Consumption of Antioxidant-Rich “Cerrado” Cashew Pseudofruit Affects Hepatic Gene Expression in Obese C57BL/6J High Fat-Fed Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vegetal Material
2.2. Mouse Diet Studies
2.3. Fasting Glucose
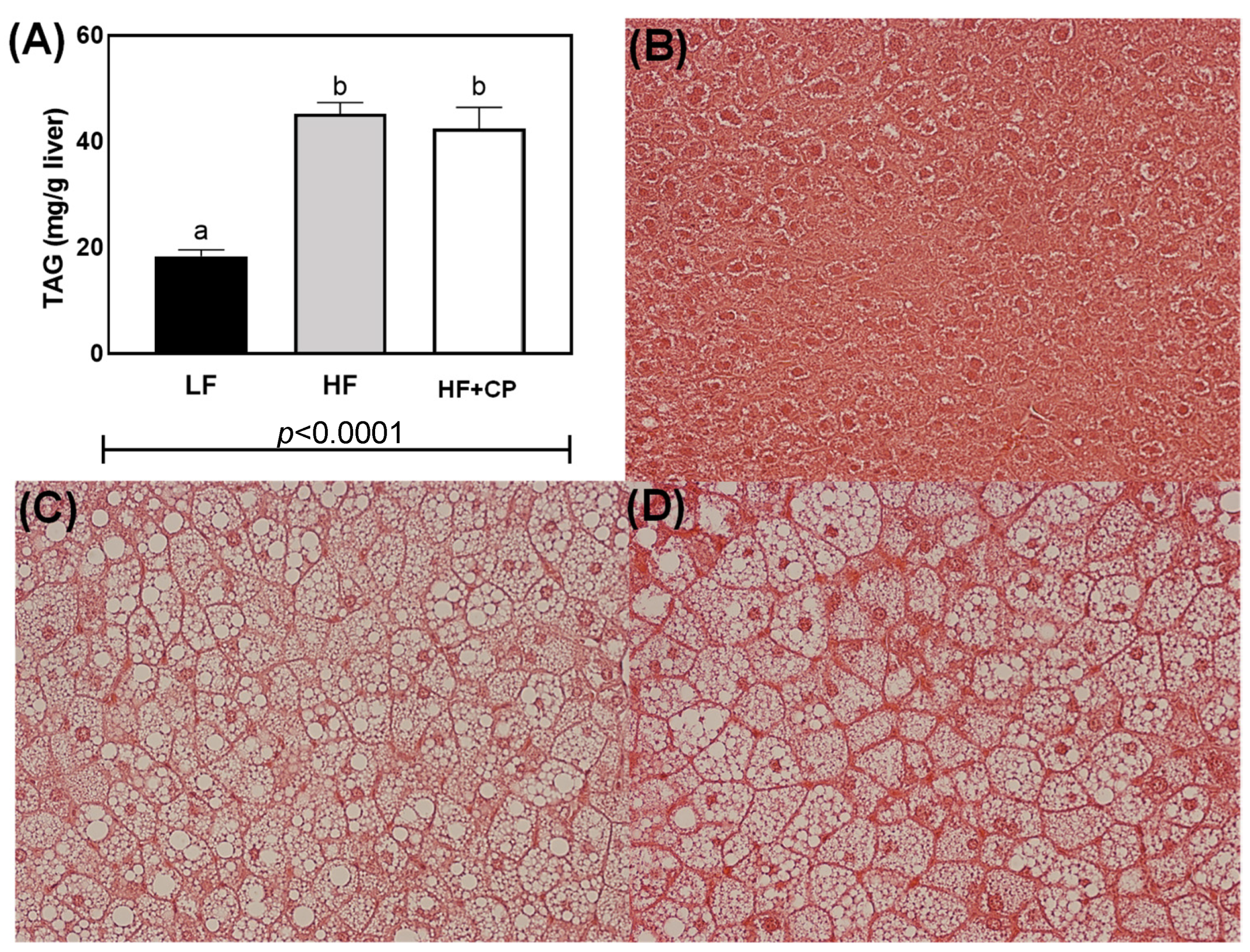
2.4. Hepatic Histological and Total Triglycerides Analysis
2.5. Serum ELISA
2.6. RNA-Sequencing
2.6.1. RNA Extraction, Library Preparation, and Sequencing
2.6.2. Differential Expression Analysis
2.7. Microbiome Profiling
2.8. Statistical Analysis
3. Results
3.1. Proximal Composition of Cashew Pulp (CP)
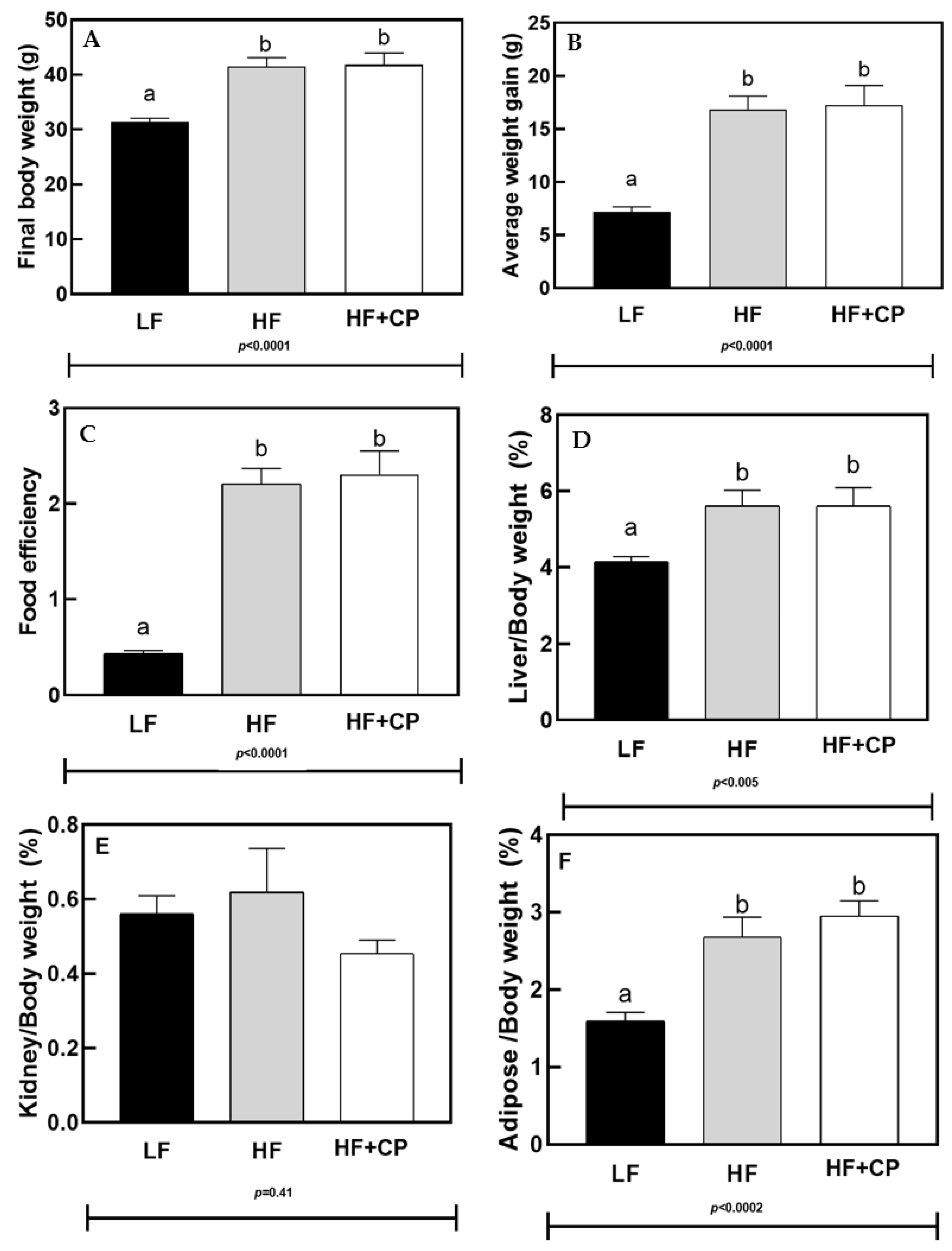
3.2. Energy Intake and Weight Gain
3.3. Biochemical Parameters
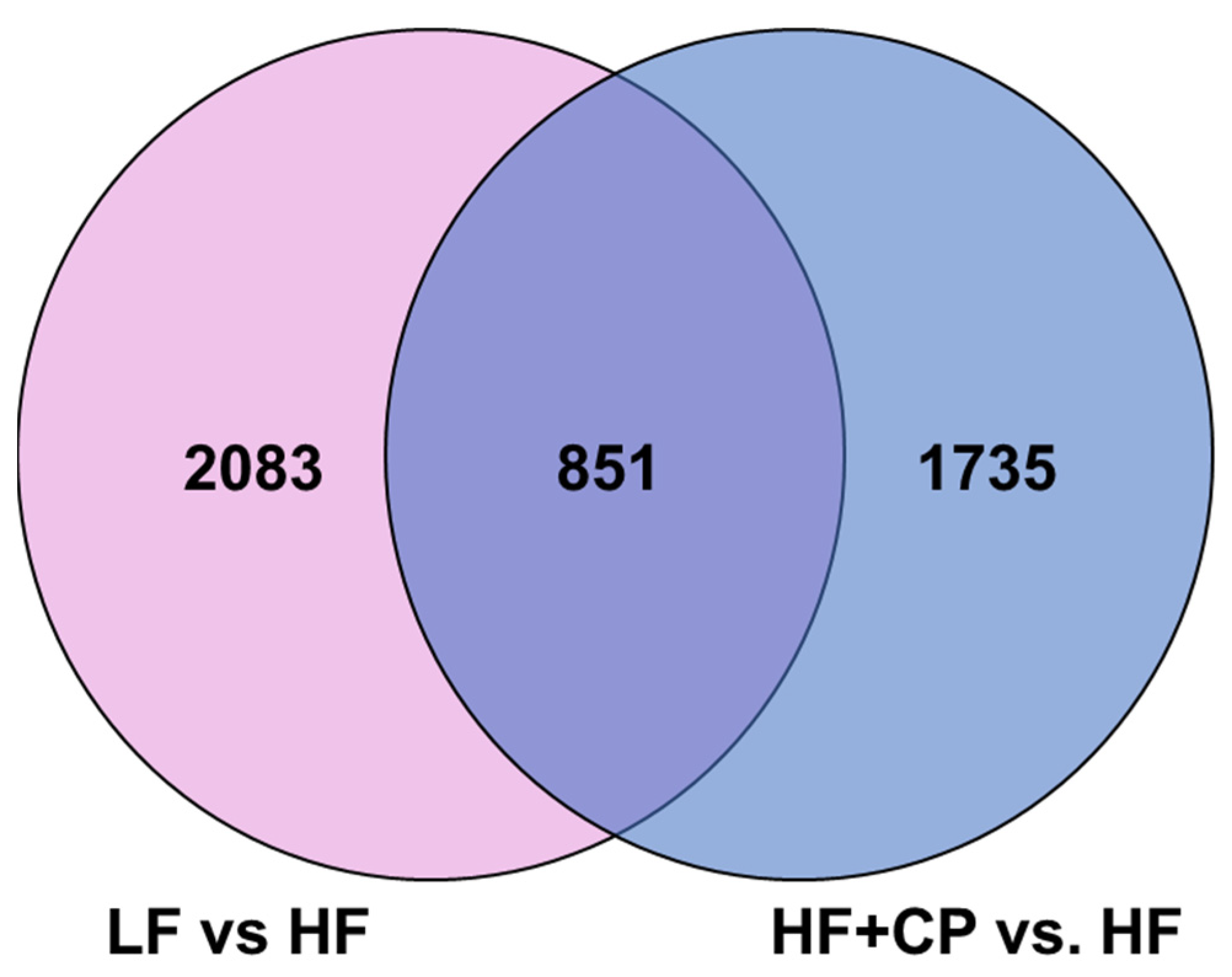
3.4. Gene Expression
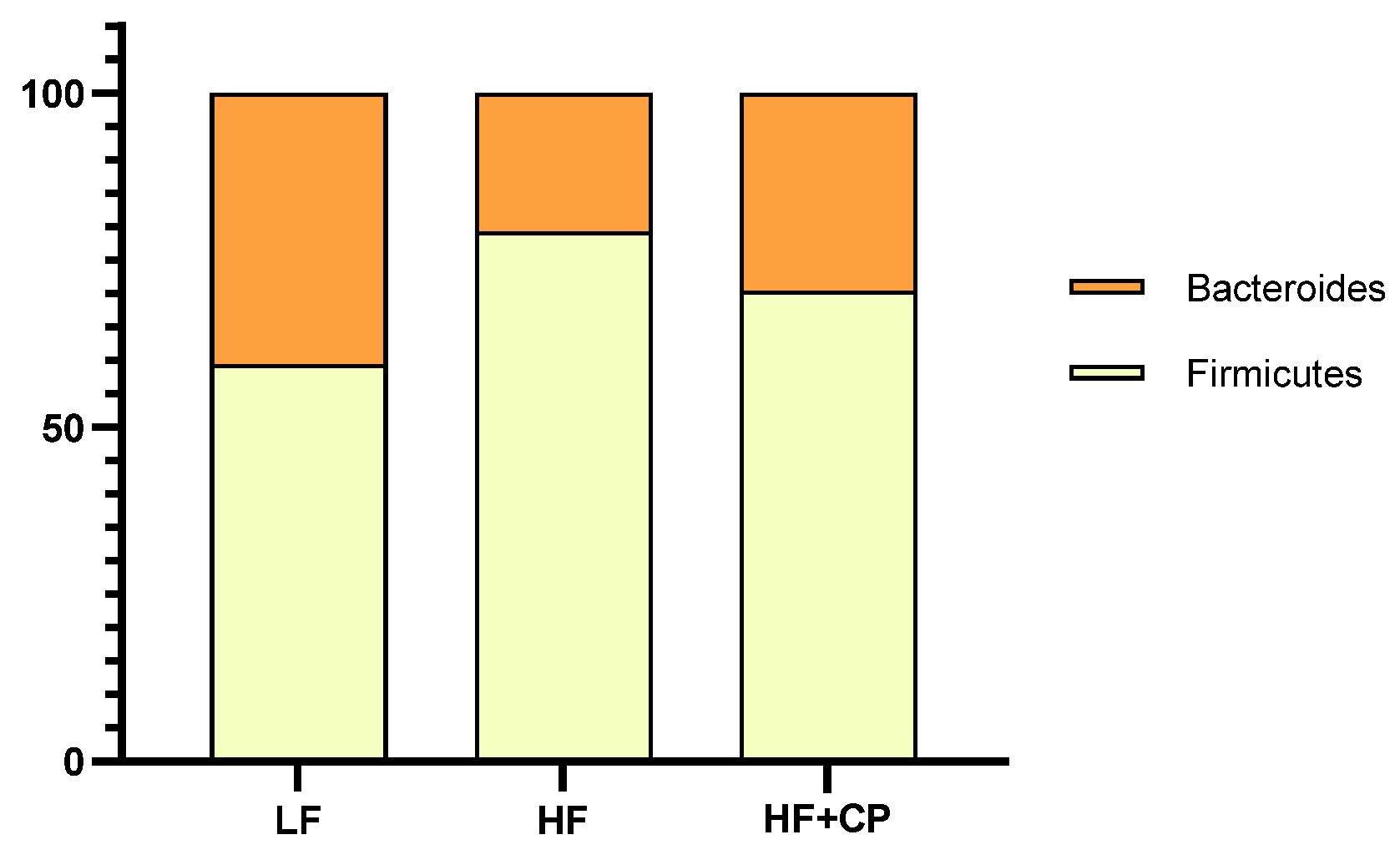
3.5. Microbiome Profile
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Finicelli, M.; Squillaro, T.; Di Cristo, F.; Di Salle, A.; Melone, M.A.B.; Galderisi, U.; Peluso, G. Metabolic syndrome, Mediterranean diet, and polyphenols: Evidence and perspectives. J. Cell. Physiol. 2019, 234, 5807–5826. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, D.M.; Snowden, J.A. Cardiovascular diseases and metabolic syndrome. In The EBMT Handbook; Springer: Cham, Switzerland, 2019; pp. 415–420. [Google Scholar]
- Dabke, K.; Hendrick, G.; Devkota, S. The gut microbiome and metabolic syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- He, Y.; Wu, W.; Wu, S.; Zheng, H.-M.; Li, P.; Sheng, H.-F.; Chen, M.-X.; Chen, Z.-H.; Ji, G.-Y.; Zheng, Z.-D.-X.; et al. Linking gut microbiota, metabolic syndrome and economic status based on a population-level analysis. Microbiome 2018, 6, 172. [Google Scholar] [CrossRef] [PubMed]
- Gildner, T.E. Links between metabolic syndrome and the microbiome. Evol. Med. Public Health 2020, 2020, 45–46. [Google Scholar] [CrossRef] [PubMed]
- Nour, M.; Lutze, S.A.; Grech, A.; Allman-Farinelli, M. The relationship between vegetable intake and weight outcomes: A systematic review of cohort studies. Nutrients 2018, 10, 1626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dos Santos, D.; de Oliveira Filho, J.; de Sousa, T.; Ribeiro, C.; Egea, M. Ameliorating effects of metabolic syndrome with the consumption of rich-bioactive compounds fruits from Brazilian Cerrado: A narrative review. Crit. Rev. Food Sci. Nutr. 2021, 1–18. [Google Scholar] [CrossRef]
