Transcriptome Analysis Reveals Potential Mechanisms of L-Serine Production by Escherichia coli Fermentation in Different Carbon–Nitrogen Ratio Medium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria, Plasmids and Medium
2.2. Determination of Bacterial Growth
2.3. Determination of Glucose Content
2.4. Shake Flask Fermentation
2.5. Determination of L-Serine Content
2.6. Transcriptome Sequencing
2.6.1. RNA Extraction and Library Construction
2.6.2. RNA Sequencing and DEGs Analysis
2.6.3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis
2.7. Data Analysis
3. Results and Discussion
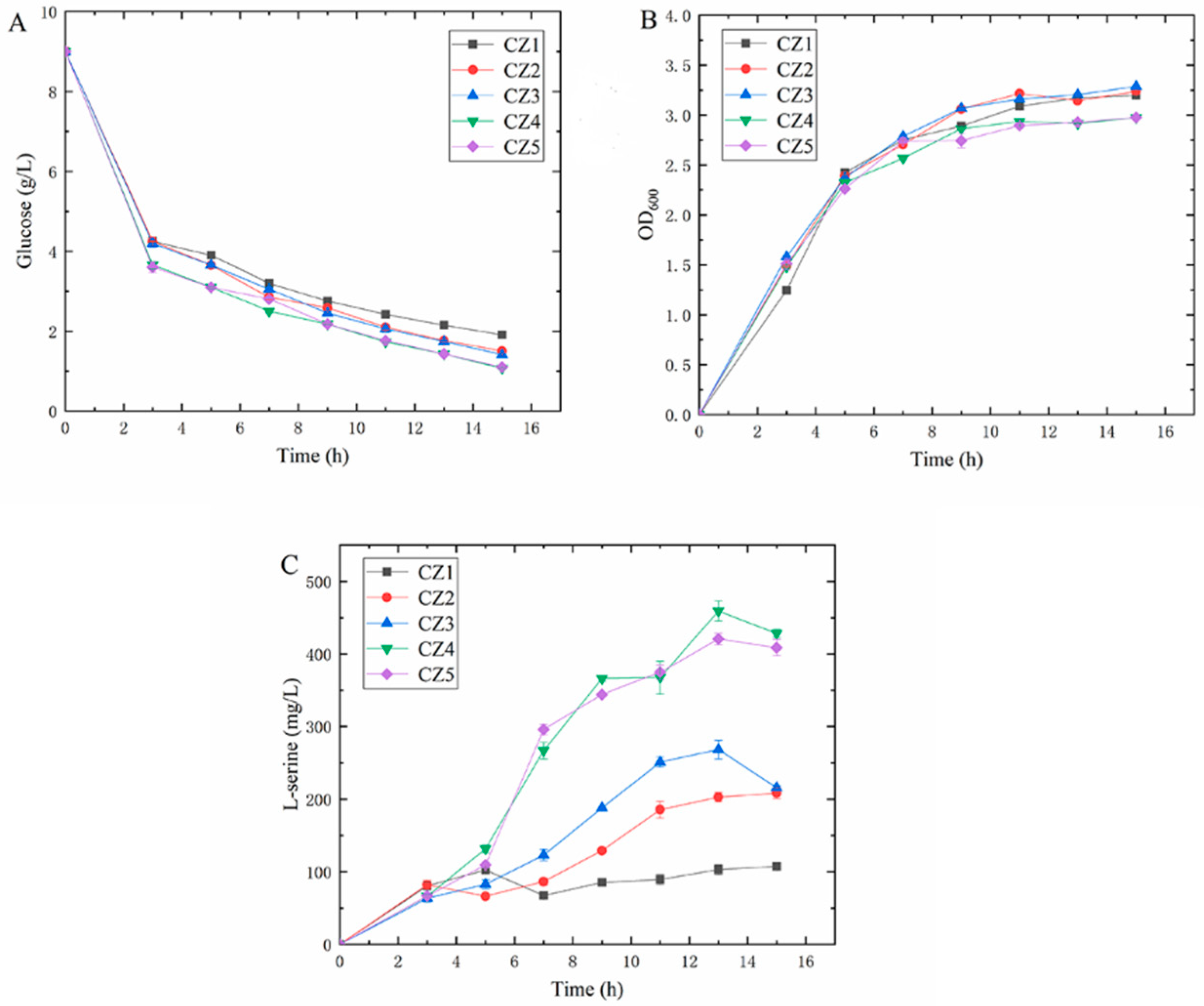
3.1. Effect of Carbon–Nitrogen Ratio Differences in Culture Medium on the Fermentation Performance of Shake Flasks
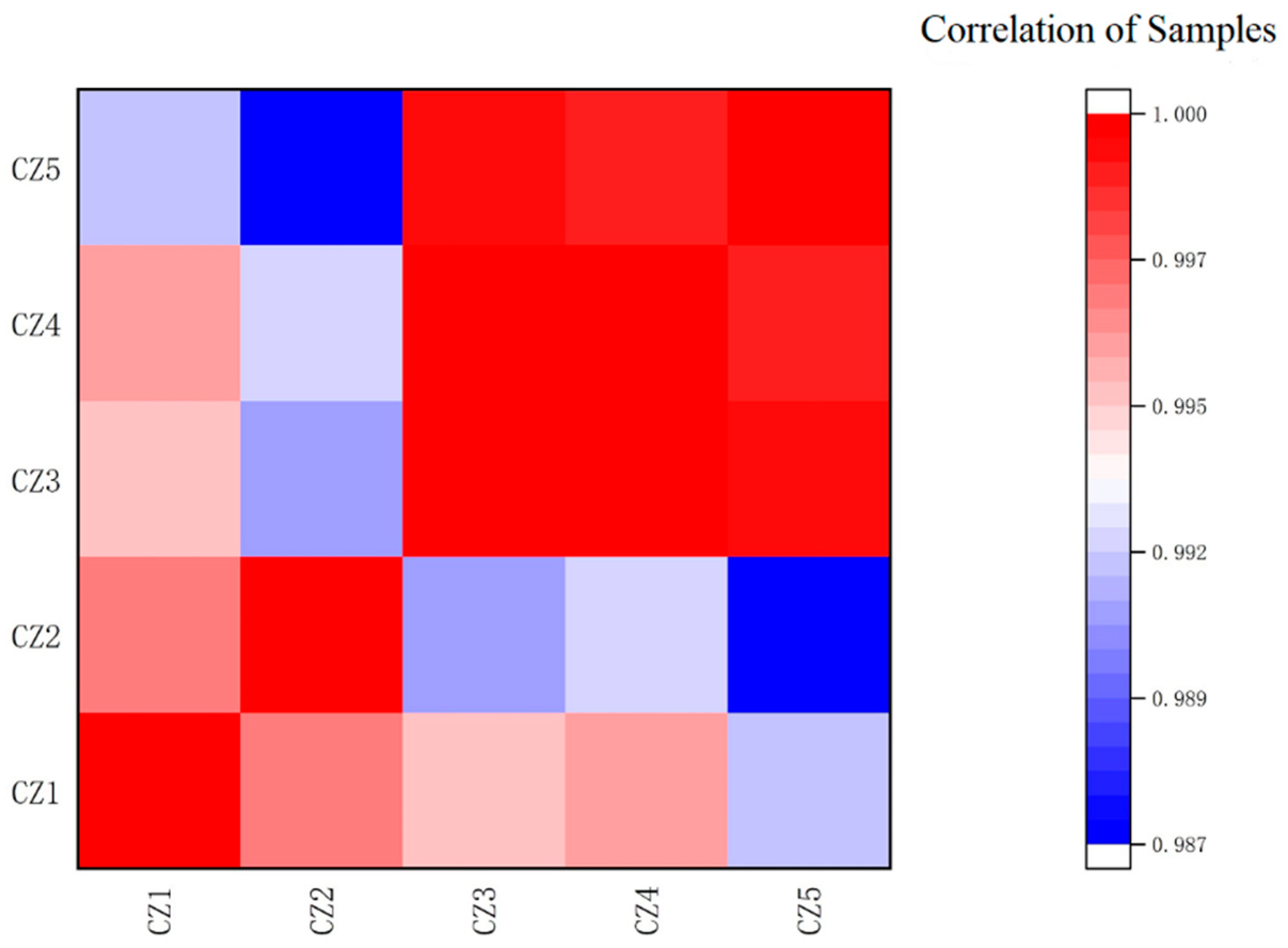
3.2. Transcriptome Analysis
3.2.1. Quality Control of Sequencing Data for Transcriptome Analysis
3.2.2. Reference Sequence Comparison Analysis
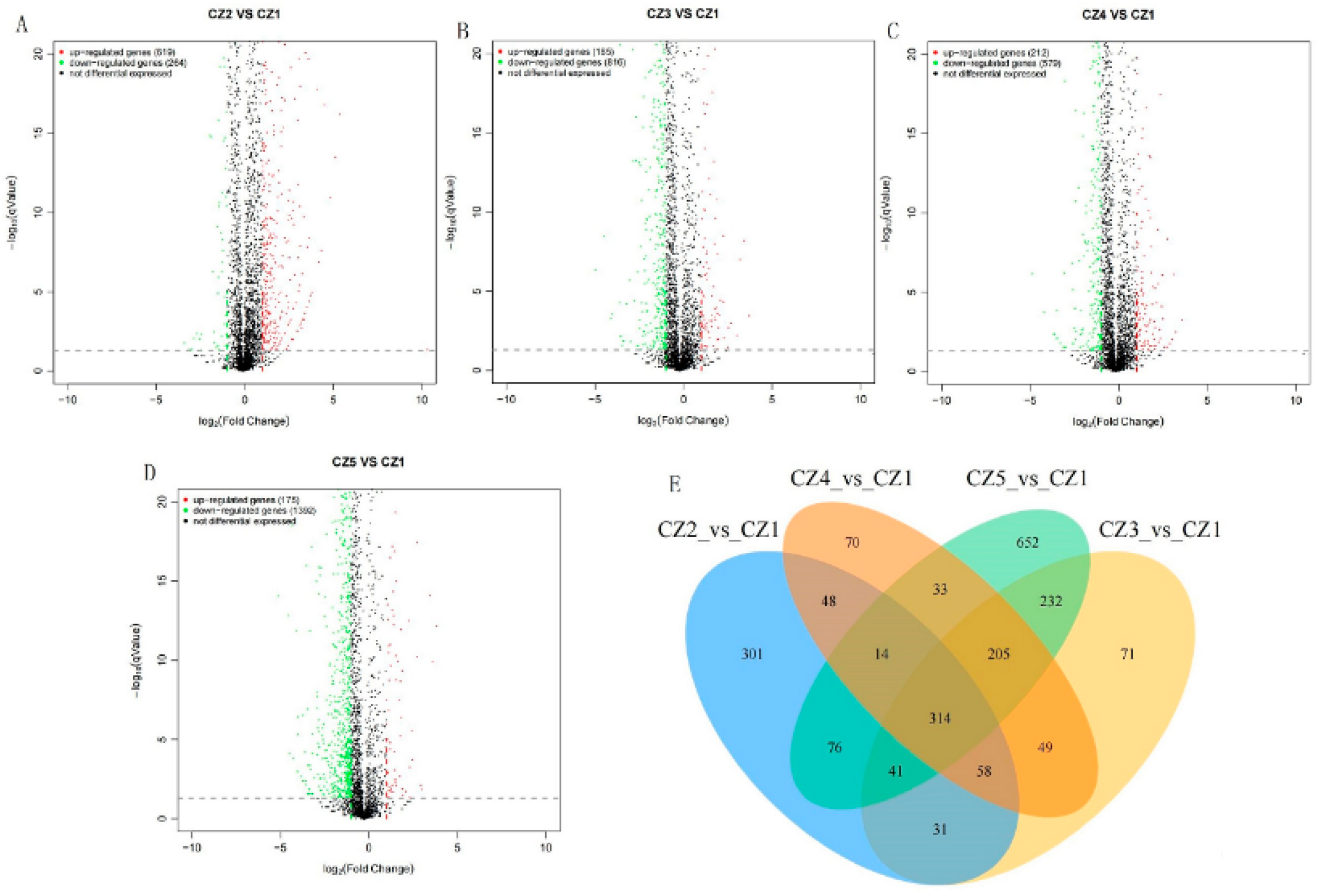
3.2.3. Screening of DEGs
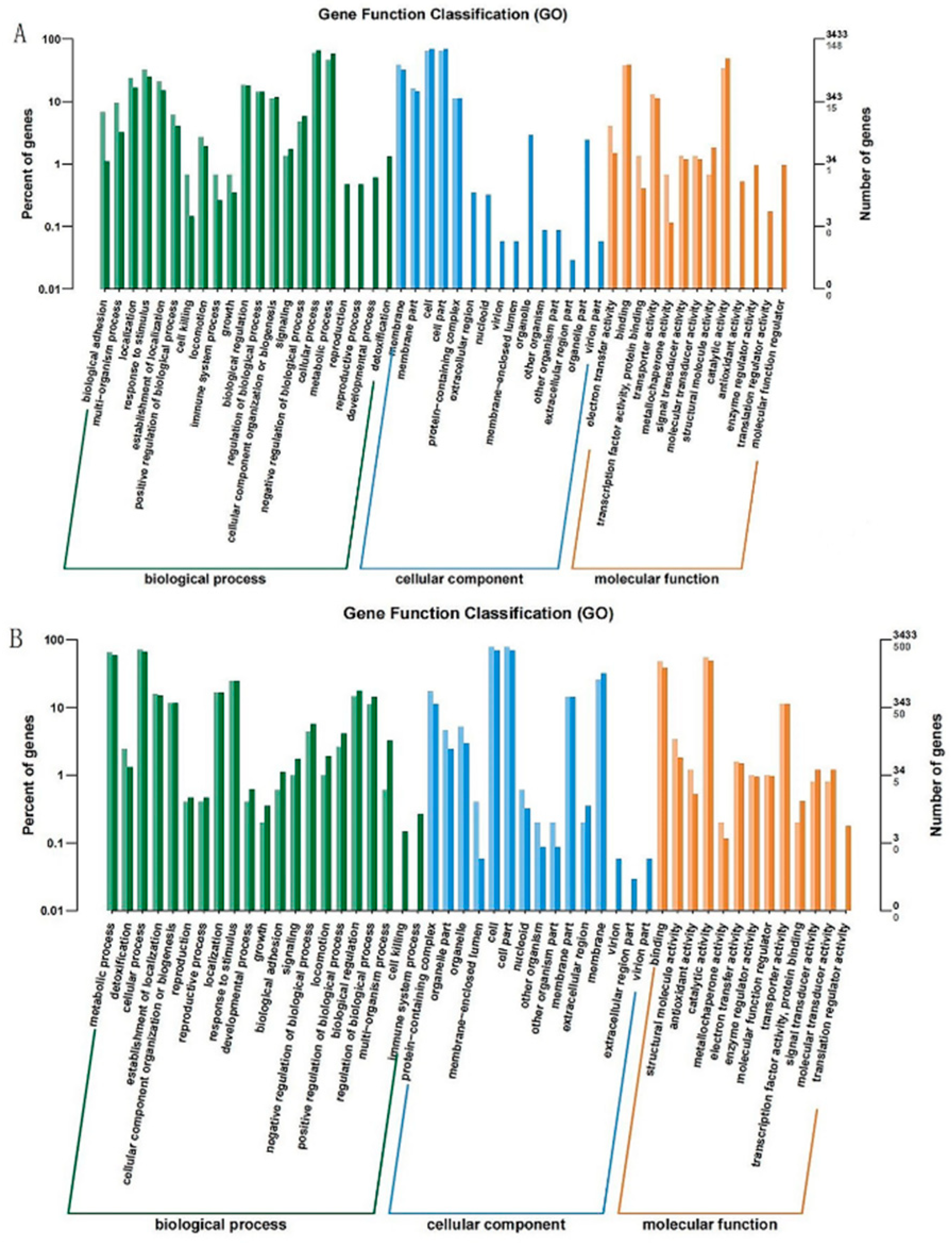
3.2.4. GO Enrichment Analysis of DEGs
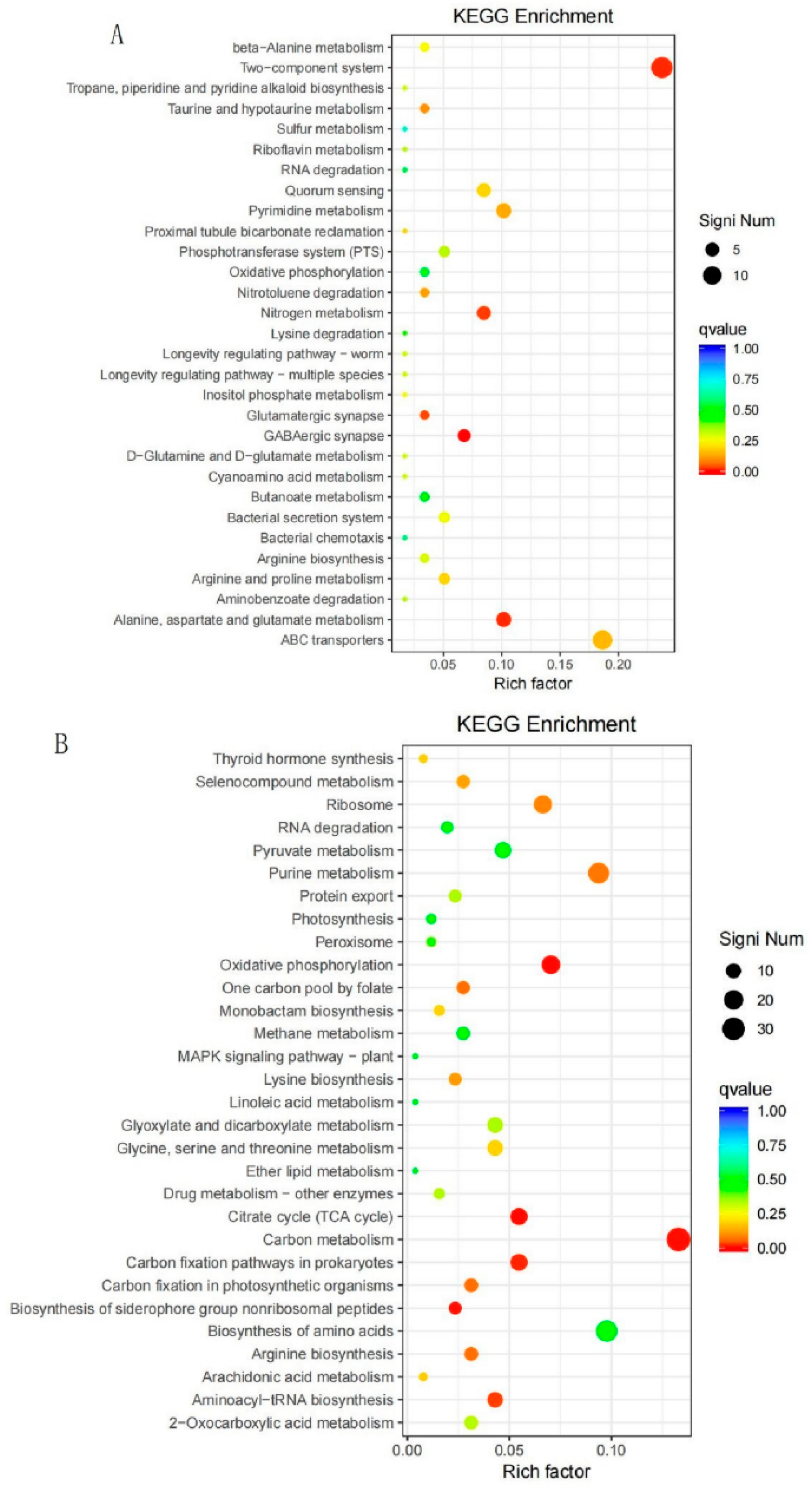
3.2.5. KEGG Metabolic Pathway Analysis of DEGs
3.3. Mining of Key Metabolic Pathways and Genes for L-Serine Production in E. coli
3.3.1. GABAergic Synapse
3.3.2. TCS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Holm, L.J.; Buschard, K. L-serine: A neglected amino acid with a potential therapeutic role in diabetes. APMIS 2019, 127, 655–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Koning, T.J.; Snell, K.; Duran, M.; Berger, R.; Poll-The, B.T.; Surtees, R. L-serine in disease and development. Biochem. J. 