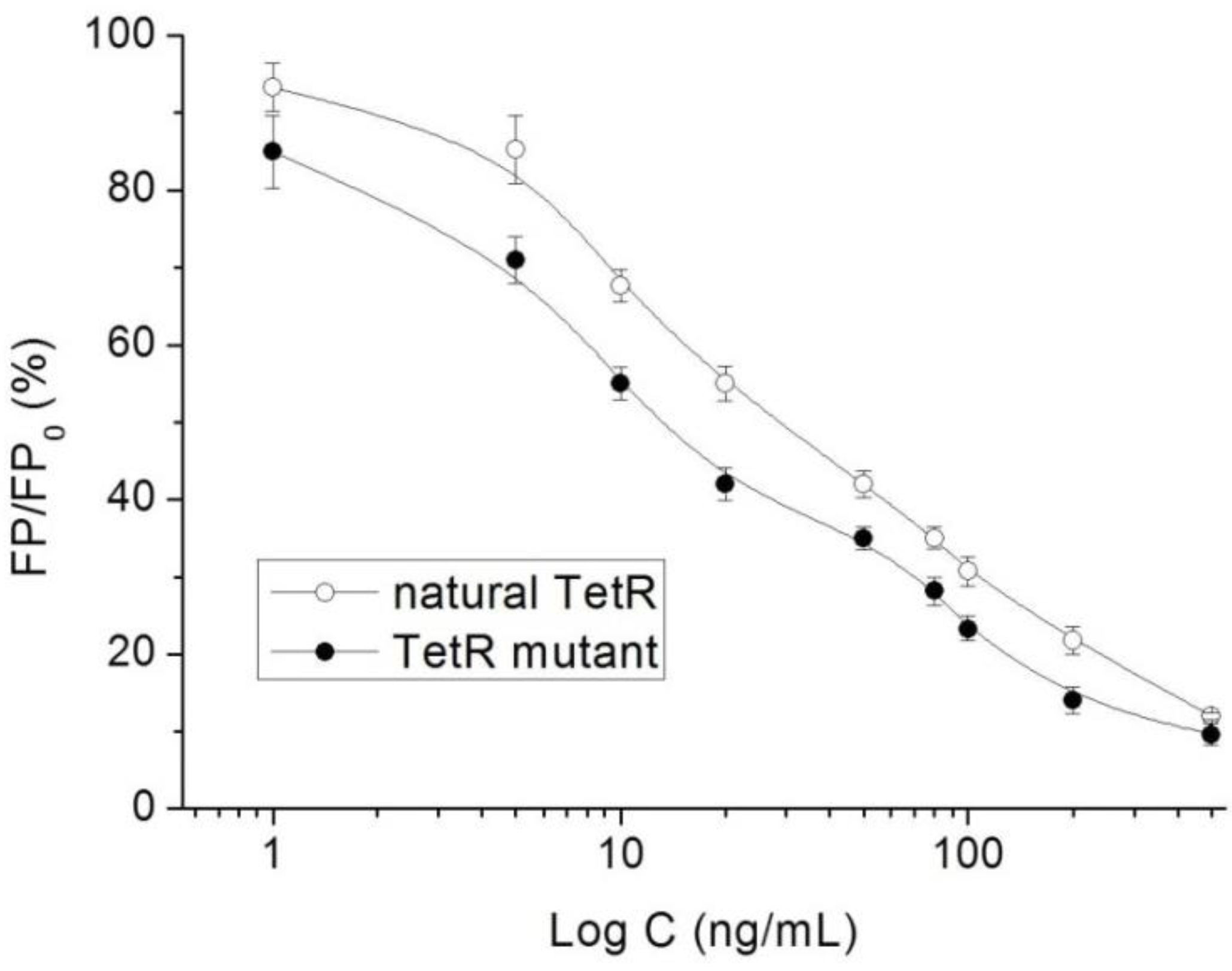
3.3. Characterization of TetR Mutant
3.4. Characterization of Fluorescent Tracers
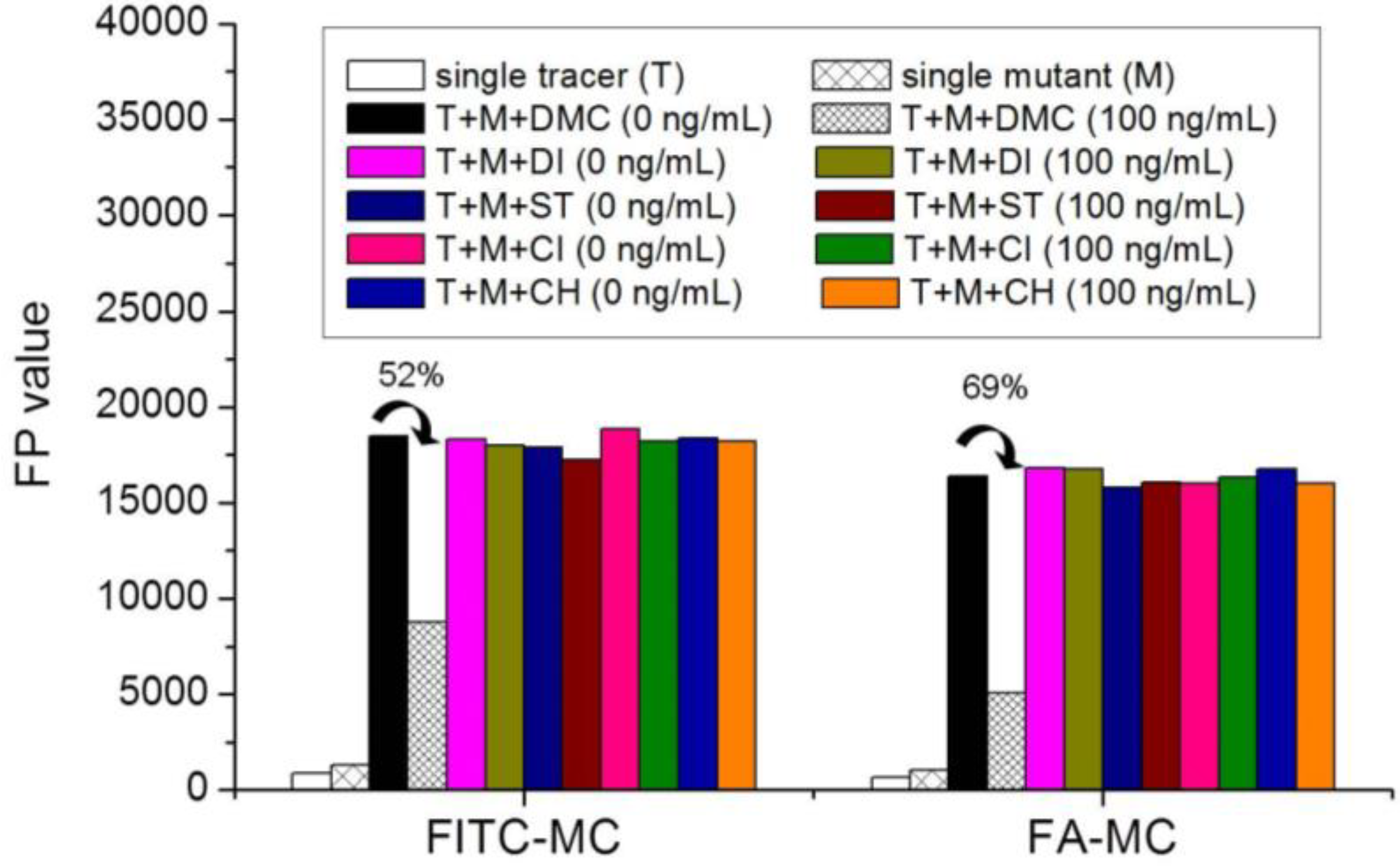
3.5. Comparison of the two Fluorescent Tracers
3.6. Optimization of FPA
3.7. Method Performances
3.8. Method Application
3.9. Comparison with Related Methods
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nelson, M.L.; Levy, S.B. The history of the tetracyclines. Ann. N. Y. Acad. Sci. 2011, 1241, 17–32. [Google Scholar] [CrossRef]
- Griffin, M.O.; Fricovsky, E.; Ceballos, G.; Villarreal, F. Tetracyclines: A pleitropic family of compounds with promising therapeutic properties. Review of the literature. Am. J. Physiol. Cell Ph. 2010, 299, C539–C548. [Google Scholar] [CrossRef]
- Levy, S.B.; McMurry, L. Detection of an inducible membrane protein associated with R-factor-mediated tetracycline resistance. Biochem. Biophys. Res. Commun. 1974, 56, 1060–1068. [Google Scholar] [CrossRef]
- Hillen, W.; Klock, G.; Kaffenberger, I.; Wray, L.V.; Reznikoff, W.S. Purification of the TET repressor and TET operator from the transposons Tn10 and characterization of their interaction. J. Biol. Chem. 1982, 252, 6605–6613. [Google Scholar] [CrossRef]
- Hinrichs, W.; Kisker, C.; Düvel, M.; Müller, A.; Tovar, K.; Hillen, W.; Saenger, W. Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 1994, 264, 418–420. [Google Scholar] [CrossRef]
- Kisker, C.; Hinrichs, W.; Tovar, K.; Hillen, W.; Saenger, W. The complex formed between Tet repressor and tetracycline-Mg2+ reveals mechanism of antibiotic resistance. J. Mol. Biol. 1995, 247, 260–280. [Google Scholar] [CrossRef] [PubMed]
- Orth, P.; Cordes, F.; Schnappinger, D.; Hillen, W.; Saenger, W.; Hinrichs, W. Conformational changes of the Tet repressor induced by tetracycline trapping. J. Mol. Biol. 1998, 279, 439–447. [Google Scholar] [CrossRef]
- Saenger, W.; Orth, P.; Kisker, C.; Hillen, W.; Hinrichs, W. The tetracycline repressor-A paradigm for a biological switch. Angew Chem. Int. Ed. Engl. 2000, 39, 2042–2052. [Google Scholar] [CrossRef] [PubMed]
