Genetic Diversity and Potential Virulence of Listeria monocytogenes Isolates Originating from Polish Artisanal Cheeses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Whole-Genome Sequencing
2.2.1. DNA Extraction
2.2.2. Library Preparation
2.2.3. Sequencing
2.2.4. Data Analysis
3. Results
3.1. Analysis of Genetic Diversity of L. monocytogenes Isolates
3.2. Analysis of Potential Virulence of L. monocytogenes Isolates
4. Discussion
4.1. Genetic Diversity of L. monocytogenes Isolates
4.2. Virulence Genes of L. monocytogenes Isolates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA and ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union One Health 2020 Zoonoses Report. EFSA J. 2021, 19, 6971. [Google Scholar] [CrossRef]
- Desai, A.N.; Anyoha, A.; Madoff, L.C.; Lassmann, B. Changing epidemiology of Listeria monocytogenes outbreaks, sporadic cases, and recalls globally: A review of ProMED reports from 1996 to 2018. Int. J. Infect. Dis. 2019, 84, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Allerberger, F.; Wagner, M. Listeriosis: A resurgent foodborne infection. Clin. Microbiol. Infect. 2010, 16, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Lewańska, M.; Godela, A.; Myga-Nowak, M. Listeriosis. Modern perception of epidemiological threat. Post. Mikrobiol. 2018, 57, 106–116. [Google Scholar] [CrossRef]
- Schiavano, G.F.; Ateba, C.N.; Petruzzelli, A.; Mele, V.; Amagliani, G.; Guidi, F.; De Santi, M.; Pomilio, F.; Blasi, G.; Gattuso, A.; et al. Whole-Genome Sequencing characterization of virulence profiles of Listeria monocytogenes food and human isolates and in vitro adhesion/invasion assessment. Microorganisms 2022, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Henri, C.; Félix, B.; Guillier, L.; Leekitcharoenphon, P.; Michelon, D.; Mariet, J.F.; Aarestrup, F.M.; Mistou, M.Y.; Hendriksen, R.S.; Roussel, S. Population genetic structure of Listeria monocytogenes strains as determined by pulsed-field gel electrophoresis and multilocus sequence typing. Appl. Environ. Microbiol. 2016, 82, 5720–5728. [Google Scholar] [CrossRef]
- Bergholz, T.M.; Shah, M.K.; Burall, L.S.; Rakic-Martinez, M.; Datta, A.R. Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 2018, 102, 3475–3485. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Chen, H.; Chen, J.; Zhang, J.; Zhang, Z.; Yang, Y.; Xu, Z.; Zhan, L.; Mei, L. Prevalence, genotypic characteristics and antibiotic resistance of Listeria monocytogenes from retail foods in bulk in Zhejiang Province, China. Front. Microbiol. 2019, 10, 1710. [Google Scholar] [CrossRef]
- Cardenas-Alvarez, M.X.; Townsend Ramsett, M.K.; Malekmohammadi, S.; Bergholz, T.M. Evidence of hypervirulence in Listeria monocytogenes clonal complex 14. J. Med. Microbiol. 2019, 68, 1677–1685. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.; Pouillot, R.; Dennis, S.; Xian, Z.; Luchansky, J.B.; Porto-Fett, A.C.S.; Lindsay, J.A.; Hammack, T.S.; Allard, M.; et al. Genetic diversity and profiles of genes associated with virulence and stress resistance among isolates from the 2010–2013 interagency Listeria monocytogenes market basket survey. PLoS ONE 2020, 15, e0231393. [Google Scholar] [CrossRef]
