Effects of Oxidation Modification by Malondialdehyde on the Structure and Functional Properties of Walnut Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Walnut Protein Isolate
2.3. Modification of Walnut Protein with MDA
2.4. The Measurement of Carbonyl Content of Walnut Protein
2.5. The Measurement of Free Sulfhydryl and Total Sulfhydryl Content
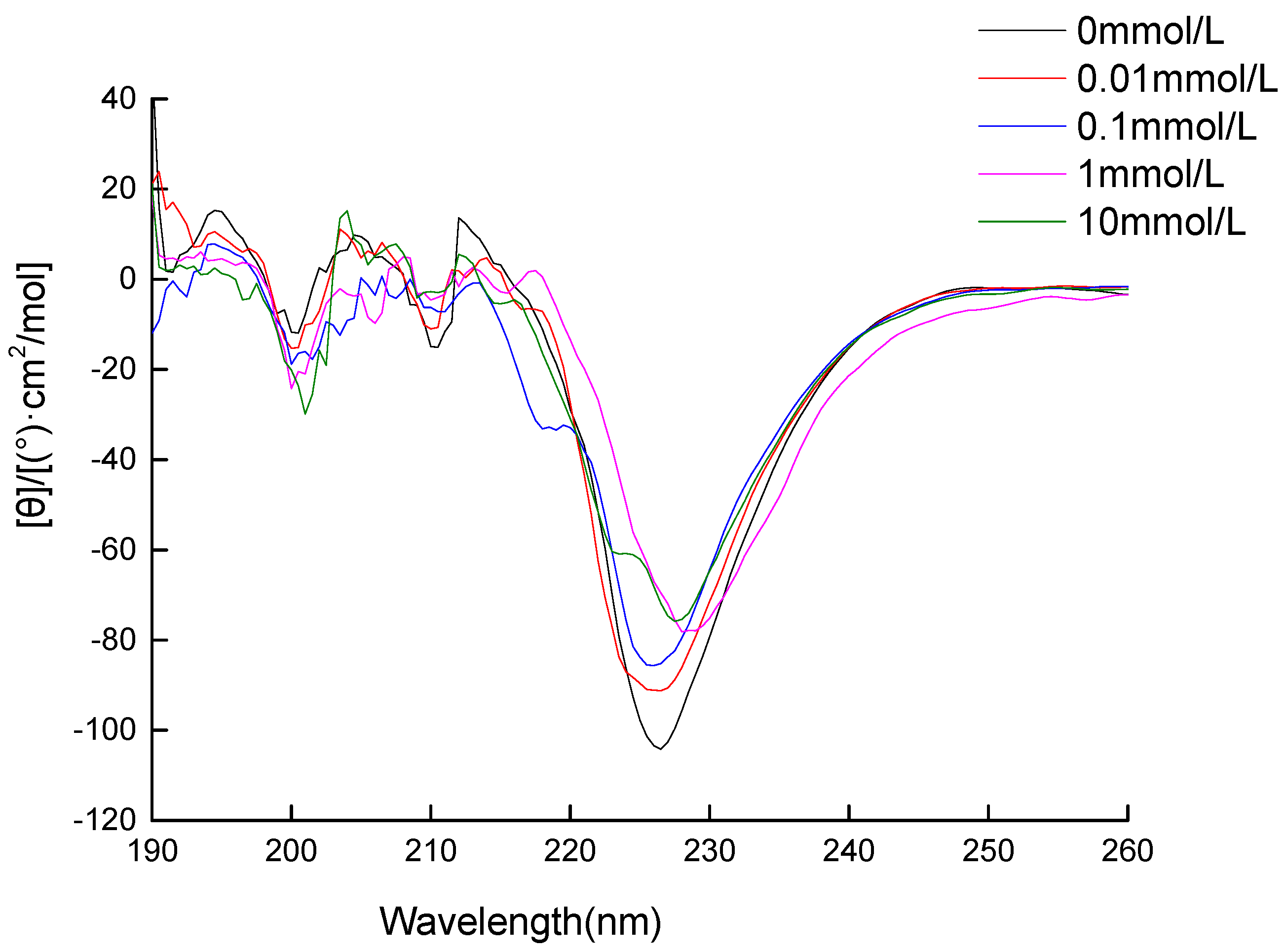
2.6. The Measurement of Circular Dichroism (CD) Spectra
2.7. Intrinsic Fluorescence
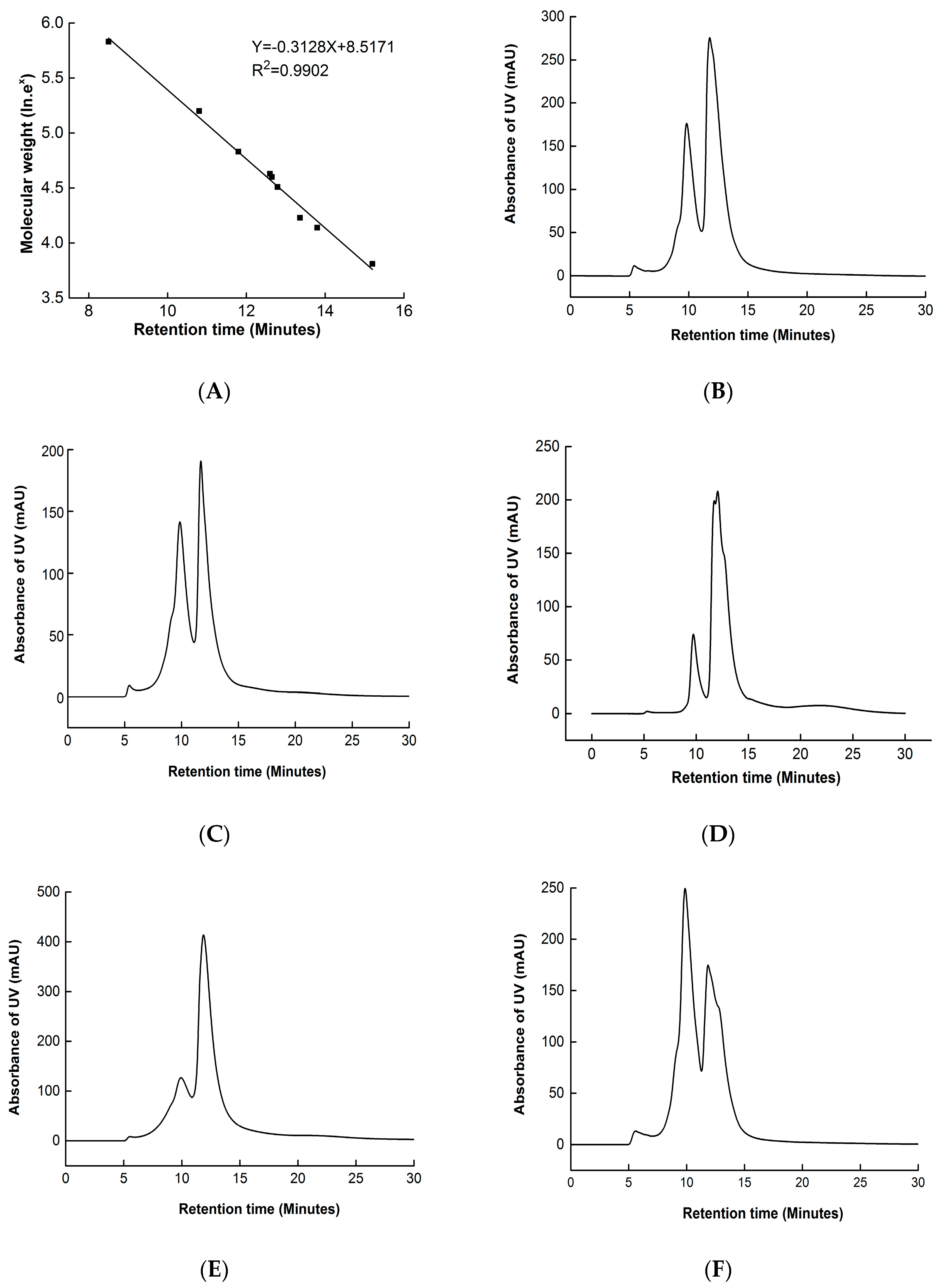
2.8. High-Performance Size Exclusion Chromatogram (HPSEC)
2.9. Foam Capacity (FC) and Foam Stability (FS)
2.10. Emulsifying Properties
2.11. Statistical Analysis
3. Results
3.1. Characterization of the Oxidation of Walnut Protein
3.2. Circular Dichroism (CD) Spectra Measurement
3.3. Intrinsic Fluorescence
3.4. High-Performance Size Exclusion Chromatogram (HPSEC)
3.5. Solubility of Proteins
3.6. Foam Capacity (FC) and Foam Stability (FS)
3.7. Emulsifying Properties
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cofrades, S.; Serrano, A.; Ayo, J.; Carballo, J.; Jimenez-Colmenero, F. Characteristics of meat batters with added native and preheated defatted walnut. Food Chem. 2008, 107, 1506–1514. [Google Scholar] [CrossRef]
- Sze-Tao, K.W.C.; Sathe, S.K. Walnuts (Juglans regia L.): Proximate composition, protein solubility, protein amino acid composition and protein in vitro digestibility. J. Sci. Food Agric. 2000, 80, 1393–1401. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, C.M.; Hua, Y.F. Structural modification of soy protein by the lipid peroxidation product malondialdehyde. J. Sci. Food Agric. 2009, 89, 1416–1423. [Google Scholar] [CrossRef]