- Orth, P.; Schnappinger, D.; Hillen, W.; Saenger, W.; Hinrichs, W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000, 7, 215–219. [Google Scholar]
- Nelson, M.L.; Levy, S. Reversal of tetracycline resistance mediated by different bacterial tetracycline resistance determinants by an inhibitor of the Tet(B) antiport protein. Antimicrob. Agents Chemother. 1999, 43, 1719–1724. [Google Scholar] [CrossRef]
- Routh, M.D.; Su, C.C.; Zhang, Q.; Yu, E.W. Structures of AcrR and CmeR: Insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. BBA-Proteins Proteom. 2009, 1794, 844–851. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Tanaka, Y.; Sakai, N.; Okada, U.; Yao, M.; Watanabe, N.; Tamura, T.; Tanaka, I. SCO4008, a putative TetR transcriptional repressor from Streptomyces coelicolor A3(2), regulates transcription of sco4007 by multidrug recognition. J. Mol. Biol. 2013, 425, 3289–3300. [Google Scholar] [CrossRef]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Teran, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Reichheld, S.E.; Savchenko, A.; Parkinson, J.; Davidson, A.R. A comprehensive analysis of structural and sequence conservation in the TetR family transcriptional regulators. J. Mol. Biol. 2010, 400, 847–864. [Google Scholar] [CrossRef] [PubMed]
- Cuthbertson, L.; Nodwell, J.R. The TetR family of regulators. Microbiol. Mol. Biol. Rev. 2013, 77, 440–475. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Watkins, W.J. Efflux pump: Their role in antibacterial drug discover. Curr. Med. Chem. 2001, 14, 1699–1711. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, T.; Jiang, L.; Wen, Y.; Song, Y.; Chen, Z.; Li, J. Characterization of SAV7471, a TetR-family transcriptional regulator involved in the regulation of coenzyme A metabolism in Streptomyces avermitilis. J. Bacteriol. 2013, 195, 4365–4372. [Google Scholar] [CrossRef] [PubMed]
- Sivaharini, S.; Smiline, G.; Vijayashree, P. Evaluation of the inhibitory effect of caffeicacid and gallic acid on tetR and tetM efflux pumps mediating tetracycline resistance in Streptococcus sp, using computational approach. J. King Saud. Univ. Sci. 2020, 32, 904–909. [Google Scholar]