- Meng, Z.; Zhang, L.; Xin, L.; Lin, K.; Yi, H.; Han, X. Technological characterization of Lactobacillus in semihard artisanal goat cheeses from different Mediterranean areas for potential use as nonstarter lactic acid bacteria. J. Dairy Sci. 2018, 101, 2887–2896. [Google Scholar] [CrossRef] [PubMed]
- Mata, G.M.; Martins, E.; Machado, S.G.; Pinto, M.S.; de Carvalho, A.F.; Vanetti, M.C. Performance of two alternative methods for Listeria detection throughout Serro Minas cheese ripening. Braz. J. Microbiol. 2016, 47, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Barría, C.; Singer, R.S.; Bueno, I.; Estrada, E.; Rivera, D.; Ulloa, S.; Fernández, J.; Mardones, F.O.; Moreno-Switt, A.I. Tracing Listeria monocytogenes contamination in artisanal cheese to the processing environments in cheese producers in southern Chile. Food Microbiol. 2020, 90, 103499. [Google Scholar] [CrossRef] [PubMed]
- Pyz-Łukasik, R.; Knysz, P.; Gondek, M. Hygiene quality and consumer safety of traditional short- and long-ripened cheeses from Poland. J. Food Qual. 2018, 2018, 8732412. [Google Scholar] [CrossRef]
- Espinosa-Mata, E.; Mejía, L.; Villacís, J.E.; Alban, V.; Zapata, S. Detection and genotyping of Listeria monocytogenes in artisanal soft cheeses from Ecuador. Rev. Argent. Microbiol. 2022, 54, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Kevenk, T.O.; Gulel, G.T. Prevalence, antimicrobial resistance and serotype distribution of Listeria monocytogenes isolated from raw milk and dairy products. J. Food Saf. 2016, 36, 11–18. [Google Scholar] [CrossRef]
- Jackson, K.A.; Gould, L.H.; Hunter, J.C.; Kucerova, Z.; Jackson, B. Listeriosis outbreaks associated with soft cheeses, United States, 1998–2014. Emerg. Infect. Dis. 2018, 24, 1116–1118. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Outbreak of listeriosis associated with homemade Mexican-style cheese--North Carolina. Morb. Mortal. Wkly Rep. 2001, 50, 560–562. [Google Scholar]
- Acciari, V.A.; Iannetti, L.; Gattuso, A.; Sonnessa, M.; Scavia, G.; Montagna, C.; Addante, N.; Torresi, M.; Zocchi, L.; Scattolini, S.; et al. Tracing sources of Listeria contamination in traditional Italian cheese associated with a US outbreak: Investigations in Italy. Epidemiol. Infect. 2016, 144, 2719–2727. [Google Scholar] [CrossRef]
- Pyz-Łukasik, R.; Gondek, M.; Winiarczyk, D.; Michalak, K.; Paszkiewicz, W.; Piróg-Komorowska, A.; Policht, A.; Ziomek, M. Occurrence of Listeria monocytogenes in artisanal cheeses from Poland and its identification by MALDI-TOF MS. Pathogens 2021, 10, 632. [Google Scholar] [CrossRef]
- PN-EN ISO 11290-1:1999; Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes—Part 1: Detection Method. PKN: Warszawa, Poland, 1999.
- PN-EN ISO 11290-1: 2017-07; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp.—Part 1: Detection Method. PKN: Warszawa, Poland, 2017.