- Oliveira, V.F.; Silva, F.G.; Resende, E.C.; Pereira, P.S.; de Le Silva, F.H.; Egea, M.B. Physicochemical characterization of ‘Cerrado’cashew (Anacardium othonianum Rizzini) fruits and pseudofruits. J. Sci. Food Agric. 2019, 99, 6199–6208. [Google Scholar] [CrossRef]
- Alves, A.M.; Dias, T.; Hassimotto, N.M.A.; Naves, M.M.V. Ascorbic acid and phenolic contents, antioxidant capacity and flavonoids composition of Brazilian Savannah native fruits. Food Sci. Technol. 2017, 37, 564–569. [Google Scholar] [CrossRef] [Green Version]
- Santana, R.V.; dos Santos, D.C.; Santana, A.C.A.; de Oliveira Filho, J.G.; de Almeida, A.B.; de Lima, T.M.; Silva, F.G.; Egea, M.B. Quality parameters and sensorial profile of clarified “Cerrado” cashew juice supplemented with Sacharomyces boulardii and different sweeteners. LWT 2020, 128, 109319. [Google Scholar] [CrossRef]
- E Silva, A.L.L.; dos Santos, D.C.; de Sousa, T.L.; Silva, F.G.; Egea, M.B. “Cerrado” cashew (Anacardium othonianum Rizz.) juice improves metabolic parameters in women: A pilot study. J. Funct. Foods 2020, 69, 103950. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis; Association of Official Analytical Chemists: Arlington, VA, USA, 2000. [Google Scholar]
- Egea, M.B.; Pierce, G.; Becraft, A.; Sturm, M.; Yu, W.; Shay, N. Intake of Watermelon and Watermelon Byproducts Male Mice Fed a Western-style Obesigenic Diet Alters Hepatic Gene Expression Patterns as Determined by RNA Sequencing. Curr. Dev. Nutr. 2020, 4, nzaa122. [Google Scholar] [CrossRef] [PubMed]
- Aleksunes, L.M.; Klaassen, C.D. Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab. Dispos. 2012, 40, 1366–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakachi, Y.; Yagi, K.; Nikaido, I.; Bono, H.; Tonouchi, M.; Schönbach, C.; Okazaki, Y. Identification of novel PPARγ target genes by integrated analysis of ChIP-on-chip and microarray expression data during adipocyte differentiation. Biochem. Biophys. Res. Commun. 2008, 372, 362–366. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Gao, J.; Xu, M.; Ren, S.; Stefanovic-Racic, M.; O’Doherty, R.M.; Xie, W. PXR ablation alleviates diet-induced and genetic obesity and insulin resistance in mice. Diabetes 2013, 62, 1876–1887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maxwell, K.N.; Soccio, R.E.; Duncan, E.M.; Sehayek, E.; Breslow, J.L. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 2003, 44, 2109–2119. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Li, L.; Yin, H. Cholesterol homeostasis and liver X receptor (LXR) in atherosclerosis. Cardiovasc. Haematol. Disord.-Drug Targets Former. Curr. Drug Targets-Cardiovasc. Hematol. Disord. 2018, 18, 27–33. [Google Scholar] [CrossRef]
- Lee, J.; Seok, S.; Yu, P.; Kim, K.; Smith, Z.; Rivas-Astroza, M.; Zhong, S.; Kemper, J.K. Genomic analysis of hepatic farnesoid X receptor binding sites reveals altered binding in obesity and direct gene repression by farnesoid X receptor in mice. Hepatology 2012, 56, 108–117. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Lee, S.I.; Rackerby, B.; Moppert, I.; McGorrin, R.; Ha, S.-D.; Park, S.H. Potential Contamination Sources on Fresh Produce Associated with Food Safety. J. Food Hyg. Saf. 2019, 34, 1–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wang, N.; Westerterp, M. ABC transporters, cholesterol efflux, and implications for cardiovascular diseases. In Lipid Transfer in Lipoprotein Metabolism and Cardiovascular Disease; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 67–83. [Google Scholar]
- Wilund, K.R.; Yu, L.; Xu, F.; Hobbs, H.H.; Cohen, J.C. High-level expression of ABCG5 and ABCG8 attenuates diet-induced hypercholesterolemia and atherosclerosis in Ldlr−/− mice. J. Lipid Res. 2004, 45, 1429–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jesch, E.D.; Carr, T.P. Food Ingredients That Inhibit Cholesterol Absorption. Prev. Nutr. Food Sci. 2017, 22, 67–80. [Google Scholar] [CrossRef] [PubMed]
- NIH. National Institute of Health. Dietary Reference Intakes (DRIs): Recommended Dietary Allowances and Adequate Intakes, Total Water and Macronutrients. Available online: https://www.ncbi.nlm.nih.gov/books/NBK545442/table/appJ_tab4/?report=objectonly (accessed on 10 October 2020).