2003, 371, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Vousden, K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 2016, 16, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Maugard, M.; Vigneron, P.A.; Bolaños, J.P.; Bonvento, G. L-Serine links metabolism with neurotransmission. Prog. Neurobiol. 2021, 197, 101896. [Google Scholar] [CrossRef] [PubMed]
- Le Douce, J.; Maugard, M.; Veran, J.; Matos, M.; Jégo, P.; Vigneron, P.A.; Faivre, E.; Toussay, X.; Vandenberghe, M.; Balbastre, M.; et al. Impairment of glycolysis-derived L-serine production in astrocytes contributes to cognitive deficits in Alzheimer’s disease. Cell Metab. 2020, 31, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Zhang, X.; Luo, Y.; Guo, W.; Xu, G.; Shi, J.; Xu, Z. l-Serine overproduction with minimization of by-product synthesis by engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2015, 99, 1665–1673. [Google Scholar] [CrossRef]
- Zhang, X.; Lai, L.; Xu, G.; Zhang, X.; Shi, J.; Koffas, M.A.; Xu, Z. Rewiring the central metabolic pathway for high-yield l-serine production in Corynebacterium glutamicum by using glucose. Biotechnol. J. 2019, 14, 1800497. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, Y.; Chen, Z.; Xu, G.; Zhang, X.; Li, H.; Shi, J.; Koffas, M.A.; Xu, Z. High-yield production of L-serine through a novel identified exporter combined with synthetic pathway in Corynebacterium glutamicum. Microb. Cell Factories 2020, 19, 115. [Google Scholar] [CrossRef]
- Pramanik, J.; Keasling, J.D. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnol. Bioeng. 1997, 56, 398–421. [Google Scholar] [CrossRef]