- Doumith, M.; Buchrieser, C.; Glaser, P.; Jacquet, C.; Martin, P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004, 42, 3819–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moura, A.; Criscuolo, A.; Pouseele, H.; Maury, M.M.; Leclercq, A.; Tarr, C.; Björkman, J.T.; Dallman, T.; Reimer, A.; Enouf, V.; et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016, 2, 16185. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Lam, T.T.; Zhu, H.; Guan, Y. Two Methods for Mapping and Visualizing Associated Data on Phylogeny Using Ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Orsi, R.H.; den Bakker, H.C.; Wiedmann, M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011, 301, 79–96. [Google Scholar] [CrossRef]
- Salcedo, C.; Arreaza, L.; Alcalá, B.; de la Fuente, L.; Vázquez, J.A. Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 2003, 41, 757–762. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, A.; Zhu, R.; Lan, R.; Jin, D.; Cui, Z.; Wang, Y.; Li, Z.; Wang, Y.; Xu, J.; et al. Genetic diversity and molecular typing of Listeria monocytogenes in China. BMC Microbiol. 2012, 12, 119. [Google Scholar] [CrossRef]
- Yin, Y.; Tan, W.; Wang, G.; Kong, S.; Zhou, X.; Zhao, D.; Jia, Y.; Pan, Z.; Jiao, X. Geographical and longitudinal analysis of Listeria monocytogenes genetic diversity reveals its correlation with virulence and unique evolution. Microbiol. Res. 2015, 175, 84–92. [Google Scholar] [CrossRef]
- Kim, S.W.; Haendiges, J.; Keller, E.N.; Myers, R.; Kim, A.; Lombard, J.E.; Karns, J.S.; Van Kessel, J.A.S.; Haley, B.J. Genetic diversity and virulence profiles of Listeria monocytogenes recovered from bulk tank milk, milk filters, and milking equipment from dairies in the United States (2002 to 2014). PLoS ONE 2018, 13, e0197053. [Google Scholar] [CrossRef]
- Caruso, M.; Fraccalvieri, R.; Pasquali, F.; Santagada, G.; Latorre, L.M.; Difato, L.M.; Miccolupo, A.; Normanno, G.; Parisi, A. Antimicrobial susceptibility and multilocus sequence typing of Listeria monocytogenes isolated over 11 years from food, humans, and the environment in Italy. Foodborne Pathog. Dis. 2020, 17, 284–294. [Google Scholar] [CrossRef]
