Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Vectors, and Reagents
2.2. Plasmid Construction
2.3. Expression of KsLX3-SIase in Bacillus subtilis
2.4. Fed-Batch Fermentation
2.5. Purification of KsLX3-SIase
2.6. Characterization of KsLX3-SIase
2.6.1. Assay of the SIase Enzyme Activity
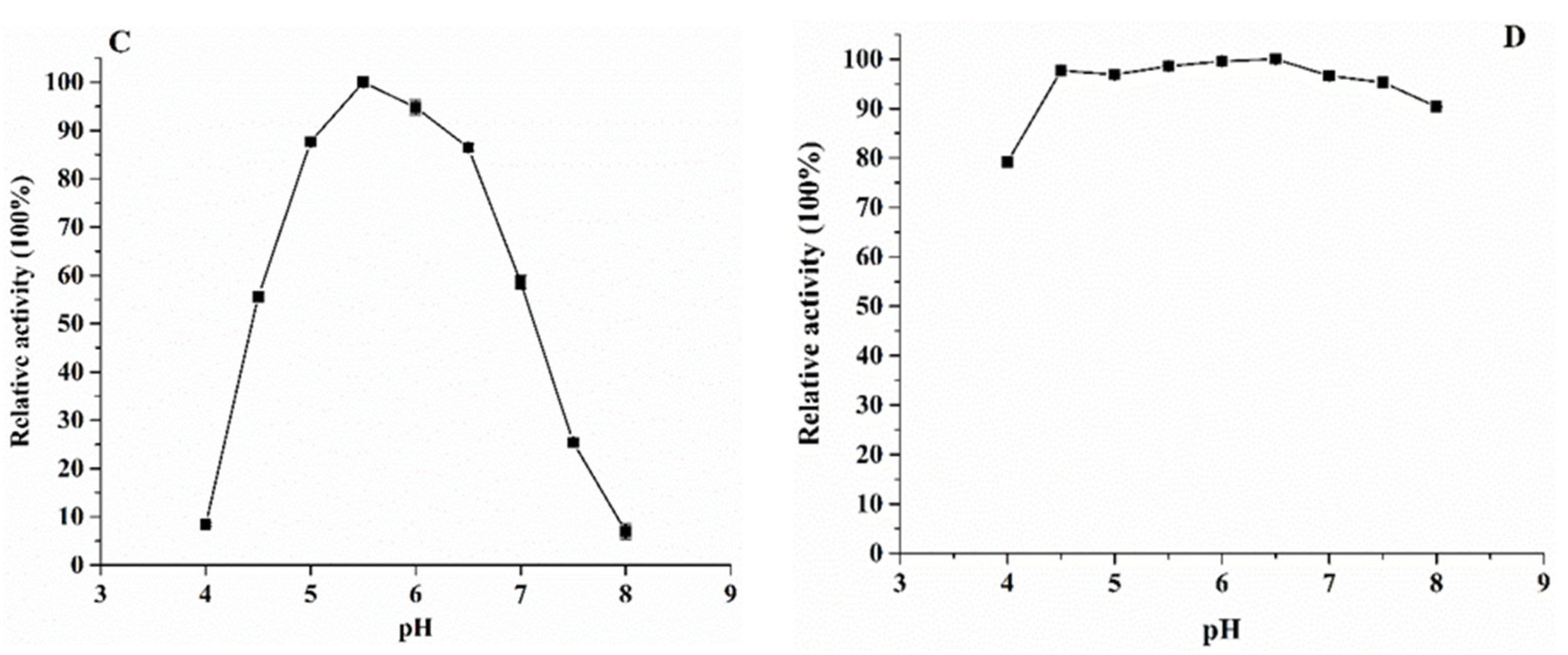
2.6.2. Effects of Temperature and pH on SIase Activity
2.6.3. Determination of Kinetic Parameters
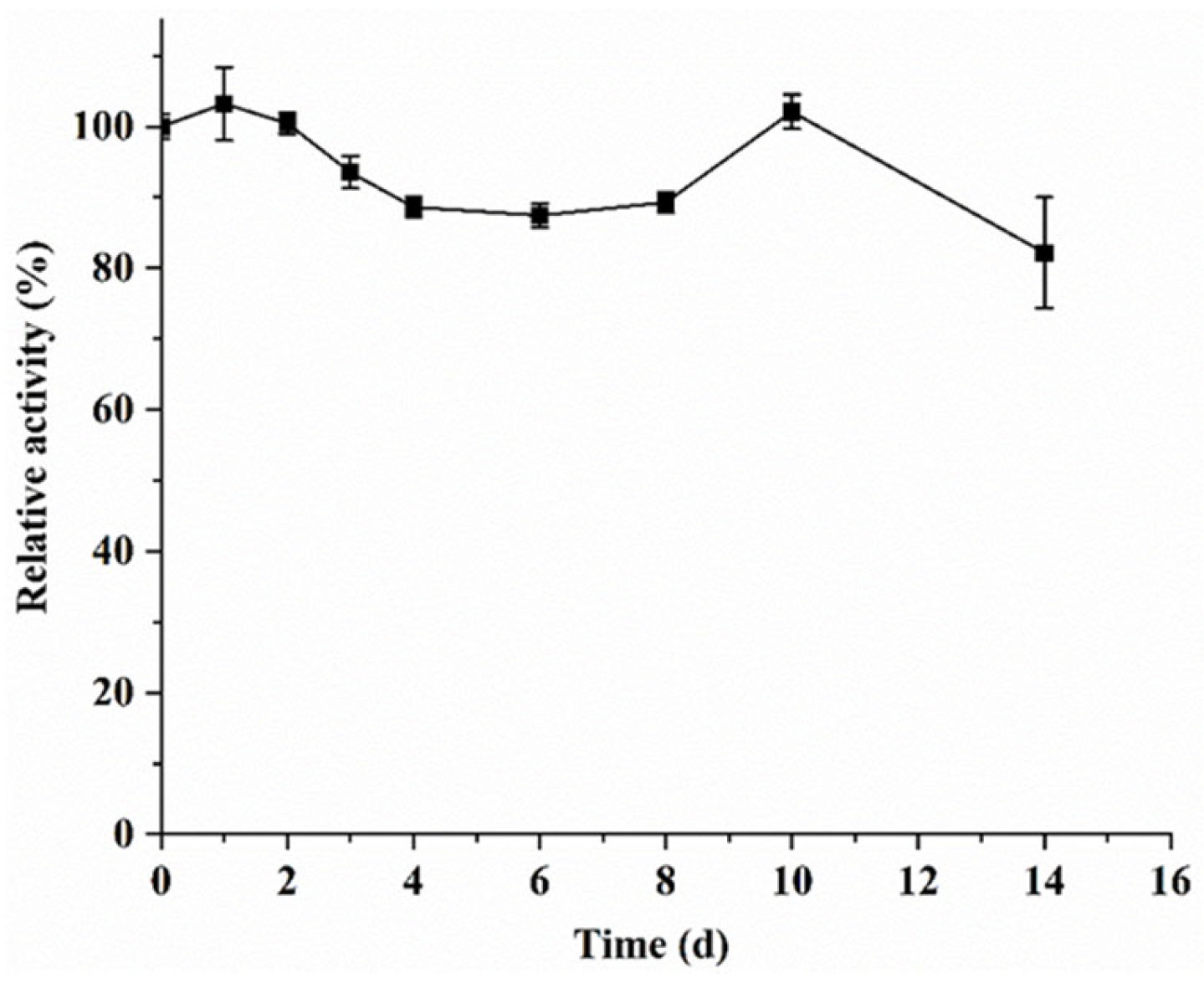
2.6.4. Storage Stability of the SIase
2.6.5. Time Course of Production of Isomaltulose
3. Results and Discussion
3.1. Optimization of Signal Peptides for SIase Expression
3.2. Production of SIase Using a Fed-Batch Strategy
3.3. Enzymatic Properties of the Purified KsLX3-SIase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sawale, P.D.; Shendurse, A.M.; Mohan, M.S.; Patil, G.R. Isomaltulose (Palatinose)-an emerging carbohydrate. Food Biosci. 2017, 18, 46–52. [Google Scholar] [CrossRef]
- Tian, Y.; Deng, Y.; Zhang, W.; Mu, W. Sucrose isomers as alternative sweeteners: Properties, production, and applications. Appl. Microbiol. Biotechnol. 2019, 103, 8677–8687. [Google Scholar] [CrossRef] [PubMed]
- Rhimi, M.; Haser, R.; Aghajari, N. Bacterial sucrose isomerases: Properties and structural studies. Biologia 2008, 63, 1020–1027. [Google Scholar] [CrossRef]
- Sokolowska, E.; Sadowska, A.; Sawicka, D.; Kotulska-Bablinska, I.; Car, H. A head-to-head comparison review of biological and toxicological studies of isomaltulose, d-tagatose, and trehalose on glycemic control. Crit. Rev. Food Sci. Nutr. 2021, 62, 5679–5704. [Google Scholar] [CrossRef]