- Shacter, E. Quantification and significance of protein oxidation in biological samples. Drug Metab. Rev. 2000, 32, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Headlam, H.A.; Davies, M.J. Markers of protein oxidation: Different oxidants give rise to variable yields of bound and released carbonyl products. Free. Radic. Biol. Med. 2004, 36, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Tironi, V.A.; Tomas, M.C.; Anon, M.C. Lipid and protein deterioration during the chilled storage of minced sea salmon (Pseudopercis semifasciata). J. Sci. Food Agric. 2007, 87, 2239–2246. [Google Scholar] [CrossRef]
- Lund, M.N.; Heinonen, M.; Baron, C.P.; Estevez, M. Protein oxidation in muscle foods: A review. Mol. Nutr. Food Res. 2011, 55, 83–95. [Google Scholar] [CrossRef]
- Ye, L.; Liao, Y.; Sun, W.Z.; Zhao, M.M. Effect of protein oxidation on the stability of peanut beverage. Cyta-J. Food 2015, 13, 49–55. [Google Scholar] [CrossRef]
- Wu, W.; Wu, X.J.; Hu, Y.F. Structural modification of soy protein by the lipid peroxidation product acrolein. Lwt-Food Sci. Technol. 2010, 43, 133–140. [Google Scholar] [CrossRef]
- Adams, A.; De Kimpe, N.; van Boekel, M. Modification of casein by the lipid oxidation product malondialdehyde. J. Agric. Food Chem. 2008, 56, 1713–1719. [Google Scholar] [CrossRef]
- Carbone, D.L.; Doorn, J.A.; Kiebler, Z.; Petersen, D.R. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem. Res. Toxicol. 2005, 18, 1324–1331. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.M.; Ahmedna, M.; Goktepe, I. Peanut protein concentrate: Production and functional properties as affected by processing. Food Chem. 2007, 103, 121–129. [Google Scholar] [CrossRef]