- Ragon, M.; Wirth, T.; Hollandt, F.; Lavenir, R.; Lecuit, M.; Le Monnier, A.; Brisse, S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008, 4, e1000146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Jiao, Y.; Lan, R.; Xu, X.; Liu, G.; Wang, X.; Zhang, L.; Pang, H.; Jin, D.; Dai, H.; et al. Characterization of Listeria monocytogenes isolated from human listeriosis cases in China. Emerg. Microbes. Infect. 2015, 4, e50. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.T.; Ko, W.C.; Chan, Y.J.; Lu, J.J.; Tsai, H.Y.; Liao, C.H.; Sheng, W.H.; Teng, L.J.; Hsueh, P.R. Disease burden of invasive listeriosis and molecular characterization of clinical isolates in Taiwan, 2000–2013. PLoS ONE 2015, 10, e0141241. [Google Scholar] [CrossRef]
- Pérez-Trallero, E.; Zigorraga, C.; Artieda, J.; Alkorta, M.; Marimón, J.M. Two outbreaks of Listeria monocytogenes infection, Northern Spain. Emerg. Infect. Dis. 2014, 20, 2155–2157. [Google Scholar] [CrossRef]
- Thomas, J.; Govender, N.; McCarthy, K.M.; Erasmus, L.K.; Doyle, T.J.; Allam, M.; Ismail, A.; Ramalwa, N.; Sekwadi, P.; Ntshoe, G.; et al. Outbreak of listeriosis in South Africa associated with processed meat. N. Engl. J. Med. 2020, 382, 632–643. [Google Scholar] [CrossRef]
- Maury, M.M.; Tsai, Y.H.; Charlier, C.; Touchon, M.; Chenal-Francisque, V.; Leclercq, A.; Criscuolo, A.; Gaultier, C.; Roussel, S.; Brisabois, A.; et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016, 48, 308–313, Erratum in: Nat. Genet. 2017, 49, 651. [Google Scholar] [CrossRef]
- Knabel, S.J.; Reimer, A.; Verghese, B.; Lok, M.; Ziegler, J.; Farber, J.; Pagotto, F.; Graham, M.; Nadon, C.A.; Canadian Public Health Laboratory Network (CPHLN). Sequence typing confirms that a predominant Listeria monocytogenes clone caused human listeriosis cases and outbreaks in Canada from 1988 to 2010. J. Clin. Microbiol. 2012, 50, 1748–1751. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs. Off. J. Eur. Union 2005, 338, 1–32. [Google Scholar]
- Vilchis-Rangel, R.E.; Espinoza-Mellado, M.D.R.; Salinas-Jaramillo, I.J.; Martinez-Peña, M.D.; Rodas-Suárez, O.R. Association of Listeria monocytogenes LIPI-1 and LIPI-3 marker llsX with invasiveness. Curr. Microbiol. 2019, 76, 637–643. [Google Scholar] [CrossRef]
- Camejo, A.; Carvalho, F.; Reis, O.; Leitão, E.; Sousa, S.; Cabanes, D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence 2011, 2, 379–394. [Google Scholar] [CrossRef]