- Liu, L.; Bilal, M.; Luo, H.; Zhao, Y.; Duan, X. Studies on biological production of isomaltulose using sucrose isomerase: Current status and future perspectives. Catal. Lett. 2021, 151, 1868–1881. [Google Scholar] [CrossRef]
- Pilak, P.; Schiefner, A.; Seiboth, J.; Oehrlein, J.; Skerra, A. Engineering a highly active sucrose isomerase for enhanced product specificity by using a “battleship” strategy. Chembiochem 2020, 21, 2161–2169. [Google Scholar] [CrossRef]
- Lee, H.C.; Kim, J.H.; Kim, S.Y.; Lee, J.K. Isomaltose Production by Modification of the Fructose-Binding Site on the Basis of the Predicted Structure of Sucrose Isomerase from “Protaminobacter rubrum”. Appl. Environ. Microb. 2008, 74, 5183–5194. [Google Scholar] [CrossRef]
- Wu, L.; Birch, R.G. Characterization of the highly efficient sucrose isomerase from Pantoea dispersa UQ68J and cloning of the sucrose isomerase gene. Appl. Environ. Microbiol. 2005, 71, 1581–1590. [Google Scholar] [CrossRef]
- Zhang, D.; Li, X.; Zhang, L.H. Isomaltulose synthase from Klebsiella sp. Strain LX3: Gene cloning and characterization and engineering of thermostability. Appl. Environ. Microbiol. 2002, 68, 2676–2682. [Google Scholar] [CrossRef]
- Wang, C.; Li, S.; Xu, H.; Wei, Y.; Cai, H. Optimization of fermentation medium of sucrose isomerase by recombinant Escherichia coli through response surface method. Chin. Biotechnol. 2011, 31, 92–97. [Google Scholar]