- Xu, P.; Zhai, Y.; Wang, J. The role of PPAR and its cross-talk with CAR and LXR in obesity and atherosclerosis. Int. J. Mol. Sci. 2018, 19, 1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schildberg, F.A.; Sharpe, A.H.; Turley, S.J. Hepatic immune regulation by stromal cells. Curr. Opin. Immunol. 2015, 32, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397. [Google Scholar] [CrossRef] [PubMed]
- Blancas-Velazquez, A.; Mendoza, J.; Garcia, A.N.; la Fleur, S.E. Diet-Induced Obesity and Circadian Disruption of Feeding Behavior. Front. Neurosci. 2017, 11, 23. [Google Scholar] [CrossRef]
- Al-Assal, K.; Martinez, A.C.; Torrinhas, R.S.; Cardinelli, C.; Waitzberg, D. Gut microbiota and obesity. Clin. Nutr. Exp. 2018, 20, 60–64. [Google Scholar] [CrossRef] [Green Version]
- Baothman, O.A.; Zamzami, M.A.; Taher, I.; Abubaker, J.; Abu-Farha, M. The role of gut microbiota in the development of obesity and diabetes. Lipids Health Dis. 2016, 15, 108. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.-H.; Cho, Y.-J.; Park, J.-H. The human symbiont Bacteroides thetaiotaomicron promotes diet-induced obesity by regulating host lipid metabolism. J. Microbiol. 2022, 60, 118–127. [Google Scholar] [CrossRef]
- Becraft, A.R.; Sturm, M.L.; Mendez, R.L.; Park, S.H.; Lee, S.I.; Shay, N.F. Intake of Watermelon or Its Byproducts Alters Glucose Metabolism, the Microbiome, and Hepatic Proinflammatory Metabolites in High-Fat–Fed Male C57BL/6 J Mice. J. Nutr. 2020, 150, 434–442. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, Q.; Ma, W.; Tian, F.; Shen, H.; Zhou, M. A combination of quercetin and resveratrol reduces obesity in high-fat diet-fed rats by modulation of gut microbiota. Food Funct. 2017, 8, 4644–4656. [Google Scholar] [CrossRef] [PubMed]
- Macchione, I.; Lopetuso, L.; Ianiro, G.; Napoli, M.; Gibiino, G.; Rizzatti, G.; Petito, V.; Gasbarrini, A.; Scaldaferri, F. Akkermansia muciniphila: Key player in metabolic and gastrointestinal disorders. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8075–8083. [Google Scholar] [PubMed]
- Jayachandran, M.; Chung, S.S.M.; Xu, B. A critical review of the relationship between dietary components, the gut microbe Akkermansia muciniphila, and human health. Crit. Rev. Food Sci. Nutr. 2020, 60, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira-a candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Zang, Y.; Zhang, L.; Igarashi, K.; Yu, C. The anti-obesity and anti-diabetic effects of kaempferol glycosides from unripe soybean leaves in high-fat-diet mice. Food Funct. 2015, 6, 834–841. [Google Scholar] [CrossRef]