- Bierne, H.; Sabet, C.; Personnic, N.; Cossart, P. Internalins: A complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 2007, 9, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Doumith, M.; Cazalet, C.; Simoes, N.; Frangeul, L.; Jacquet, C.; Kunst, F.; Martin, P.; Cossart, P.; Glaser, P.; Buchrieser, C. New aspects regarding evolution and virulence of Listeria monocytogenes revealed by comparative genomics and DNA arrays. Infect. Immun. 2004, 72, 1072–1083. [Google Scholar] [CrossRef] [Green Version]
- Dortet, L.; Mostowy, S.; Cossart, P. Listeria and autophagy escape: Involvement of InlK, an internalin-like protein. Autophagy 2012, 8, 132–134. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Halvorsen, E.M.; Ammendolia, D.A.; Mor-Vaknin, N.; O’Riordan, M.X.D.; Brumell, J.H.; Markovitz, D.M.; Higgins, D.E. Invasion of the brain by Listeria monocytogenes is mediated by InlF and host cell vimentin. mBio 2018, 9, e00160-18. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.; Gianfelice, A.; Gray-Owen, S.D.; Ireton, K. Impact of the Listeria monocytogenes protein InlC on infection in mice. Infect. Immun. 2013, 81, 1334–1340. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Cao, G.; Zhang, J.; Pan, H.; Zhang, D.; Kuang, D.; Yang, X.; Xu, X.; Shi, X.; Meng, J. Characterization of internalin genes in Listeria monocytogenes from food and humans, and their association with the invasion of Caco-2 cells. Gut Pathog. 2019, 11, 30. [Google Scholar] [CrossRef]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; González-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef] [PubMed]
- Milohanic, E.; Glaser, P.; Coppée, J.Y.; Frangeul, L.; Vega, Y.; Vázquez-Boland, J.A.; Kunst, F.; Cossart, P.; Buchrieser, C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 2003, 47, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Osborne, S.E.; Brumell, J.H. Listeriolysin O: From bazooka to Swiss army knife. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160222. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Sun, J.; Yu, H.; Ma, T.; Guan, C.; Zeng, H.; Zhang, X.; Chen, Z.; Song, H. Listeriolysin O pore-forming activity is required for ERK1/2 phosphorylation during Listeria monocytogenes infection. Front. Immunol. 2020, 11, 1146. [Google Scholar] [CrossRef] [PubMed]