- Zhang, F.; Cheng, F.; Jia, D.X.; Gu, Y.H.; Liu, Z.Q.; Zheng, Y.G. Characterization of a recombinant sucrose isomerase and its application to enzymatic production of isomaltulose. Biotechnol. Lett. 2021, 43, 261–269. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Z.; Liu, S.; Wang, Y.; Zhang, Z.; Liu, X.; Du, Y.; Yuan, X. Overexpression of secreted sucrose isomerase in Yarrowia lipolytica and its application in isomaltulose production after immobilization. Int. J. Biol. Macromol. 2019, 121, 97–103. [Google Scholar] [CrossRef]
- Park, J.; Jung, J.; Seo, D.; Ha, S.; Yoon, J.; Kim, Y.; Shim, J.; Park, C. Microbial production of palatinose through extracellular expression of a sucrose isomerase from Enterobacter sp. FMB-1 in Lactococcus lactis MG1363. Bioresour. Technol. 2010, 101, 8828–8833. [Google Scholar] [CrossRef]
- Wu, L.; Wu, S.; Qiu, J.; Xu, C.; Li, S.; Xu, H. Green synthesis of isomaltulose from cane molasses by Bacillus subtilis WB800-pHA01-palI in a biologic membrane reactor. Food Chem. 2017, 229, 761–768. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Z.P.; Sheng, J.; Zheng, Y.; Ji, X.F.; Zhou, H.X.; Liu, X.Y.; Chi, Z.M. High and efficient isomaltulose production using an engineered Yarrowia lipolytica strain. Bioresour. Technol. 2018, 265, 577–580. [Google Scholar] [CrossRef]