- Remely, M.; Ferk, F.; Sterneder, S.; Setayesh, T.; Roth, S.; Kepcija, T.; Noorizadeh, R.; Rebhan, I.; Greunz, M.; Beckmann, J.; et al. EGCG Prevents High Fat Diet-Induced Changes in Gut Microbiota, Decreases of DNA Strand Breaks, and Changes in Expression and DNA Methylation of Dnmt1 and MLH1 in C57BL/6J Male Mice. Oxid. Med. Cell. Longev. 2017, 2017, 3079148. [Google Scholar] [CrossRef] [Green Version]
- Cremonini, E.; Daveri, E.; Mastaloudis, A.; Oteiza, P.I. (–)-Epicatechin and Anthocyanins Modulate GLP-1 Metabolism: Evidence from C57BL/6J Mice and GLUTag Cells. J. Nutr. 2021, 151, 1497–1506. [Google Scholar] [CrossRef]
- Daveri, E.; Cremonini, E.; Mastaloudis, A.; Hester, S.N.; Wood, S.M.; Waterhouse, A.L.; Anderson, M.; Fraga, C.G.; Oteiza, P.I. Cyanidin and delphinidin modulate inflammation and altered redox signaling improving insulin resistance in high fat-fed mice. Redox Biol. 2018, 18, 16–24. [Google Scholar] [CrossRef]

| Pathway | #Total | #Up | #Down | Significance | p-Value | |
|---|---|---|---|---|---|---|
| 1 | Complement Activation, Classical Pathway (part of Immune system) | 8 | 7 | 1 | 3.46 | 0.000 |
| 2 | Cytoplasmic Ribosomal Proteins | 21 | 4 | 17 | 3.45 | 0.000 |
| 3 | G Protein-coupled Receptor (GPCR) | 11 | 3 | 8 | 2.77 | 0.002 |
| 4 | Complement and Coagulation Cascades | 16 | 14 | 2 | 2.64 | 0.002 |
| 5 | MAPK Cascade | 9 | 5 | 4 | 2.3 | 0.005 |
| 6 | Odorant GPCR | 14 | 8 | 6 | 2.17 | 0.007 |
| 7 | Non-odorant GPCR | 18 | 8 | 10 | 2.13 | 0.007 |
| 8 | MAPK signaling pathway | 29 | 18 | 11 | 1.74 | 0.018 |
| 9 | Metapathway biotransformation | 8 | 6 | 2 | 1.72 | 0.019 |
| 10 | Nuclear receptors in lipid metabolism and toxicity | 8 | 7 | 1 | 1.69 | 0.021 |
| 11 | TYROBP Causal Network | 13 | 12 | 1 | 1.65 | 0.022 |
| 12 | MAPK signaling pathway | 30 | 21 | 9 | 1.64 | 0.023 |
| 13 | Microglia Pathogen Phagocytosis Pathway | 10 | 10 | 0 | 1.61 | 0.025 |
| 14 | Robo4 and VEGF Signaling Pathways Crosstalk | 3 | 3 | 0 | 1.6 | 0.025 |
| 15 | miRNAs involved in DNA damage response | 1 | 1 | 0 | 1.59 | 0.026 |
| 16 | Glycolysis and Gluconeogenesis | 1 | 1 | 0 | 1.58 | 0.026 |