- Quereda, J.J.; Andersson, C.; Cossart, P.; Johansson, J.; Pizarro-Cerdá, J. Role in virulence of phospholipases, listeriolysin O and listeriolysin S from epidemic Listeria monocytogenes using the chicken embryo infection model. Vet. Res. 2018, 49, 13. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Agaisse, H. The metalloprotease Mpl supports Listeria monocytogenes dissemination through resolution of membrane protrusions into vacuoles. Infect. Immun. 2016, 84, 1806–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillich, H.; Puri, M.; Chakraborty, T. ActA of Listeria monocytogenes and its manifold activities as an important listerial virulence factor. Curr. Top. Microbiol. Immunol. 2017, 399, 113–132. [Google Scholar] [CrossRef]
- Cotter, P.D.; Draper, L.A.; Lawton, E.M.; Daly, K.M.; Groeger, D.S.; Casey, P.G.; Ross, R.P.; Hill, C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008, 4, e1000144. [Google Scholar] [CrossRef] [PubMed]
- Milohanic, E.; Pron, B.; The European Listeria Genome Consortium; Berche, P.; Gaillard, J.L. Identification of new loci involved in adhesion of Listeria monocytogenes to eukaryotic cells. European Listeria Genome Consortium. Microbiology 2000, 146, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Rismondo, J.; Haddad, T.F.M.; Shen, Y.; Loessner, M.J.; Gründling, A. GtcA is required for LTA glycosylation in Listeria monocytogenes serovar 1/2a and Bacillus subtilis. Cell Surf. 2020, 6, 100038. [Google Scholar] [CrossRef]
- Faith, N.; Kathariou, S.; Cheng, Y.; Promadej, N.; Neudeck, B.L.; Zhang, Q.; Luchansky, J.; Czuprynski, C. The role of L. monocytogenes serotype 4b gtcA in gastrointestinal listeriosis in A/J mice. Foodborne Pathog. Dis. 2009, 6, 39–48. [Google Scholar] [CrossRef]
- Cabanes, D.; Dussurget, O.; Dehoux, P.; Cossart, P. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 2004, 51, 1601–1614. [Google Scholar] [CrossRef]
- Cabanes, D.; Sousa, S.; Cebriá, A.; Lecuit, M.; García-del Portillo, F.; Cossart, P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 2005, 24, 2827–2838. [Google Scholar] [CrossRef]
- Alonzo, F., 3rd; Port, G.C.; Cao, M.; Freitag, N.E. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 2009, 77, 2612–2623. [Google Scholar] [CrossRef]
- Lebreton, A.; Job, V.; Ragon, M.; Le Monnier, A.; Dessen, A.; Cossart, P.; Bierne, H. Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA. mBio 2014, 5, e00775-13. [Google Scholar] [CrossRef] [PubMed]
- de las Heras, A.; Cain, R.J.; Bielecka, M.K.; Vázquez-Boland, J.A. Regulation of Listeria virulence: PrfA master and commander. Curr. Opin. Microbiol. 2011, 14, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Gaillot, O.; Pellegrini, E.; Bregenholt, S.; Nair, S.; Berche, P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 2000, 35, 1286–1294. [Google Scholar] [CrossRef] [PubMed]