- Wang, C.; Wu, J.; Shi, B.; Shi, J.; Zhao, Z. Improving L-serine formation by Escherichia coli by reduced uptake of produced L-serine. Microb. Cell Factories 2020, 19, 66. [Google Scholar] [CrossRef] [Green Version]
- Tran, K.N.T.; Eom, G.T.; Hong, S.H. Improving L-serine production in Escherichia coli via synthetic protein scaffold of SerB, SerC, and EamA. Biochem. Eng. J. 2019, 148, 138–142. [Google Scholar] [CrossRef]
- Rennig, M.; Mundhada, H.; Wordofa, G.G.; Gerngross, D.; Wulff, T.; Worberg, A.; Nielsen, A.T.; Nørholm, M.H. Industrializing a bacterial strain for l-serine production through translation initiation optimization. ACS Synth. Biol. 2019, 8, 2347–2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mundhada, H.; Schneider, K.; Christensen, H.B.; Nielsen, A.T. Engineering of high yield production of L-serine in Escherichia coli. Biotechnol. Bioeng. 2016, 113, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, T.; Zhao, Q.; Xie, X.; Li, Y.; Chen, Q.; Cheng, F.; Tian, J.; Gu, H.; Huang, J. Comparative Transcriptome Analysis of the Accumulation of Anthocyanins Revealed the Underlying Metabolic and Molecular Mechanisms of Purple Pod Coloration in Okra (Abelmoschus esculentus L.). Foods 2021, 10, 2180. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, B.; Zheng, L.; Peng, Y.; Tian, R.; Yuan, Y.; Zhu, L.; Su, J.; Ma, F.; Li, M.; et al. Combined Profiling of Transcriptome and DNA Methylome Reveal Genes Involved in Accumulation of Soluble Sugars and Organic Acid in Apple Fruits. Foods 2021, 10, 2198. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Godana, E.A.; Xu, M.; Yang, Q.; Dhanasekaran, S.; Zhang, H. Transcriptome Characterization and Expression Profiles of Disease Defense-Related Genes of Table Grapes in Response to Pichia anomala Induced with Chitosan. Foods 2021, 10, 1451. [Google Scholar] [CrossRef]