| 17 | Novel Jun-Dmp1 Pathway | 7 | 5 | 2 | 1.55 | 0.028 |
| 18 | Mitochondrial LC-Fatty Acid Beta-Oxidation | 5 | 3 | 2 | 1.48 | 0.033 |
| 19 | PPAR signaling pathway | 16 | 11 | 5 | 1.41 | 0.039 |
| 20 | Fatty Acid Beta Oxidation (streamlined) | 8 | 5 | 3 | 1.32 | 0.047 |
| 21 | Toll-like receptor signaling pathway | 18 | 13 | 5 | 1.23 | 0.058 |
| 22 | Steroid Biosynthesis | 4 | 4 | 0 | 1.23 | 0.059 |
| 23 | Focal Adhesion | 32 | 22 | 10 | 1.19 | 0.064 |
| 24 | GPCRs, Class A Rhodopsin-like | 18 | 9 | 9 | 1.19 | 0.065 |
| 25 | Eicosanoid Synthesis | 5 | 4 | 1 | 1.18 | 0.066 |
| No | Upregulated Genes | Downregulated Genes | ||||||
|---|---|---|---|---|---|---|---|---|
| FC | p-Value | Gene | Function | FC | p-Value | Gene | Function | |
| 1 | 12.00 | 4.82 × 10−2 | Myc | cell cycle progression, apoptosis, and cellular transformation | −12.98 | 1.50 × 10−3 | Cyp2b9 | synthesis of cholesterol, steroids and other lipids |
| 2 | 7.87 | 3.91 × 10−2 | Foxq1 | cell cycle regulation | −4.46 | 4.15 × 10−2 | Gdf15 | TGF-beta ligant |
| 3 | 5.79 | 4.16 × 10−2 | Usp2 | TNF signaling (REACTOME) and Regulation of TP53 Activity | −3.58 | 2.80 × 10−3 | Spp1 | the protein encoded by this gene is involved in the attachment of osteoclasts to the mineralized bone matrix |
| 4 | 5.66 | 3.79 × 10−2 | Phlda1 | anti-apoptotic effects of insulin-like growth factor-1 | −3.19 | 3.00 × 10−4 | G0s2 | regulation of lipid metabolism |
| 5 | 5.29 | 6.50 × 10−3 | Wee1 | involved in DNA replication | −2.72 | 6.80 × 10−3 | Cyp2b10 | steroid, fatty acid, and xenobiotic compounds oxidation |
| 6 | 4.24 | 3.51 × 10−7 | Ehd3 | megakaryocyte development and platelet production and response to elevated platelet cytosolic Ca2+ | −2.59 | 1.20 × 10−3 | Leap2 | xenobiotic metabolism |
| 7 | 4.12 | 5.54 × 10−5 | Dnase1l3 | breakdown of DNA during apoptosis | −2.51 | 3.90 × 10−3 | Psen2 | intracellular signaling |
| 8 | 4.01 | 6.13 × 10−6 | Clec4g | receptor with an affinity for mannose and fucose | −2.51 | 1.01 × 10−7 | Ucn | activation of cAMP-Dependent PKA (glucose homeostasis) pathways |
| 9 | 3.95 | 2.56 × 10−5 | Kdr | vascular endothelial growth factor (VEGF) | −2.49 | 1.59 × 10−2 | Cyr61 | cell grown and differentiation, and extracellular matrix formation. |