- Pan, T.; Huang, P.; Xiong, G.; Maser, E. Isolation and identification of a repressor TetR for 3,17b-HSD expressional regulation in Comamonas testosterone. Chem.-Biol. Interact. 2015, 234, 205–212. [Google Scholar] [CrossRef]
- Pooja, M.; Tanaya, B.; Pooja, S.; Sabita, R.; Preeti, S. Isolation and identification of a TetR familyprotein that regulates the biodesulfurization operon. AMB Express. 2019, 9, 71. [Google Scholar]
- Murarka, P.; Srivastava, P. Characterization of DNA binding and ligand binding properties of theTetR family protein involved in regulation of dsz operon in Gordoniasp. IITR100. Int. J. Biol. Macromol. 2019, 141, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Beliakova, M.M.; Anokhina, M.M.; Spiridonova, V.A.; Dobrov, E.N.; Egorov, T.A.; Wittmann-Liebold, B.; Orth, P.; Saenger, W.; Kopylov, A.M. A direct photo-activated affinity modification of tetracycline transcription repressor protein TetR(D) with tetracycline. FEBS Lett. 2000, 477, 263–267. [Google Scholar] [CrossRef]
- Werten, S.; Dalm, D.; Palm, G.J.; Grimm, C.C.; Hinrichs, W. Tetracycline repressor allostery does not depend on divalent metal recognition. Biochemistry 2014, 53, 7990–7998. [Google Scholar] [CrossRef]
- Werten, S.; Schneider, J.; Palm, G.J.; Hinrichs, W. Modular organisation of inducer recognition and allostery in the tetracycline repressor. FEBS J. 2016, 283, 2102–2114. [Google Scholar] [CrossRef]
- Kedracka-Krok, S.; Gorecki, A.; Bonarek, P.; Wasylewski, Z. Kinetic and thermodynamic studies of Tet repressor-tetracycline interaction. Biochemistry 2005, 44, 1037–1046. [Google Scholar] [CrossRef]
- Alpeeva, I.S.; Anokhina, M.M.; Tashlitskii, V.N.; Kopylov, A.M. Determination of a fraction of the transcription repressor protein TetR(D) active for binding of tetracycline by a competitive binding method. Mosc. Univ. Chem. Bull. 2005, 60, 52–56. [Google Scholar]