- Rouquette, C.; de Chastellier, C.; Nair, S.; Berche, P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol. Microbiol. 1998, 27, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Derré, I.; Msadek, T.; Gaillot, O.; Berche, P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol. Microbiol. 2000, 35, 800–811. [Google Scholar] [CrossRef]
- Nair, S.; Frehel, C.; Nguyen, L.; Escuyer, V.; Berche, P. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol. Microbiol. 1999, 31, 185–196. [Google Scholar] [CrossRef]
- Aubry, C.; Goulard, C.; Nahori, M.A.; Cayet, N.; Decalf, J.; Sachse, M.; Boneca, I.G.; Cossart, P.; Dussurget, O. OatA, a peptidoglycan O-acetyltransferase involved in Listeria monocytogenes immune escape, is critical for virulence. J. Infect. Dis. 2011, 204, 731–740. [Google Scholar] [CrossRef]
- Réglier-Poupet, H.; Frehel, C.; Dubail, I.; Beretti, J.L.; Berche, P.; Charbit, A.; Raynaud, C. Maturation of lipoproteins by type II signal peptidase is required for phagosomal escape of Listeria monocytogenes. J. Biol. Chem. 2003, 278, 49469–49477. [Google Scholar] [CrossRef]
- Réglier-Poupet, H.; Pellegrini, E.; Charbit, A.; Berche, P. Identification of LpeA, a PsaA-like membrane protein that promotes cell entry by Listeria monocytogenes. Infect. Immun. 2003, 71, 474–482. [Google Scholar] [CrossRef]
- Keeney, K.M.; Stuckey, J.A.; O’Riordan, M.X. LplA1-dependent utilization of host lipoyl peptides enables Listeria cytosolic growth and virulence. Mol. Microbiol. 2007, 66, 758–770. [Google Scholar] [CrossRef]
- Poimenidou, S.V.; Dalmasso, M.; Papadimitriou, K.; Fox, E.M.; Skandamis, P.N.; Jordan, K. Virulence gene sequencing highlights similarities and differences in sequences in Listeria monocytogenes Serotype 1/2a and 4b strains of clinical and food origin from 3 different geographic locations. Front. Microbiol. 2018, 9, 1103. [Google Scholar] [CrossRef] [PubMed]
- Bechtel, T.D.; Gibbons, J.G. Population genomic analysis of Listeria monocytogenes from food reveals substrate-specific genome variation. Front. Microbiol. 2021, 12, 620033. [Google Scholar] [CrossRef] [PubMed]
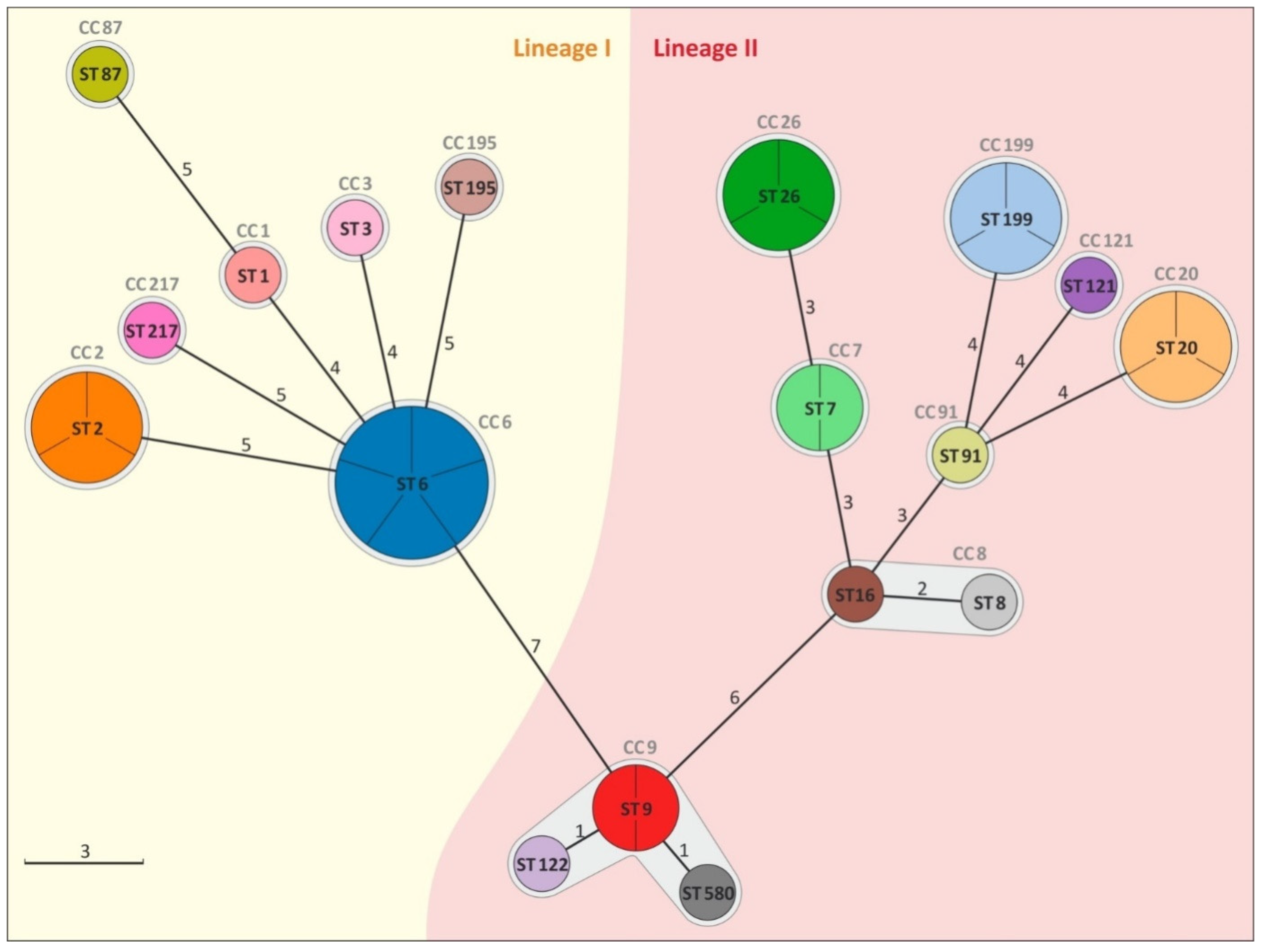
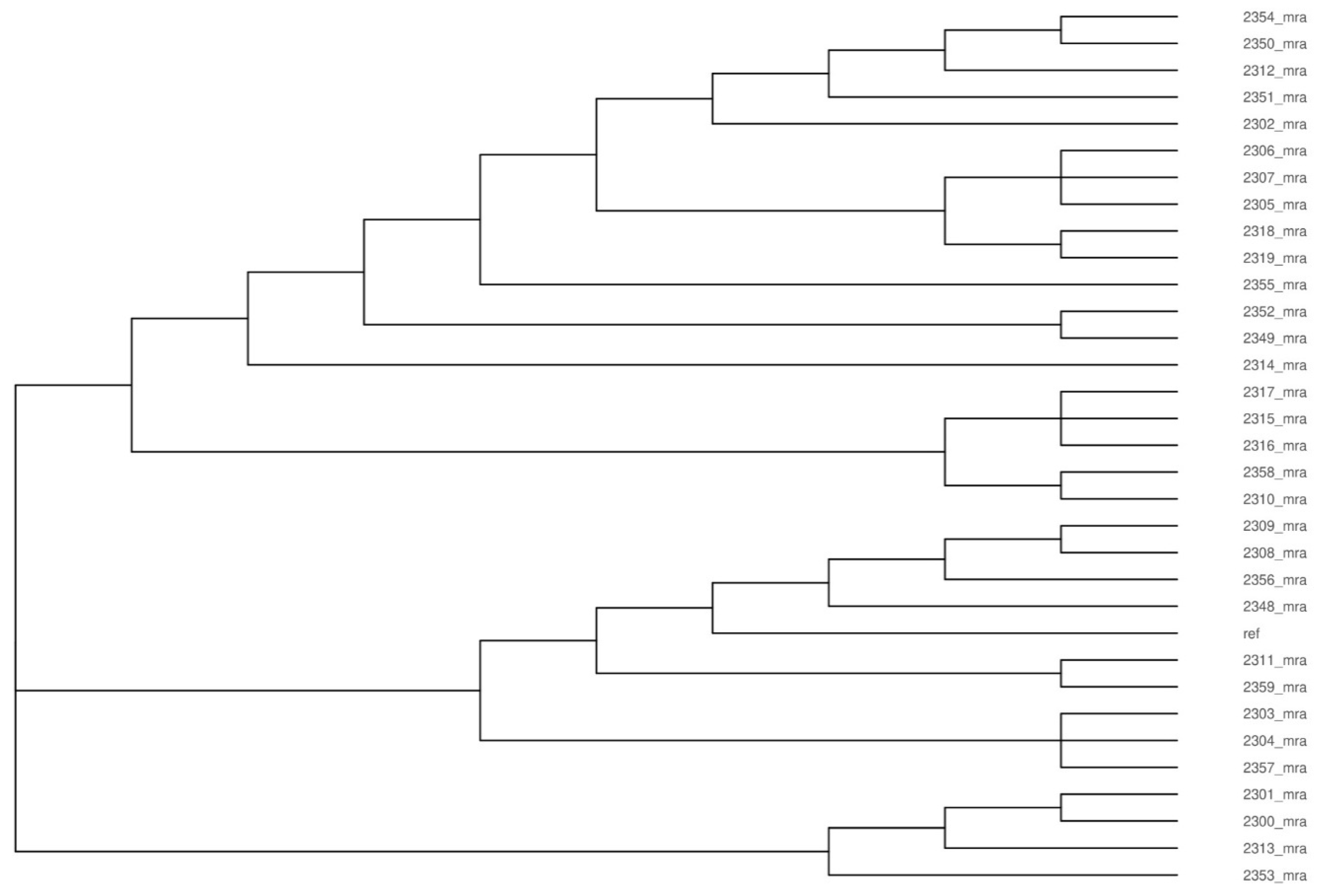