| 10 | 3.57 | 4.00 × 10−4 | Fcgr2b | involved in the phagocytosis of immune complexes and in the regulation of antibody production by B-cells | −2.47 | 9.30 × 10−3 | Cyp2b13 | arachidonic acid epoxygenase activity |
| 11 | 3.49 | 2.00 × 10−4 | Oit3 | downregulation in hepatocarcinom | −2.3 | 2.66 × 10−2 | Pde9a | cAMP and cGMP regulation |
| 12 | 3.34 | 2.80 × 10−3 | Por | synthesis of bile acids and bile salts and cytochrome P450 | −2.29 | 4.72 × 10−2 | Col3a1 | extracellular matrix |
| 13 | 3.34 | 2.06 × 10−2 | Mt2 | steroid biosynthetic process | −2.21 | 1.48 × 10−2 | Sco2 | cell growth |
| 14 | 3.29 | 2.29 × 10−2 | Ciart | circadian rhythm | −2.18 | 1.71 × 10−2 | Golm1 | involved with liver disease and autoimmune hepatitis |
| 15 | 3.14 | 3.04 × 10−2 | Tsku | among its related pathways are ectoderm differentiation | −2.18 | 3.00 × 10−4 | Rab1b | cell transportation |
| 16 | 3.05 | 6.80 × 10−3 | H2-Ab1 | adaptive immunity | −2.09 | 2.10 × 10−3 | Tbca | hormone regulation |
| 17 | 3.04 | 1.90 × 10−3 | Slc6a9 | transportation of molecules and NRF2 pathway | −2.02 | 2.30 × 10−3 | Fabp2 | lipid homeostasis |
| 18 | 3 | 1.00 × 10−4 | Fcgr3 | immune response | −2.01 | 2.49 × 10−2 | Acly | acetyl-CoA synthesis |
| 19 | 2.96 | 2.00 × 10−3 | Eng | TGF-beta component | −1.99 | 2.57 × 10−2 | Gldc | glucose homeostasis |
| 20 | 2.96 | 1.00 × 10−3 | Sdc3 | organization and signaling (sugar-dependent mechanism) cellular | −1.98 | 1.10 × 10−3 | Atf5 | IL-2 Pathway/Imune sistem |
| 21 | 2.86 | 1.40 × 10−3 | Clec4f | receptor with an affinity for galactose and fucose | −1.96 | 2.00 × 10−4 | Prelid2 | phosphatidic acid transfer activity |
| 22 | 2.82 | 3.85 × 10−5 | Wdr46 | galactose and carbohydrate binding | −1.95 | 6.70 × 10−3 | Kdm4a | nuclear protein |
| 23 | 2.74 | 2.00 × 10−3 | Rasgrp3 | activates the oncogenes HRAS and RAP1A. | −1.93 | 2.27 × 10−2 | Rnd1 | cell regulation |
| 24 | 2.73 | 3.00 × 10−3 | Ica1 | autoantigen in T2DM (or IDDM) | −1.92 | 3.40 × 10−3 | Faap20 | fatty acid synthesis |
| 25 | 2.7 | 9.00 × 10−4 | Wfdc17 | activated macrophage/microglia WAP domain protein | −1.92 | 4.31 × 10−2 | Fgfr4 | lipid homeostasis |
| Phyla | LF | HF | HF+CP | p-Value |
|---|---|---|---|---|