- Morales, F.J.; Jimenez-Perez, S. Effect of malondialdehyde on the determination of furosine in milk-based products. J. Agric. Food Chem. 2000, 48, 680–684. [Google Scholar] [CrossRef][Green Version]
- Mao, X.Y.; Hua, Y.F. Composition, Structure and Functional Properties of Protein Concentrates and Isolates Produced from Walnut (Juglans regia L.). Int. J. Mol. Sci. 2012, 13, 1561–1581. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.R.; Hua, Y.F.; Qiu, A.Y. Soybean protein aggregation induced by lipoxygenase catalyzed linoleic acid oxidation. Food Res. Int. 2006, 39, 240–249. [Google Scholar] [CrossRef]
- Beveridge, T.; Toma, S.J.; Nakai, S. Determination of SH-and SS-groups in some food proteins using Ellman’s reagent. J. Food Sci. 1974, 39, 49–51. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, C.M.; Kong, X.Z.; Hua, Y.F. Oxidative modification of soy protein by peroxyl radicals. Food Chem. 2009, 116, 295–301. [Google Scholar] [CrossRef]
- Ye, L.; Liao, Y.; Zhao, M.M.; Sun, W.Z. Effect of Protein Oxidation on the Conformational Properties of Peanut Protein Isolate. J. Chem. 2013, 2013, 423254. [Google Scholar] [CrossRef]
- Zhang, H.J.; Zhang, H.; Wang, L.; Guo, X.N. Preparation and functional properties of rice bran proteins from heat-stabilized defatted rice bran. Food Res. Int. 2012, 47, 359–363. [Google Scholar] [CrossRef]
- Pearce, K.N.; Kinsella, J.E. Emulsifying properties of proteins: Evaluation of a turbidimetric technique. J. Agric. Food Chem. 1978, 26, 716–723. [Google Scholar] [CrossRef]