- Zhang, H.; Zhao, X.; Zhao, C.; Zhang, J.; Liu, Y.; Yao, M.; Liu, J. Effects of glycerol and glucose on docosahexaenoic acid synthesis in Aurantiochyrium limacinum SFD-1502 by transcriptome analysis. Prep. Biochem. Biotechnol. 2022, 0, 1–12. [Google Scholar] [CrossRef]
- Lv, J.; Zhao, F.; Feng, J.; Liu, Q.; Nan, F.; Liu, X.; Xie, S. Transcriptomic analysis reveals the mechanism on the response of Chlorococcum sp. GD to glucose concentration in mixotrophic cultivation. Bioresour. Technol. 2019, 288, 121568. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, G.; Zhu, C.; Wang, C.; Wang, D.; Wei, G. Metabolic flux and transcriptome analyses provide insights into the mechanism underlying zinc sulfate improved β-1, 3-D-glucan production by Aureobasidium pullulans. Int. J. Biol. Macromol. 2020, 164, 140–148. [Google Scholar] [CrossRef]
- Hirasawa, T.; Saito, M.; Yoshikawa, K.; Furusawa, C.; Shmizu, H. Integrated Analysis of the Transcriptome and Metabolome of Corynebacterium glutamicum during Penicillin-Induced Glutamic Acid Production. Biotechnol. J. 2018, 13, 1700612. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, Z.; Wen, Z.; Lu, M.; Wang, Z.; Zhang, Y.; Zhou, H.; Jin, M. Combined adaptive evolution and transcriptomic profiles reveal aromatic aldehydes tolerance mechanisms in Yarrowia lipolytica. Bioresour. Technol. 2021, 329, 124910. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Sun, Z.G.; Zhang, C.Y.; Zhang, Q.Z.; Guo, X.W.; Xiao, D.G. Comparative transcriptome analysis reveals the key regulatory genes for higher alcohol formation by yeast at different α-amino nitrogen concentrations. Food Microbiol. 2021, 95, 103713. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, R.; Lu, W.; Zhou, Z.; Jiang, X.; Zhao, H.; Yang, B.; Lü, S. Transcriptome analysis identifies key genes involved in the regulation of epidermal lupeol biosynthesis in Ricinus communis. Ind. Crops Prod. 2021, 160, 113100. [Google Scholar] [CrossRef]
- Dang, B.T.; Nguyen, T.T.; Ngo, H.H.; Pham, M.D.T.; Le, L.T.; Nguyen, N.K.Q.; Vo, T.D.H.; Varjani, S.; You, S.J.; Lin, K.; et al. Influence of C/N ratios on treatment performance and biomass production during co-culture of microalgae and activated sludge. Sci. Total Environ. 2022, 837, 155832. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Glucose biosensors: 40 years of advances and challenges. Electroanalysis 2001, 13, 983–988. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, Q.; Wei, G.; Liang, Q.; Qi, Q. Production in Escherichia coli of poly (3-hydroxybutyrate-co-3-hydroxyvalerate) with differing monomer compositions from unrelated carbon sources. Appl. Environ. Microbiol. 2011, 77, 4886–4893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Filipič, J.; Kraigher, B.; Tepuš, B.; Kokol, V.; Mandic-Mulec, I. Effects of low-density static magnetic fields on the growth and activities of wastewater bacteria Escherichia coli and Pseudomonas putida. Bioresour. Technol. 2012, 120, 225–232. [Google Scholar] [CrossRef]