- Yang, H.; Qu, J.; Zou, W.; Shen, W.; Chen, X. An overview and future prospects of recombinant protein production in Bacillus subtilis. Appl. Microbiol. Biot. 2021, 105, 6607–6626. [Google Scholar] [CrossRef] [PubMed]
- Zocca, V.; Correa, G.G.; Lins, M.; de Jesus, V.N.; Tavares, L.F.; Amorim, L.; Kundlatsch, G.E.; Pedrolli, D.B. The CRISPR toolbox for the gram-positive model bacterium Bacillus subtilis. Crit. Rev. Biotechnol. 2022, 42, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gong, J.; Qin, J.; Li, H.; Lu, Z.; Shi, J.; Xu, Z. Improving the intensity of integrated expression for microbial production. ACS Synth. Biol. 2021, 10, 2796–2807. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Dai, Y.; Zhang, T. Characterization of Feruloyl Esterase from Bacillus pumilus SK52.001 and its Application in Ferulic Acid Production from De-Starched Wheat Bran. Foods 2021, 10, 1229. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.; Liu, Q.; Sun, J.; Secundo, F.; Mao, X. Construction of a Super-Folder fluorescent Protein-Guided secretory expression system for the production of phospholipase d in Bacillus subtilis. J. Agric. Food Chem. 2021, 69, 6842–6849. [Google Scholar] [CrossRef]
- Zhang, K.; Su, L.; Duan, X.; Liu, L.; Wu, J. High-level extracellular protein production in Bacillus subtilis using an optimized dual-promoter expression system. Microb. Cell Fact. 2017, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, F.; Lin, L.; He, D.; Chen, J.; Wei, W.; Wei, D. Biochemical characterization of a novel thermostable type I pullulanase produced recombinantly in Bacillus subtilis. Starch-Stärke 2018, 70, 1700179. [Google Scholar] [CrossRef]