- Uchida, K.; Kanematsu, M.; Sakai, K.; Matsuda, T.; Hattori, N.; Mizuno, Y.; Suzuki, D.; Miyata, T.; Noguchi, N.; Niki, E.; et al. Protein-bound acrolein: Potential markers for oxidative stress. Proc. Natl. Acad. Sci. USA 1998, 95, 4882–4887. [Google Scholar] [CrossRef]
- Burcham, P.C.; Kuhan, Y.T. Introduction of carbonyl groups into proteins by the lipid peroxidation product, malondialdehyde. Biochem. Biophys. Res. Commun. 1996, 220, 996–1001. [Google Scholar] [CrossRef]
- Uchida, K.; Sakai, K.; Itakura, K.; Osawa, T.; Toyokuni, S. Protein modification by lipid peroxidation products: Formation of malondialdehyde-derived N epsilon-(2-propenal)lysine in proteins. Arch. Biochem. Biophys. 1997, 346, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, M.; Fang, Z.X.; Bhandari, B. Gelation properties of myofibrillar protein under malondialdehyde-induced oxidative stress. J. Sci. Food Agric. 2017, 97, 50–57. [Google Scholar] [CrossRef]
- Zhang, S.B.; Lu, Q.Y. Characterizing the structural and surface properties of proteins isolated before and after enzymatic demulsification of the aqueous extract emulsion of peanut seeds. Food Hydrocoll. 2015, 47, 51–60. [Google Scholar] [CrossRef]
- Alleoni, A.C.C. Albumen protein and functional properties of gelation and foaming. Sci. Agric. 2006, 63, 291–298. [Google Scholar] [CrossRef]
- Sun, W.Z.; Zhou, F.B.; Sun, D.W.; Zhao, M.M. Effect of Oxidation on the Emulsifying Properties of Myofibrillar Proteins. Food Bioprocess Technol. 2013, 6, 1703–1712. [Google Scholar] [CrossRef]