- Palm, G.J.; Buchholz, I.; Werten, S.; Girbardt, B.; Berndt, L.; Delcea, M.; Hinrichs, W. Thermodynamics, cooperativity and stability of the tetracycline repressor(TetR) upon tetracycline binding. BBA-Proteins Proteom. 2020, 1868, 140404. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Huang, D.; Lai, C.; Zeng, G.; Qin, L.; Zhang, C.; Yi, H.; Li, B.; Deng, R.; Liu, S.; et al. Recent advances in sensors for tetracycline antibiotics and their applications. TrAC-Trend Anal. Chem. 2018, 109, 260–274. [Google Scholar] [CrossRef]
- Wang, G.; Xia, W.Q.; Liu, J.X.; Wang, J.P.; Liu, J. Directional evolution of TetR protein and development of a fluoroimmunoassay for screening of tetracyclines in egg. Microchem. J. 2019, 150, 104184. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.C.; Liu, J.; Wang, J.P. A receptor-based chemiluminescence enzyme linked immunosorbent assay for determination of tetracyclines in milk. Anal. Biochem. 2019, 564–565, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.C.; Link, N.; Fux, C.; Zisch, A.H.; Weber, W.; Fussenegger, M. Broad-spectrum protein biosensors for class-specific detection of antibiotics. Biotechnol. Bioeng. 2005, 89, 9–17. [Google Scholar] [CrossRef]
- Link, N.; Weber, W.; Fussenegger, M. A novel generic dipstick-based technology for rapid and precise detection of tetracycline, streptogramin and macrolide antibiotics in food samples. J. Biotechnol. 2007, 128, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Kling, A.; Chatelle, C.; Armbrecht, L.; Qelibari, E.; Kieninger, J.; Dincer, C.; Weber, W.; Urban, G. Multianalyte antibiotic detection on an electrochemical microfluidic platform. Anal. Chem. 2016, 88, 10036–10043. [Google Scholar] [CrossRef] [PubMed]
- Meyer, V.K.; Chatelle, C.V.; Weber, W.; Niessner, R.; Seidel, M. Flow-based regenerable chemiluminescence receptor assay for the detection of tetracyclines. Anal. Bioanal. Chem. 2020, 412, 3467–3476. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, S.; Ruyck, K.D.; Beloglazova, N.; Eremin, S.A.; DeSaeger, S.; Zhang, S.; Shen, J.; Wang, Z. Fluorescence polarization assays for chemical contaminants in food and environmental analyses. TrAC-Trend Anal. Chem. 2019, 114, 293–313. [Google Scholar] [CrossRef]