- Zhang, K.; Su, L.; Wu, J. Recent Advances in Recombinant Protein Production by Bacillus subtilis. Annu. Rev. Food Sci. Technol. 2020, 11, 295–318. [Google Scholar] [CrossRef]
- Zhang, J.; Kang, Z.; Ling, Z.; Cao, W.; Liu, L.; Wang, M.; Du, G.; Chen, J. High-level extracellular production of alkaline polygalacturonate lyase in Bacillus subtilis with optimized regulatory elements. Bioresour. Technol. 2013, 146, 543–548. [Google Scholar] [CrossRef]
- Khadye, V.S.; Sawant, S.; Shaikh, K.; Srivastava, R.; Chandrayan, S.; Odaneth, A.A. Optimal secretion of thermostable Beta-glucosidase in Bacillus subtilis by signal peptide optimization. Protein Expres. Purif. 2021, 182, 105843. [Google Scholar] [CrossRef]
- Russell, D.W.; Sambrook, J. Molecular Cloning. A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001. [Google Scholar]
- Xue, G.; Johnson, J.S.; Dalrymple, B.P. High osmolarity improves the electro-transformation efficiency of the gram-positive bacteria Bacillus subtilis and Bacillus licheniformis. J. Microbiol. Meth. 1999, 34, 183–191. [Google Scholar] [CrossRef]
- Tjalsma, H.; Bolhuis, A.; Jongbloed, J.D.; Bron, S.; van Dijl, J.M. Signal peptide-dependent protein transport in Bacillus subtilis: A genome-based survey of the secretome. Microbiol. Mol. Biol. Rev. 2000, 64, 515–547. [Google Scholar] [CrossRef]