- Chen, N.N.; Zhao, M.M.; Sun, W.Z.; Ren, J.Y.; Cui, C. Effect of oxidation on the emulsifying properties of soy protein isolate. Food Res. Int. 2013, 52, 26–32. [Google Scholar] [CrossRef]
- King, A.J.; Li, S.J. Association of Malonaldehyde with Rabbit Myosin Subfragment 1. In Quality Attributes of Muscle Foods; Springer: New York, NY, USA, 1999. [Google Scholar]
- Li, S.J.; King, A.J. Structural changes of rabbit myosin subfragment 1 altered by malonaldehyde, a byproduct of lipid oxidation. J. Agric. Food Chem. 1999, 47, 3124–3129. [Google Scholar] [CrossRef]
- Kong, B.H.; Xiong, Y.L.L.; Cui, X.H.; Zhao, X. Hydroxyl Radical-Stressed Whey Protein Isolate: Functional and Rheological Properties. Food Bioprocess Technol. 2013, 6, 169–176. [Google Scholar] [CrossRef]
- Wang, W.; Li, J.S.; Yan, L.J.; Huang, G.X.; Dong, Z. Effect of oxidization and chitosan on the surface activity of soy protein isolate. Carbohydr. Polym. 2016, 151, 700–706. [Google Scholar] [CrossRef] [PubMed]
- German, J.B.; O’Neill, T.E.; Kinsella, J.E. Film forming and foaming behavior of food proteins. J. Am. Oil Chem. Soc. 1985, 62, 1358–1362. [Google Scholar] [CrossRef]
- Lawal, O.S.; Adebowale, K.O.; Ogunsanwo, B.M.; Sosanwo, O.A.; Bankole, S.A. On the functional properties of globulin and albumin protein fractions and flours of African locust bean (Parkia biglobossa). Food Chem. 2005, 92, 681–691. [Google Scholar] [CrossRef]
- Zhao, Q.; Selomulya, C.; Xiong, H.; Chen, X.D.; Ruan, X.; Wang, S.Q.; Xie, J.H.; Peng, H.L.; Sun, W.J.; Zhou, Q. Comparison of functional and structural properties of native and industrial process-modified proteins from long-grain indica rice. J. Cereal Sci. 2012, 56, 568–575. [Google Scholar] [CrossRef]