- Xia, W.Q.; Cui, P.L.; Wang, J.P.; Liu, J. Synthesis of photoaffinity labeled activity-based protein profiling probe and production of natural TetR protein for immunoassay of tetracyclines in milk. Microchem. J. 2021, 170, 106779. [Google Scholar] [CrossRef]
- He, T.; Cui, P.L.; Liu, J.; Feng, C.; Wang, J.P. Production of a natural dihydropteroate synthase and development of a signal amplified pseudo immunoassay for determination of sulfonamides in pork. J. Agric. Food Chem. 2022, 70, 3023–3032. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Liu, J.; Wang, J.P. Development of a dihydropteroate synthase based fluorescence polarization assay for detection of sulfonamides and studying its recognition mechanism. J. Agric. Food Chem. 2021, 69, 13953–13963. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Feng, C.; Qu, P.; Wang, G.; Liu, J.; Liu, J.X.; Wang, J.P. Determination of tetracyclines in chicken bydispersive solid phase microextraction based on metal-organic frameworks/molecularly imprinted nano-polymer and ultra performance liquid chromatography. Food Anal. Methods 2020, 13, 1211–1219. [Google Scholar] [CrossRef]

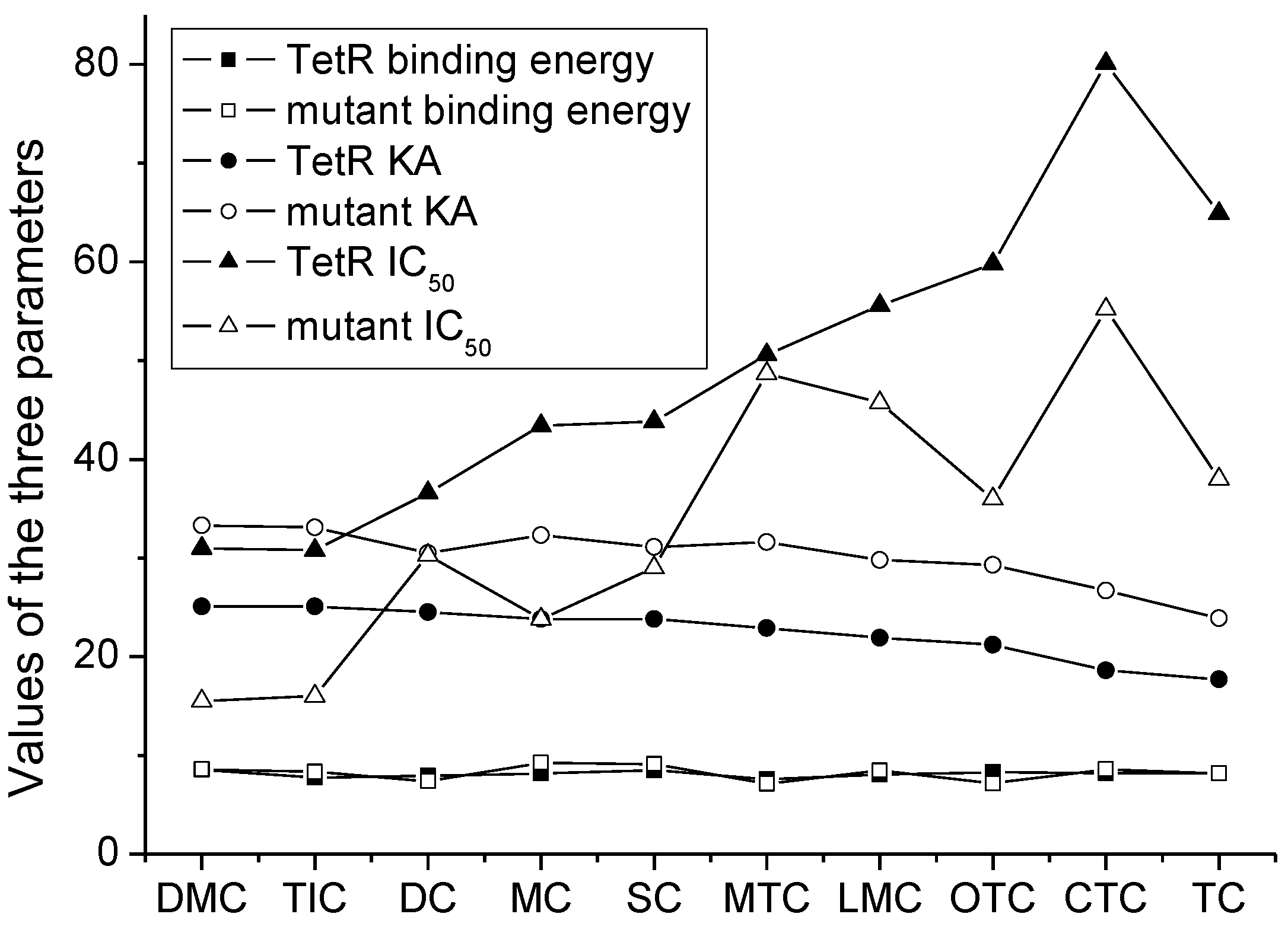
| Drug | Natural TetR | TetR Mutant | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Binding Sites | Contact Amino Acid | Binding Energy (kcal/mol) | KA | IC50 (ng/mL) | Binding Sites | Contact Amino Acid | Binding Energy (kcal/mol) | KA | IC50 (ng/mL) | LOD (ng/mL) | |
| MC | 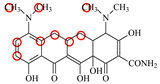 | PRO105 PHE86 LEU113 LEU131 | 8.16 | 23.8 | 43.4 | 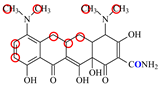 | PRO105 PHE86 LEU113 LEU131 SER135 | 9.26 | 32.3 | 23.8 | 0.5 |
| OTC | 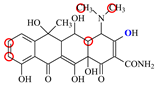 | PRO105 PHE86 LEU113 | 8.31 | 21.2 | 59.8 |  | PHE86 LEU113 LEU131 SER135TRP139 | 7.16 | 29.3 | 36.0 | 0.8 |
| TC | 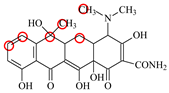 | PRO105 PHE86 LEU113 LEU131 | 8.16 | 17.7 | 64.9 | 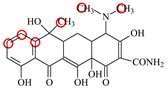 | PRO105 PHE86 LEU113 LEU131 | 8.16 | 23.9 | 38.0 | 1.3 |