- Rahman, K.H.A.; Najimudin, N.; Ismail, K.S.K. Transcriptomes analysis of Pichia kudriavzevii UniMAP 3–1 in response to acetic acid supplementation in glucose and xylose medium at elevated fermentation temperature. Process Biochem. 2022, 118, 41–51. [Google Scholar] [CrossRef]
- Hama, H.; Sumita, Y.; Kakutani, Y.; Tsuda, M.; Tsuchiya, T. Target of serine inhibition in Escherichia coli. Biochem. Biophys. Res. Commun. 1990, 168, 1211–1216. [Google Scholar] [CrossRef]
- Ham, S.; Bhatia, S.K.; Gurav, R.; Choi, Y.K.; Jeon, J.M.; Yoon, J.J.; Choi, K.K.; Ahn, J.; Kim, H.; Yang, Y.H. Gamma aminobutyric acid (GABA) production in Escherichia coli with pyridoxal kinase (pdxY) based regeneration system. Enzym. Microb. Technol. 2022, 155, 109994. [Google Scholar] [CrossRef]
- Xue, C.; Ng, I.S. Sustainable production of 4-aminobutyric acid (GABA) and cultivation of Chlorella sorokiniana and Chlorella vulgaris as circular economy. Bioresour. Technol. 2022, 343, 126089. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Lee, I.S.; Frey, J.; Slonczewski, J.L.; Foster, J.W. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 1995, 177, 4097–4104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, M.L.; Ferreri, M.C.; Farb, D.H.; Gravielle, M.C. GABA-induced uncoupling of GABA/benzodiazepine site interactions is associated with increased phosphorylation of the GABAA receptor. J. Neurosci. Res. 2014, 92, 1054–1061. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Xie, M.; Yang, Y.; Feng, J.; Tan, L.; Chen, C. The role of L-alanine metabolism revealed by transcriptome analysis in Vibrio alginolyticus. Gene Rep. 2018, 10, 184–187. [Google Scholar] [CrossRef]