- Cao, Y.G.; Ai, N.S.; True, A.D.; Xiong, Y.L. Effects of (-)-epigallocatechin-3-gallate incorporation on the physicochemical and oxidative stability of myofibrillar protein-soybean oil emulsions. Food Chem. 2018, 245, 439–445. [Google Scholar] [CrossRef]
- Liu, Q.; Lu, Y.; Han, J.C.; Chen, Q.; Kong, B.H. Structure-modification by moderate oxidation in hydroxyl radical-generating systems promote the emulsifying properties of soy protein isolate. Food Struct. 2015, 6, 21–28. [Google Scholar] [CrossRef]

 , and the FS is represented by the gray column
, and the FS is represented by the gray column  . The different letters in the same indicator indicate significant difference at p < 0.05).
. The different letters in the same indicator indicate significant difference at p < 0.05).
 , and the FS is represented by the gray column
, and the FS is represented by the gray column  . The different letters in the same indicator indicate significant difference at p < 0.05).
. The different letters in the same indicator indicate significant difference at p < 0.05).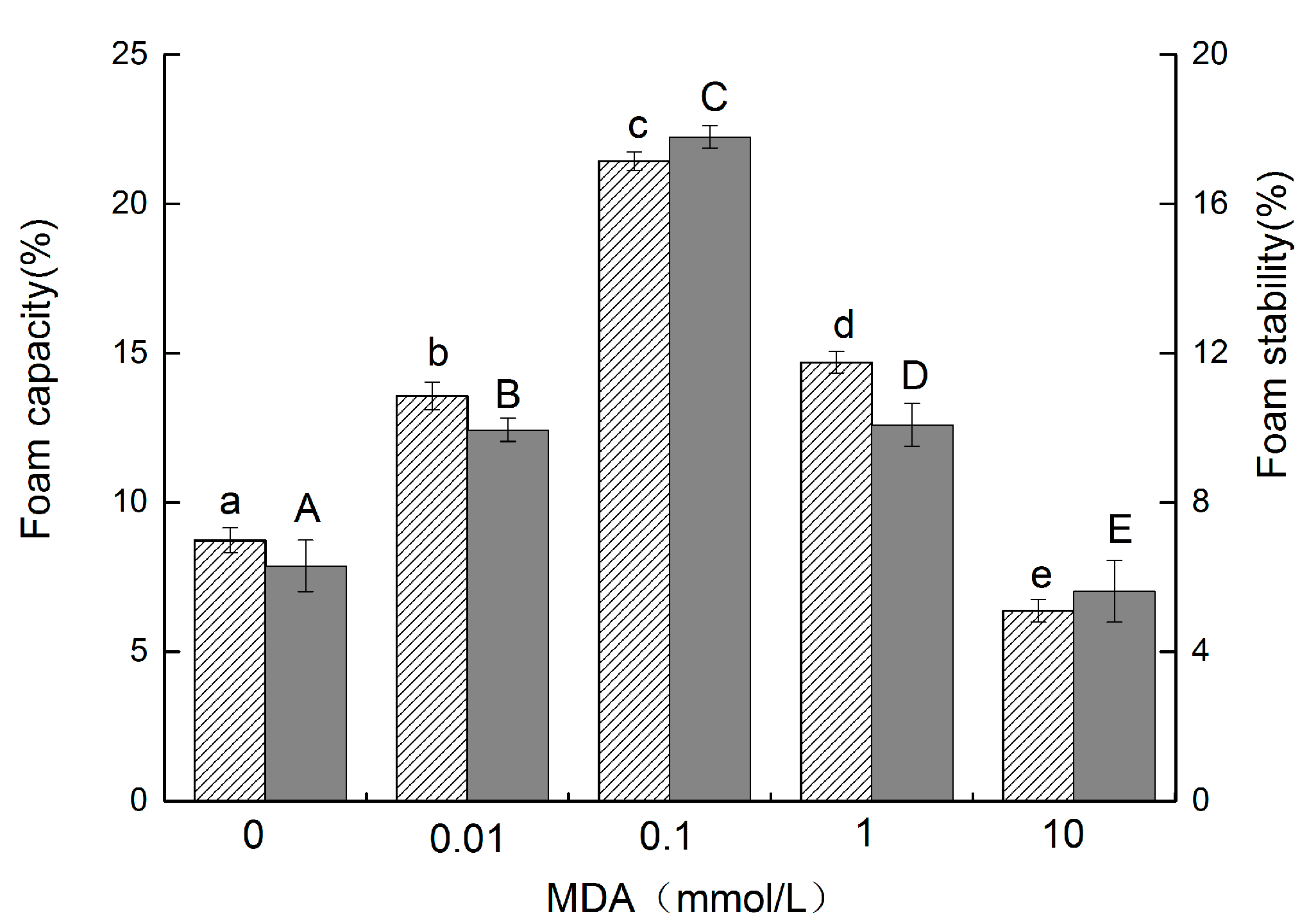
 , and the ESI is represented by the gray column
, and the ESI is represented by the gray column  . The different letters in the same indicator indicate significant difference at p < 0.05).
. The different letters in the same indicator indicate significant difference at p < 0.05).
 , and the ESI is represented by the gray column
, and the ESI is represented by the gray column  . The different letters in the same indicator indicate significant difference at p < 0.05).