| DC | 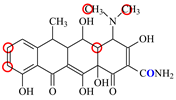 | PRO105 PHE86 LEU113 | 7.91 | 24.5 | 36.6 |  | PRO105 PHE86 LEU113 LEU131 TRP139 | 7.4 | 30.5 | 30.3 | 1.0 |
| DMC |  | PRO105 PHE86 | 8.56 | 25.1 | 31 | 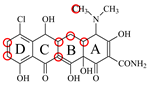 | PRO105 PHE86 LEU113 SER135 TRP139 | 8.57 | 33.3 | 15.5 | 0.4 |
| CTC | 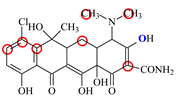 | PRO105 PHE86 LEU131 | 8.22 | 18.6 | 80.1 | 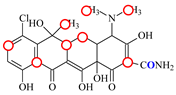 | PRO105 PHE86 LEU113 LEU131 SER135 | 8.57 | 26.7 | 55.2 | 1.4 |
| MTC | 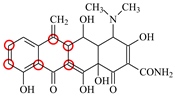 | PRO105 LEU113 LEU131 | 7.58 | 22.9 | 50.6 | 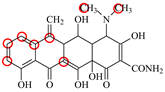 | PRO105 LEU113 LEU131 TRP139 | 7.13 | 31.6 | 48.7 | 1.5 |
| SC |  | PRO105 PHE86 LEU113 LEU131 | 8.48 | 23.8 | 43.8 | 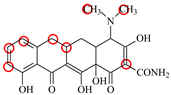 | PRO105 PHE86 LEU113 LEU131 TRP139 | 9.11 | 31.1 | 29.0 | 0.7 |
| TIC | 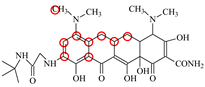 | PRO105 LEU113 LEU131 | 7.77 | 25.1 | 30.8 | 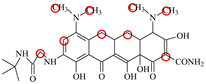 | PRO105 PHE86 LEU113 LEU131 | 8.33 | 33.1 | 16.0 | 0.4 |
| LMC | 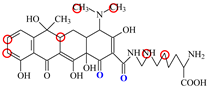 | PRO105 PHE86 LEU113 LEU131 | 8.06 | 21.9 | 55.6 |  | PRO105 PHE86 LEU113 LEU131 SER135 TRP139 | 8.47 | 29.8 | 45.7 | 1.0 |
| Drug | Added (ng/g) | Intra-Assay | Inter-Assay | ||
|---|---|---|---|---|---|
| Recovery (%) | Coefficient of Variation (%) | Recovery (%) | Coefficient of Variation (%) | ||
| MC | 10 | 85.2 | 7.5 | 73.5 | 6.2 |
| 100 | 93.3 | 8.5 | 86.2 | 8.5 | |
| TC | 10 | 88.1 | 7.3 | 81.2 | 11.6 |
| 100 | 87.2 | 7.1 | 93.5 | 9.2 | |
| OTC | 10 | 91.3 | 9.5 | 75.7 | 10.6 |
| 100 | 86.5 | 5.1 | 79.8 | 13.4 | |
| CTC | 10 | 78.6 | 11.2 | 81.2 | 9.3 |
| 100 | 81.2 | 9.4 | 87.4 | 8.7 | |
| TIC | 10 | 85.3 | 8.7 | 76.8 | 16.3 |
| 100 | 84.2 | 8.0 | 84.3 | 7.6 | |
| DC | 10 | 90.7 | 6.9 | 71.5 | 6.6 |
| 100 | 95.1 | 7.1 | 92.4 | 7.2 | |
| MTC | 10 | 79.2 | 8.5 | 83.1 | 8.5 |
| 100 | 78.5 | 9.4 | 91.2 | 9.4 | |
| LMC | 10 | 82.4 | 9.3 | 90.2 | 8.6 |
| 100 | 82.5 | 8.5 | 95.4 | 8.0 | |
| DMC | 10 | 77.4 | 8.7 | 79.4 | 6.2 |