| Administrative Divisions | Cheese Dairy | Number of Isolates n = 32 | Year of Isolation | Molecular Group | Lineage | ST | CC |
|---|---|---|---|---|---|---|---|
| 2 | D1, D2, I | 5 | 2016, 2018 | IV b 4ab-4b-4d-4e | I | 6 | 6 |
| 2 | D3, G1 | 3 | 2014, 2018 | II a 1/2a-3a | II | 20 | 20 |
| 2, 3 | G3, J | 3 | 2015, 2017, 2018 | IV b 4ab-4b-4d-4e | I | 2 | 2 |
| 2 | I | 3 | 2015 | II a 1/2a-3a | II | 26 | 26 |
| 2 | H | 3 | 2018 | II a 1/2a-3a | II | 199 | 199 |
| 2 | G2 | 2 | 2016 | II c 1/2c-3c | II | 9 | 9 |
| 2 | F | 2 | 2017 | II a 1/2a-3a | II | 7 | 7 |
| 2 | G1 | 1 | 2014 | IV b 4ab-4b-4d-4e | I | 1 | 1 |
| 2 | F | 1 | 2018 | II a 1/2a-3a | II | 16 | 8 |
| 4 | K | 1 | 2018 | II a 1/2a-3a | II | 121 | 121 |
| 2 | I | 1 | 2018 | II c 1/2c-3c | II | 122 | 9 |
| 2 | D4 | 1 | 2014 | IIb 1/2b-3b-7 | I | 87 | 87 |
| 1 | A | 1 | 2016 | IV b 4ab-4b-4d-4e | I | 217 | 217 |
| 1 | C | 1 | 2016 | II b 1/2b-3b-7 | I | 195 | 195 |
| 2 | E | 1 | 2018 | II a 1/2a-3a | II | 91 | 14 |
| 2 | G2 | 1 | 2017 | II b 1/2b-3b-7 | I | 3 | 3 |
| 1 | C | 1 | 2017 | II c 1/2c-3c | II | 580 | 9 |
| 1 | B | 1 | 2018 | II a 1/2a-3a | II | 8 | 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pyz-Łukasik, R.; Paszkiewicz, W.; Kiełbus, M.; Ziomek, M.; Gondek, M.; Domaradzki, P.; Michalak, K.; Pietras-Ożga, D. Genetic Diversity and Potential Virulence of Listeria monocytogenes Isolates Originating from Polish Artisanal Cheeses. Foods 2022, 11, 2805. https://doi.org/10.3390/foods11182805
Pyz-Łukasik R, Paszkiewicz W, Kiełbus M, Ziomek M, Gondek M, Domaradzki P, Michalak K, Pietras-Ożga D. Genetic Diversity and Potential Virulence of Listeria monocytogenes Isolates Originating from Polish Artisanal Cheeses. Foods. 2022; 11(18):2805. https://doi.org/10.3390/foods11182805
Chicago/Turabian StylePyz-Łukasik, Renata, Waldemar Paszkiewicz, Michał Kiełbus, Monika Ziomek, Michał Gondek, Piotr Domaradzki, Katarzyna Michalak, and Dorota Pietras-Ożga. 2022. "Genetic Diversity and Potential Virulence of Listeria monocytogenes Isolates Originating from Polish Artisanal Cheeses" Foods 11, no. 18: 2805. https://doi.org/10.3390/foods11182805
APA StylePyz-Łukasik, R., Paszkiewicz, W., Kiełbus, M., Ziomek, M., Gondek, M., Domaradzki, P., Michalak, K., & Pietras-Ożga, D. (2022). Genetic Diversity and Potential Virulence of Listeria monocytogenes Isolates Originating from Polish Artisanal Cheeses. Foods, 11(18), 2805. https://doi.org/10.3390/foods11182805