. The different letters in the same indicator indicate significant difference at p < 0.05).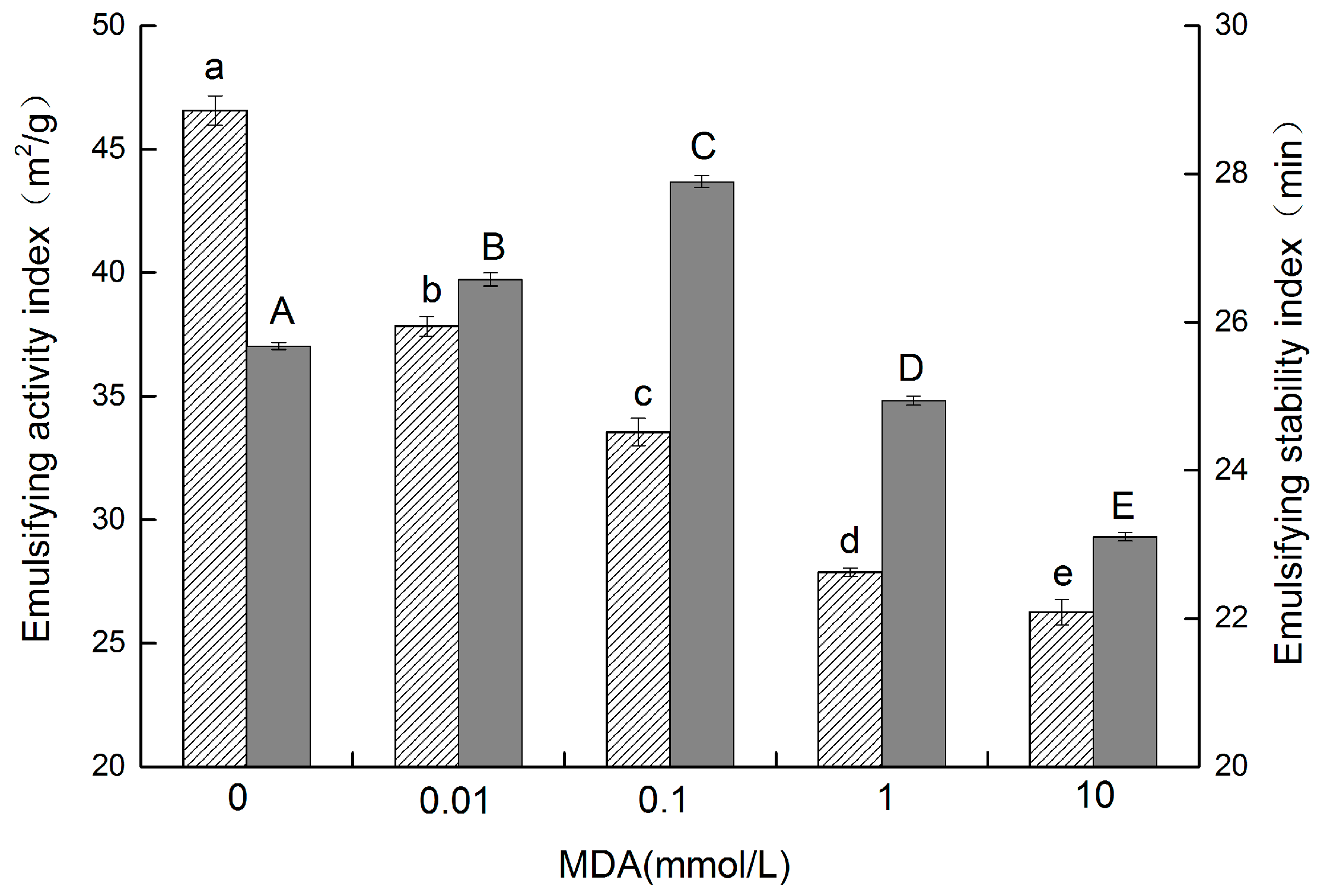
| MDA | Protein Carbonyl | Free Sulfydryl | Total Sulfydryl | Solubility |
|---|---|---|---|---|
| (mM) | (nmole/mg) | (nmole/mg) | (nmole/mg) | (%) |
| 0 | 3.12 ± 0.11 a | 34.84 ± 0.90 a | 167.49 ± 3.35 a | 11.78 ± 0.14 a |
| 0.01 | 4.64 ± 0.14 b | 32.17 ± 0.91 a | 148.06 ± 3.69 b | 9.51 ± 0.24 b |
| 0.1 | 5.26 ± 0.15 c | 23.02 ± 1.23 b | 126.73 ± 1.95 c | 6.73 ± 0.21 c |
| 1 | 9.09 ±0.17 d | 20.79 ± 0.53 c | 110.61 ± 1.73 d | 4.51 ± 0.44 d |
| 10 | 20.61 ± 0.15 e | 9.85 ± 1.62 d | 83.12 ± 4.04 e | 3.50 ± 0.39 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Wu, Q.; Mao, X. Effects of Oxidation Modification by Malondialdehyde on the Structure and Functional Properties of Walnut Protein. Foods 2022, 11, 2432. https://doi.org/10.3390/foods11162432
Sun L, Wu Q, Mao X. Effects of Oxidation Modification by Malondialdehyde on the Structure and Functional Properties of Walnut Protein. Foods. 2022; 11(16):2432. https://doi.org/10.3390/foods11162432
Chicago/Turabian StyleSun, Lingge, Qingzhi Wu, and Xiaoying Mao. 2022. "Effects of Oxidation Modification by Malondialdehyde on the Structure and Functional Properties of Walnut Protein" Foods 11, no. 16: 2432. https://doi.org/10.3390/foods11162432
APA StyleSun, L., Wu, Q., & Mao, X. (2022). Effects of Oxidation Modification by Malondialdehyde on the Structure and Functional Properties of Walnut Protein. Foods, 11(16), 2432. https://doi.org/10.3390/foods11162432