- Wu, L.; Luo, S.; Ma, S.; Liang, Z.; Wu, J.R. Construction of light-sensing two-component systems in Escherichia coli. Sci. Bull. 2017, 62, 813–815. [Google Scholar] [CrossRef] [Green Version]
- Jacob-Dubuisson, F.; Mechaly, A.; Betton, J.M.; Antoine, R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018, 16, 585–593. [Google Scholar] [CrossRef]
- Lu, P.; Ma, D.; Chen, Y.; Guo, Y.; Chen, G.Q.; Deng, H.; Shi, Y. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013, 23, 635–644. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Lu, P.; Yan, C.; Fan, C.; Yin, P.; Wang, J.; Shi, Y. Structure and mechanism of a glutamate–GABA antiporter. Nature 2012, 483, 632–636. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
| Sample | Total Reads Count | Total Bases Count (bp) | Average Read Length (bp) | Q20 Bases Ratio (%) | Q30 Bases Ratio (%) | GC Bases Ratio (%) |
|---|---|---|---|---|---|---|
| CZ1 | 27,860,276 | 4,127,583,116 | 148.15 | 98.96 | 96.08 | 52.94 |
| CZ2 | 65,073,878 | 9,459,591,029 | 145.37 | 99.02 | 96.31 | 52.76 |
| CZ3 | 31,583,064 | 4,686,203,693 | 148.38 | 98.96 | 96.03 | 52.60 |
| CZ4 | 30,706,250 | 4,560,411,727 | 148.52 | 99.01 | 96.16 | 52.63 |
| CZ5 | 30,088,160 | 4,463,605,237 | 148.35 | 98.98 | 96.09 | 52.47 |
| Sample | Total Reads | Total Mapped | Mutiple Mapped | Uniquely Mapped | Reads Mapped to ‘+’ | Reads Mapped to ‘−’ | Non-Spliced Reads | Reads Mapped in Proper Pairs |
|---|---|---|---|---|---|---|---|---|
| CZ1 | 27,844,844 | 27,433,999 (98.52%) | 857,821 (3.08%) | 26,576,178 (95.44%) | 13,288,453 | 13,287,725 | 26,576,178 | 26,181,602 |
| CZ2 | 64,908,882 | 63,563,985 (97.93%) | 8,423,620 (12.98%) | 55,140,365 (84.95%) | 27,571,051 | 27,569,314 | 55,140,365 | 50,666,474 |