| p__Actinobacteria | 6.45 ± 0.64 b | 16.59 ± 0.43 a | 7.02 ± 0.60 b | 0.001 |
| p__Bacteroidetes | 29.18 ± 0.28 a | 13.76 ± 0.66 c | 19.41 ± 0.47 b | 0.0001 |
| p__Firmicutes | 44.26 ± 4.19 | 52.75 ± 0.66 | 46.13 ± 0.43 | 0.46 |
| p__Proteobacteria | 1.77 ± 0.38 | 1.68 ± 0.03 | 2.20 ± 0.03 | 0.73 |
| p__Verrucomicrobia | 18.34 ± 2.99 | 15.22 ± 0.57 | 25.23 ± 0.33 | 0.25 |
| Genera | ||||
| g__Akkermansia | 18.31 ± 2.97 | 15.22 ± 0.57 | 25.23 ± 0.49 | 0.25 |
| o__Clostridiales | 16.90 ± 6.04 | 11.98 ± 2.17 | 20.53 ± 0.27 | 0.15 |
| g__Bacteroides | 9.66 ± 0.49 | 8.22 ± 0.45 | 11.99 ± 1.06 | 0.11 |
| o__Bacteroidales;S24-7 | 19.48 ± 0.24 a | 5.54 ± 0.25 b | 7.42 ± 0.35 b | <0.0001 |
| f__Coriobacteriaceae | 6.38 ± 0.64 b | 16.45 ± 0.43 a | 6.86 ± 0.87 b | 0.001 |
| g__Oscillospira | 4.08 ± 0.26 b | 4.63 ± 0.24 b | 7.18 ± 0.61 a | 0.025 |
| g__Lactococcus | 4.01 ± 0.41 b | 6.84 ± 0.27 a | 2.54 ± 0.08 b | 0.008 |
| f__Ruminococcaceae | 1.91 ± 0.14 | 3.26 ± 0.28 | 1.93 ± 0.36 | 0.10 |
| g__Sutterella | 1.77 ± 0.38 | 1.68 ± 0.03 | 2.20 ± 0.04 | 0.73 |
| Others | 17.51 ± 4.08 | 26.18 ± 0.74 | 14.11 ± 0.16 | 0.28 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Egea, M.B.; Pierce, G.; Park, S.-H.; Lee, S.-I.; Heger, F.; Shay, N. Consumption of Antioxidant-Rich “Cerrado” Cashew Pseudofruit Affects Hepatic Gene Expression in Obese C57BL/6J High Fat-Fed Mice. Foods 2022, 11, 2543. https://doi.org/10.3390/foods11172543
Egea MB, Pierce G, Park S-H, Lee S-I, Heger F, Shay N. Consumption of Antioxidant-Rich “Cerrado” Cashew Pseudofruit Affects Hepatic Gene Expression in Obese C57BL/6J High Fat-Fed Mice. Foods. 2022; 11(17):2543. https://doi.org/10.3390/foods11172543
Chicago/Turabian StyleEgea, Mariana Buranelo, Gavin Pierce, Si-Hong Park, Sang-In Lee, Fabienne Heger, and Neil Shay. 2022. "Consumption of Antioxidant-Rich “Cerrado” Cashew Pseudofruit Affects Hepatic Gene Expression in Obese C57BL/6J High Fat-Fed Mice" Foods 11, no. 17: 2543. https://doi.org/10.3390/foods11172543
APA StyleEgea, M. B., Pierce, G., Park, S.-H., Lee, S.-I., Heger, F., & Shay, N. (2022). Consumption of Antioxidant-Rich “Cerrado” Cashew Pseudofruit Affects Hepatic Gene Expression in Obese C57BL/6J High Fat-Fed Mice. Foods, 11(17), 2543. https://doi.org/10.3390/foods11172543