- Zanen, G.; Houben, E.N.; Meima, R.; Tjalsma, H.; Jongbloed, J.D.; Westers, H.; Oudega, B.; Luirink, J.; van Dijl, J.M.; Quax, W.J. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J. 2005, 272, 4617–4630. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.T.; Tran, L.T.; Schumann, W.; Nguyen, H.D. Development of Pgrac100-based expression vectors allowing high protein production levels in Bacillus subtilis and relatively low basal expression in Escherichia coli. Microb. Cell Fact. 2015, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cai, H.; Qing, Y.; Ren, B.; Xu, H.; Zhu, H.; Yao, J. Cloning and characterization of a sucrose isomerase from Erwinia rhapontici NX-5 for isomaltulose hyperproduction. Appl. Biochem. Biotechnol. 2011, 163, 52–63. [Google Scholar] [CrossRef]
- Aroonnual, A.; Nihira, T.; Seki, T.; Panbangred, W. Role of several key residues in the catalytic activity of sucrose isomerase from Klebsiella pneumoniae NK33-98-8. Enzyme Microb. Technol. 2007, 40, 1221–1227. [Google Scholar] [CrossRef]
- Zhang, F.; Cheng, F.; Jia, D.; Liu, Q.; Liu, Z.; Zheng, Y. Tuning the catalytic performances of a sucrose isomerase for production of isomaltulose with high concentration. Appl. Microbiol. Biot. 2022, 106, 2493–2501. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Jung, J.; Seo, D.; Hansin, J.; Ha, S.; Cha, J.; Kim, Y.; Park, C. Isomaltulose production via yeast surface display of sucrose isomerase from Enterobacter sp. FMB-1 on Saccharomyces cerevisiae. Bioresour. Technol. 2011, 102, 9179–9184. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhu, P.; Liang, J.; Xu, Z.; Feng, X.; Liu, Y.; Xu, H.; Li, S. Economical production of isomaltulose from agricultural residues in a system with sucrose isomerase displayed on Bacillus subtilis spores. Bioproc. Biosyst. Eng. 2020, 43, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, H.; Cheng, H.; Deng, Z. Isomaltulose production by yeast surface display of sucrose isomerase from Pantoea dispersa on Yarrowia lipolytica. J. Funct. Foods 2017, 32, 208–217. [Google Scholar] [CrossRef]
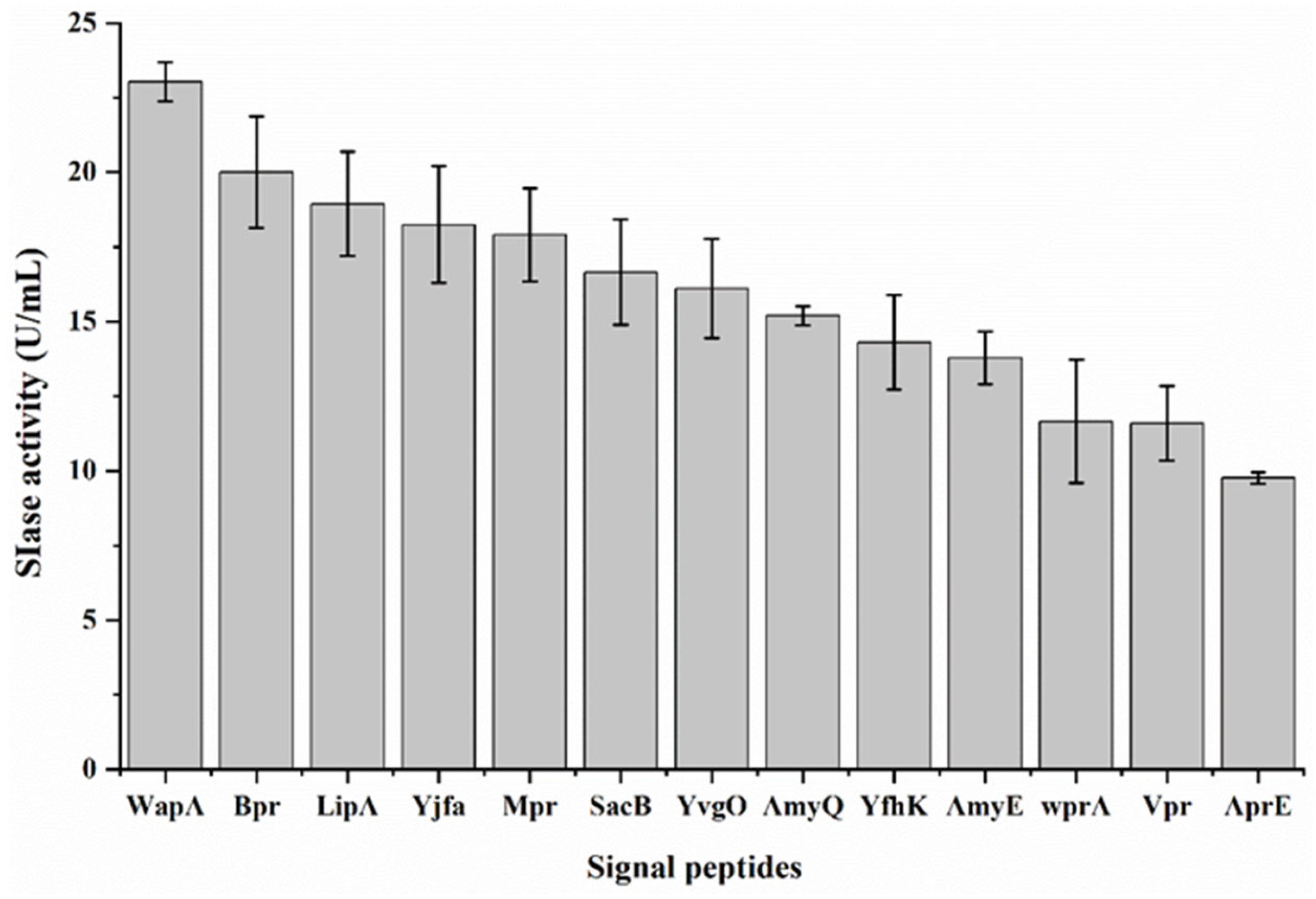
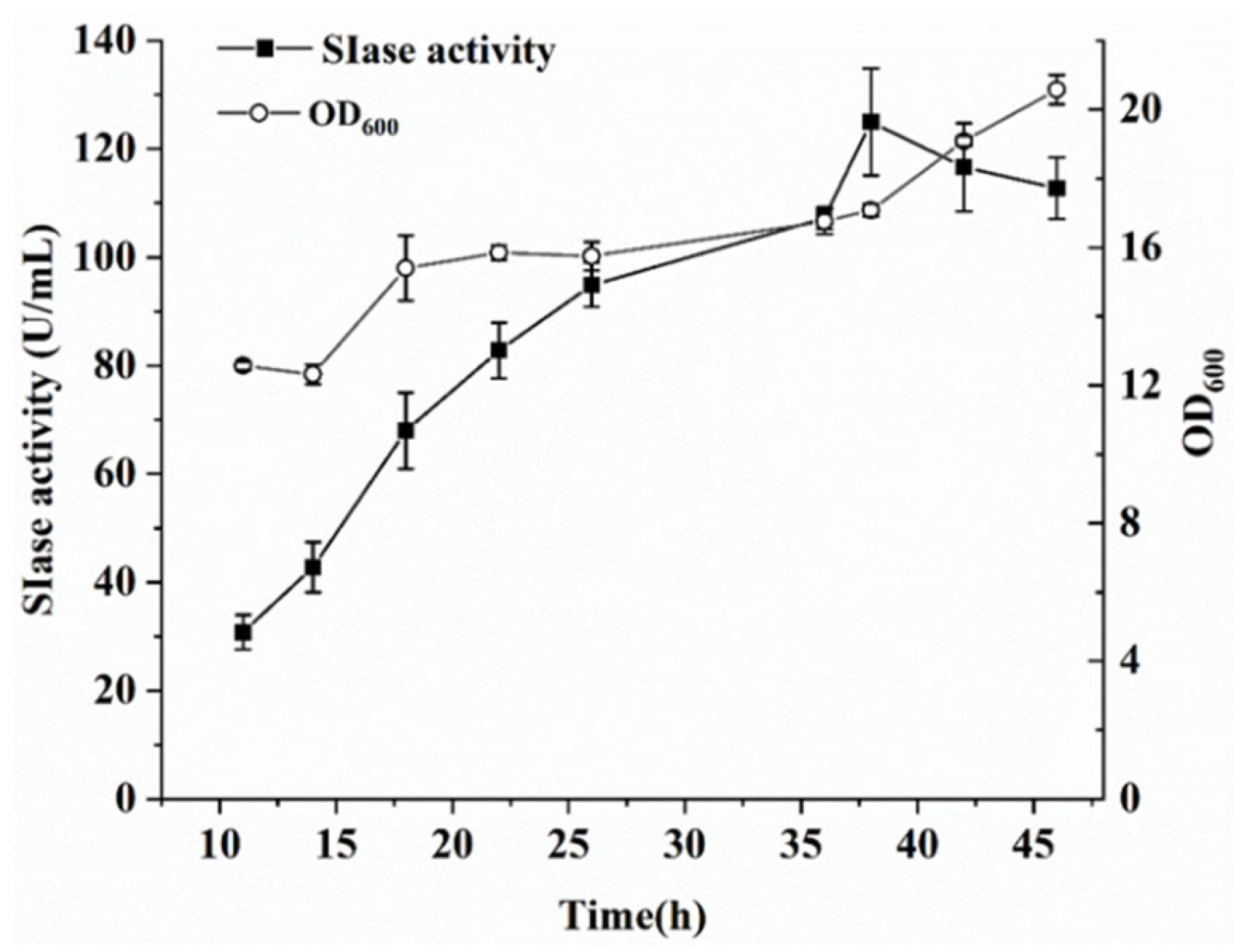

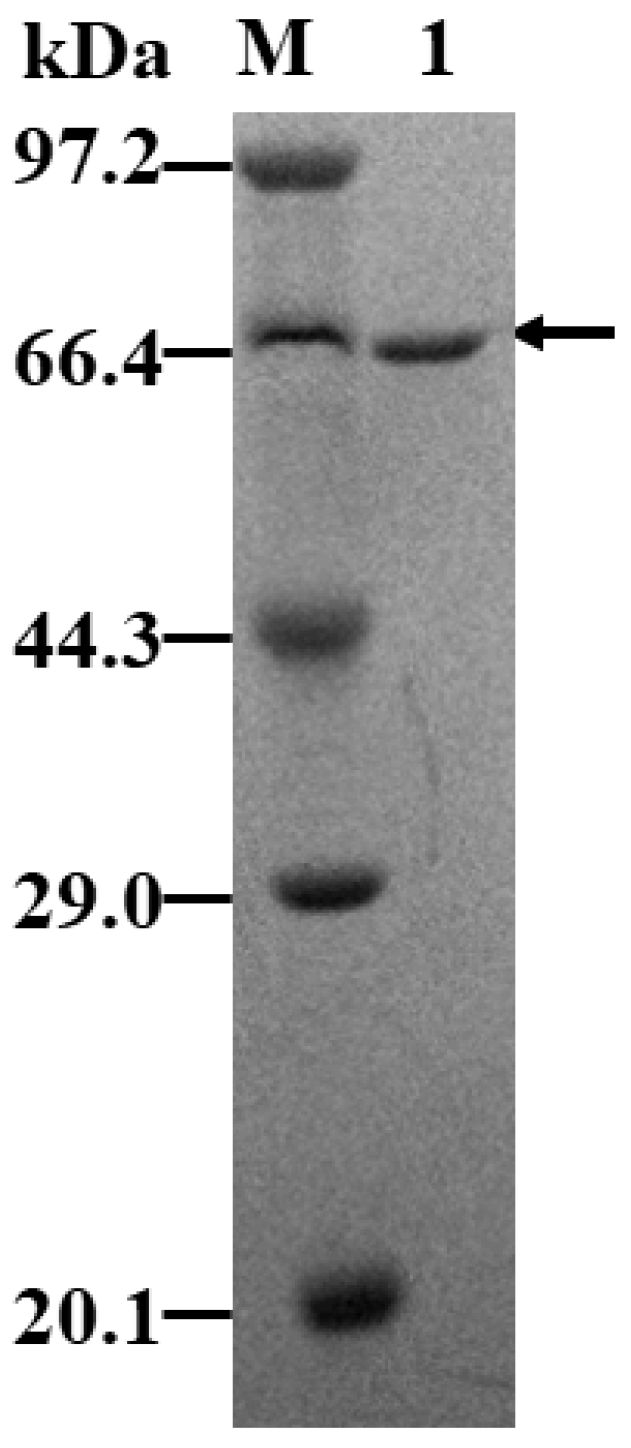
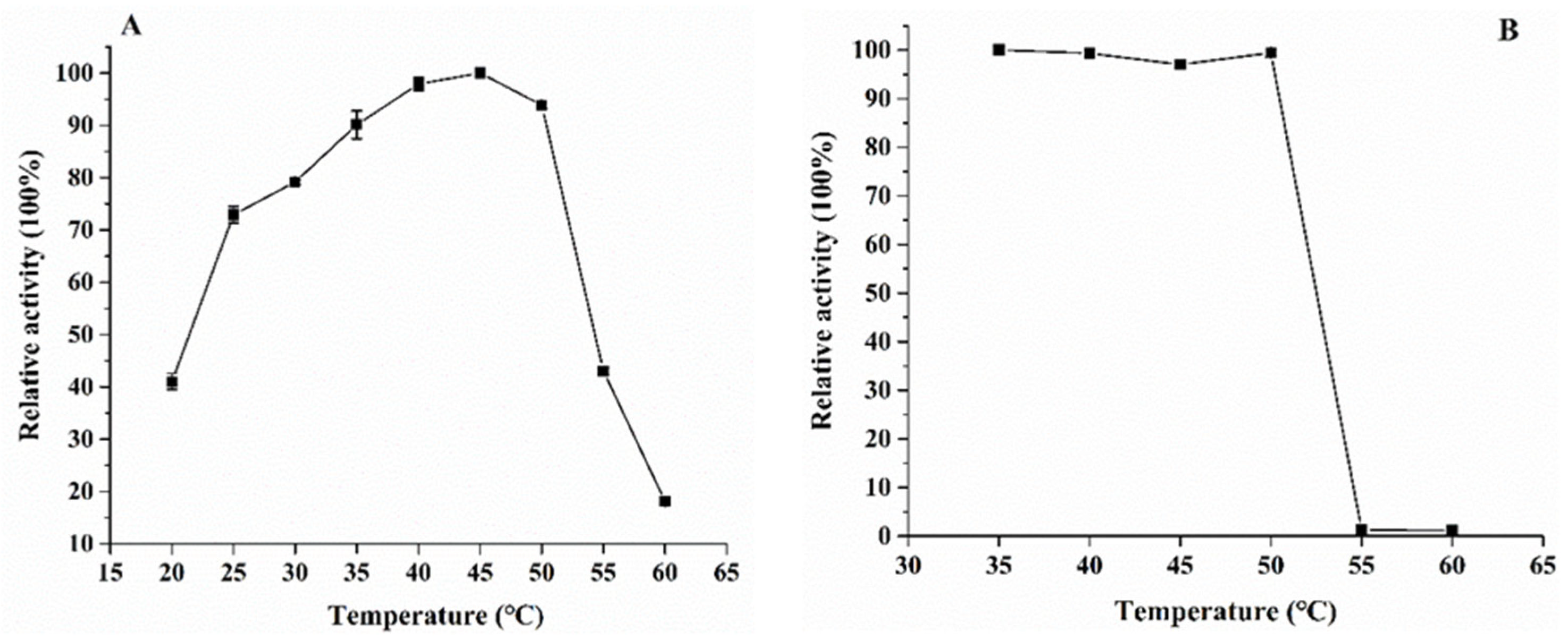

| Signal Peptide | Sequence | Secretory Pathway | Charged N-Region | Hydrophobic Amino Acid |
|---|---|---|---|---|
| AmyE | MFAKRFKTSLLPLFAGFLLLFHLVLAGPAAASA | Sec | 3 | 20 |
| AprE | MRSKKLWISLLFALTLIFTMAFSNMSVQA | Sec | 3 | 15 |
| AmyQ | MIQKRKRTVSFRLVLMCTLLFVSLPITKTSA | Sec | 4 | 12 |
| Bpr | MRKKTKNRLISSVLSTVVISSLLFPGAAGA | Sec | 5 | 13 |
| Mpr | MKLVPRFRKQWFAYLTVLCLALAAAVSFGVPAKA | Sec | 4 | 17 |
| Vpr | MKKGIIRFLLVSFVLFFALSTGITGVQAAPA | Sec | 3 | 16 |
| YjfA | MKRLFMKASLVLFAVVFVFAVKGAPAKA | Sec | 3 | 16 |
| YvgO | MKRIRIPMTLALGAALTIAPLSFASA | Sec | 3 | 15 |
| SacB | MNIKKFAKQATVLTFTTALLAGGATQAFA | Sec | 3 | 11 |
| YfhK | MKKKQVMLALTAAAGLGLTALHSAPAAKA | Sec | 3 | 17 |
| LipA | MKFVKRRIIALVTILMLSVTSLFALQPSAKAA | Tat | 2 | 15 |
| WapA | MKKRKRRNFKRFIAAFLVLALMISLVPADVLA | Tat | 6 | 16 |