| CZ3 | 31,569,790 | 31,033,431 (98.30%) | 844,243 (2.67%) | 30,189,188 (95.63%) | 15,097,174 | 15,092,014 | 30,189,188 | 29,876,526 |
| CZ4 | 30,683,164 | 30,085,238 (98.05%) | 822,624 (2.68%) | 29,262,614 (95.37%) | 14,633,739 | 14,628,875 | 29,262,614 | 28,974,756 |
| CZ5 | 30,064,744 | 29,597,379 (98.45%) | 977,979 (3.25%) | 28,619,400 (95.19%) | 14,310,590 | 14,308,810 | 28,619,400 | 28,259,990 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Chen, X.; Li, Q.; Zhou, P.; Zhao, Z.; Li, B. Transcriptome Analysis Reveals Potential Mechanisms of L-Serine Production by Escherichia coli Fermentation in Different Carbon–Nitrogen Ratio Medium. Foods 2022, 11, 2092. https://doi.org/10.3390/foods11142092
Chen Z, Chen X, Li Q, Zhou P, Zhao Z, Li B. Transcriptome Analysis Reveals Potential Mechanisms of L-Serine Production by Escherichia coli Fermentation in Different Carbon–Nitrogen Ratio Medium. Foods. 2022; 11(14):2092. https://doi.org/10.3390/foods11142092
Chicago/Turabian StyleChen, Zheng, Xiaojia Chen, Qinyu Li, Peng Zhou, Zhijun Zhao, and Baoguo Li. 2022. "Transcriptome Analysis Reveals Potential Mechanisms of L-Serine Production by Escherichia coli Fermentation in Different Carbon–Nitrogen Ratio Medium" Foods 11, no. 14: 2092. https://doi.org/10.3390/foods11142092
APA StyleChen, Z., Chen, X., Li, Q., Zhou, P., Zhao, Z., & Li, B. (2022). Transcriptome Analysis Reveals Potential Mechanisms of L-Serine Production by Escherichia coli Fermentation in Different Carbon–Nitrogen Ratio Medium. Foods, 11(14), 2092. https://doi.org/10.3390/foods11142092