| 100 | 86.8 | 7.9 | 84.4 | 10.5 | |
| SC | 10 | 83.4 | 9.4 | 78.2 | 12.6 |
| 100 | 90.5 | 10.3 | 91.2 | 14.3 | |
| Recognition Reagent | Method | Analyte | Limit of Detection (ng/mL) | Suitable for Screening? | Assay Time (from Add Sample) | Ref. |
|---|---|---|---|---|---|---|
| recombinant TetR | Chemiluminescence ELISA | 5 TCs | 0.005–0.016 | Yes | 40 min | 29 |
| TetR mutant | fluoroimmunoassay | 7 TCs | 0.3–0.58 | Yes | 30 min | 30 |
| recombinant TetR | ELISA | 8 TCs | 0.1–0.72 | Yes | >2 h | 31 |
| recombinant TetR | dipstick | 6 TCs | 10 | Yes | 35 min | 32 |
| recombinant TetR | electrochemical chip | TC | 6.33 | No | 15 min | 33 |
| recombinant TetR | flow-based chemiluminescence | TC | 0.1 | No | 30 min | 34 |
| natural TetR | chemiluminescence ELISA | 10 TCs | 0.002–0.009 | Yes | 40 min | 36 |
| TetR mutant | FPIA | 10 TCs | 0.4–1.5 | Yes | 2 min | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Liu, J.; Wang, J. Identification and Evolution of a Natural Tetr Protein Based on Molecular Docking and Development of a Fluorescence Polari-Zation Assay for Multi-Detection of 10 Tetracyclines in Milk. Foods 2022, 11, 3850. https://doi.org/10.3390/foods11233850
Xia W, Liu J, Wang J. Identification and Evolution of a Natural Tetr Protein Based on Molecular Docking and Development of a Fluorescence Polari-Zation Assay for Multi-Detection of 10 Tetracyclines in Milk. Foods. 2022; 11(23):3850. https://doi.org/10.3390/foods11233850
Chicago/Turabian StyleXia, Wanqiu, Jing Liu, and Jianping Wang. 2022. "Identification and Evolution of a Natural Tetr Protein Based on Molecular Docking and Development of a Fluorescence Polari-Zation Assay for Multi-Detection of 10 Tetracyclines in Milk" Foods 11, no. 23: 3850. https://doi.org/10.3390/foods11233850
APA StyleXia, W., Liu, J., & Wang, J. (2022). Identification and Evolution of a Natural Tetr Protein Based on Molecular Docking and Development of a Fluorescence Polari-Zation Assay for Multi-Detection of 10 Tetracyclines in Milk. Foods, 11(23), 3850. https://doi.org/10.3390/foods11233850