| WprA | MKRRKFSSVVAAVLIFALIFSLFSPGTKAAA | Tat | 4 | 16 |
| SIase Source | Expression System | Conversion Conditions | Yield of Isomaltulose (%) | Reference |
|---|---|---|---|---|
| Enterobacter sp. FMB-1 | Lactococcus lactis MG1363 | pH 6.0, 30 °C | 72 | [13] |
| Enterobacter sp. FMB-1 | Saccharomyces cerevisiae EBY100 | pH 6.0, 45 °C | 7.4 | [34] |
| Erwinia rhapontici NX-5 | Bacillus subtilis 168 | pH 6.0, 30 °C | 92 | [35] |
| Pantoea dispersa UQ68J | Yarrowia lipolytica | pH 6.0, 30 °C | 93 | [36] |
| Klebsiella sp.LX3 | Bacillus subtilis WB800N | pH 5.5, 25 °C | 87.8 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, D.; Li, M.; Jiang, M.; Cong, G.; Liu, Y.; Wang, C.; Li, X. Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides. Foods 2022, 11, 2468. https://doi.org/10.3390/foods11162468
Guo D, Li M, Jiang M, Cong G, Liu Y, Wang C, Li X. Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides. Foods. 2022; 11(16):2468. https://doi.org/10.3390/foods11162468
Chicago/Turabian StyleGuo, Dan, Mingyu Li, Mengtong Jiang, Guilong Cong, Yuxin Liu, Conggang Wang, and Xianzhen Li. 2022. "Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides" Foods 11, no. 16: 2468. https://doi.org/10.3390/foods11162468
APA StyleGuo, D., Li, M., Jiang, M., Cong, G., Liu, Y., Wang, C., & Li, X. (2022). Enhanced Extracellular Production and Characterization of Sucrose Isomerase in Bacillus subtilis with Optimized Signal Peptides. Foods, 11(16), 2468. https://doi.org/10.3390/foods11162468





