NMR-Based Metabolomic Approach for Evaluation of the Harvesting Time and Cooking Characteristics of Different Cassava Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Lignin Extraction
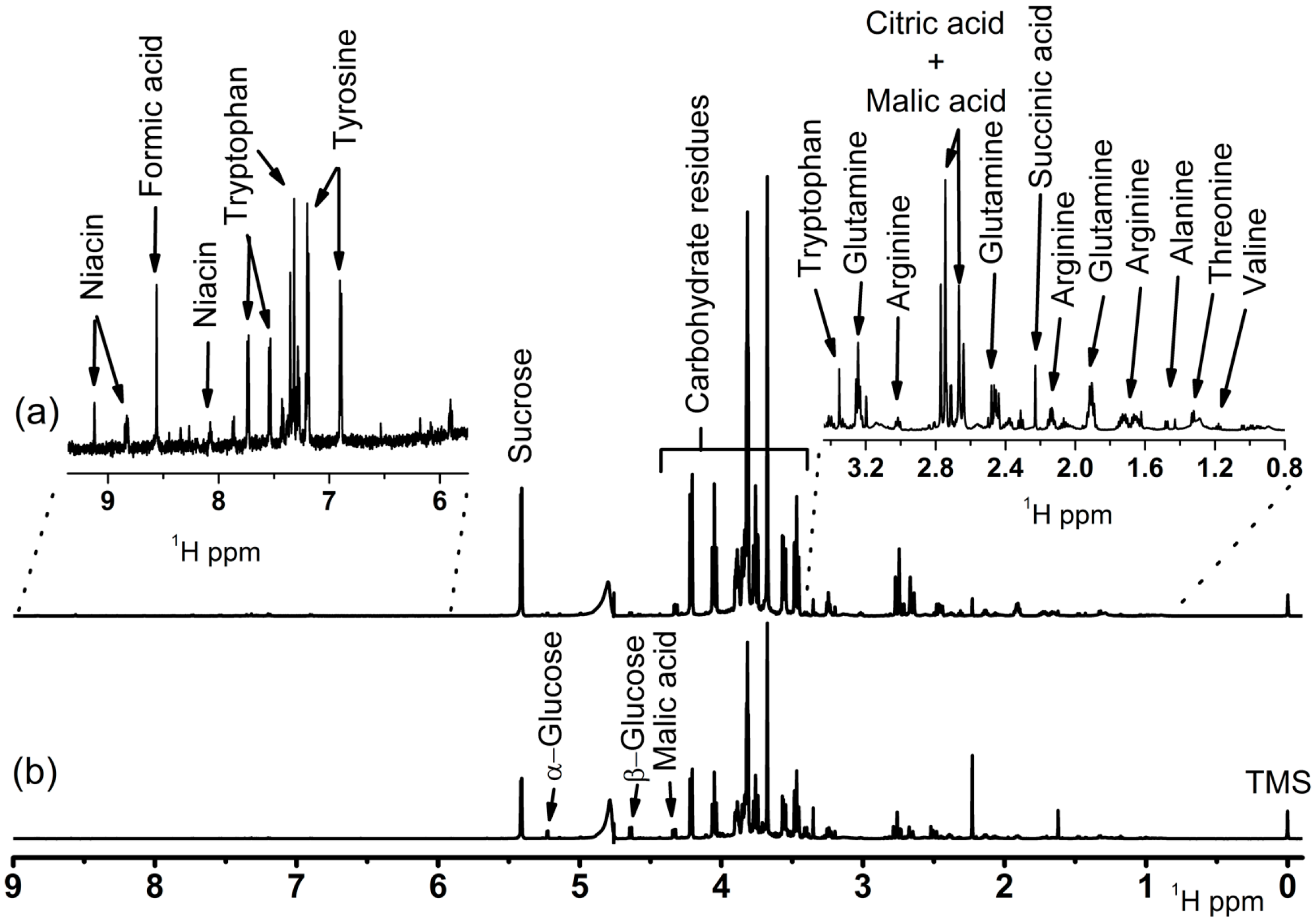
2.3. NMR Spectroscopy
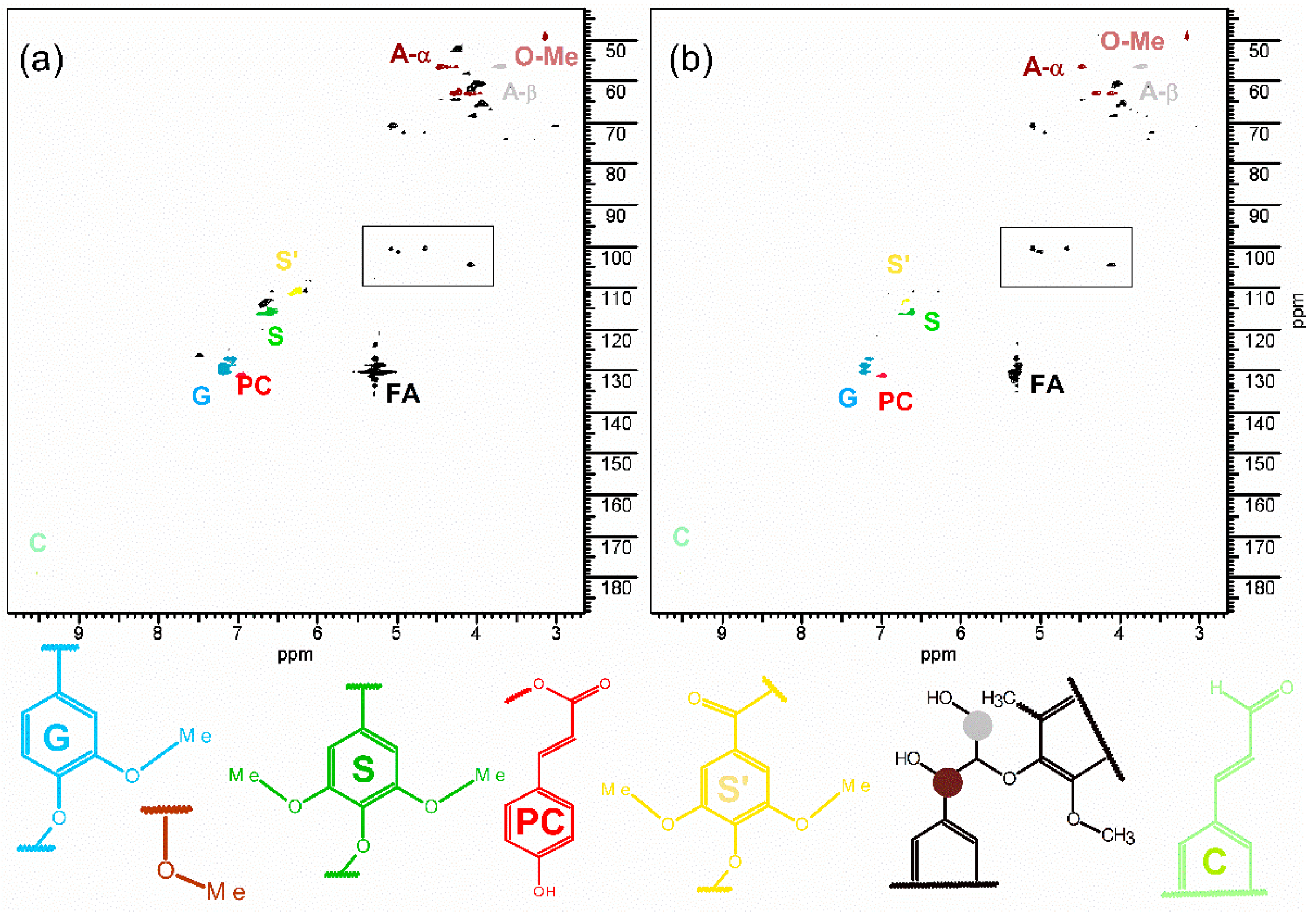
2.4. Relative Quantitative 1H-13C HSQC NMR Analysis of the Lignin
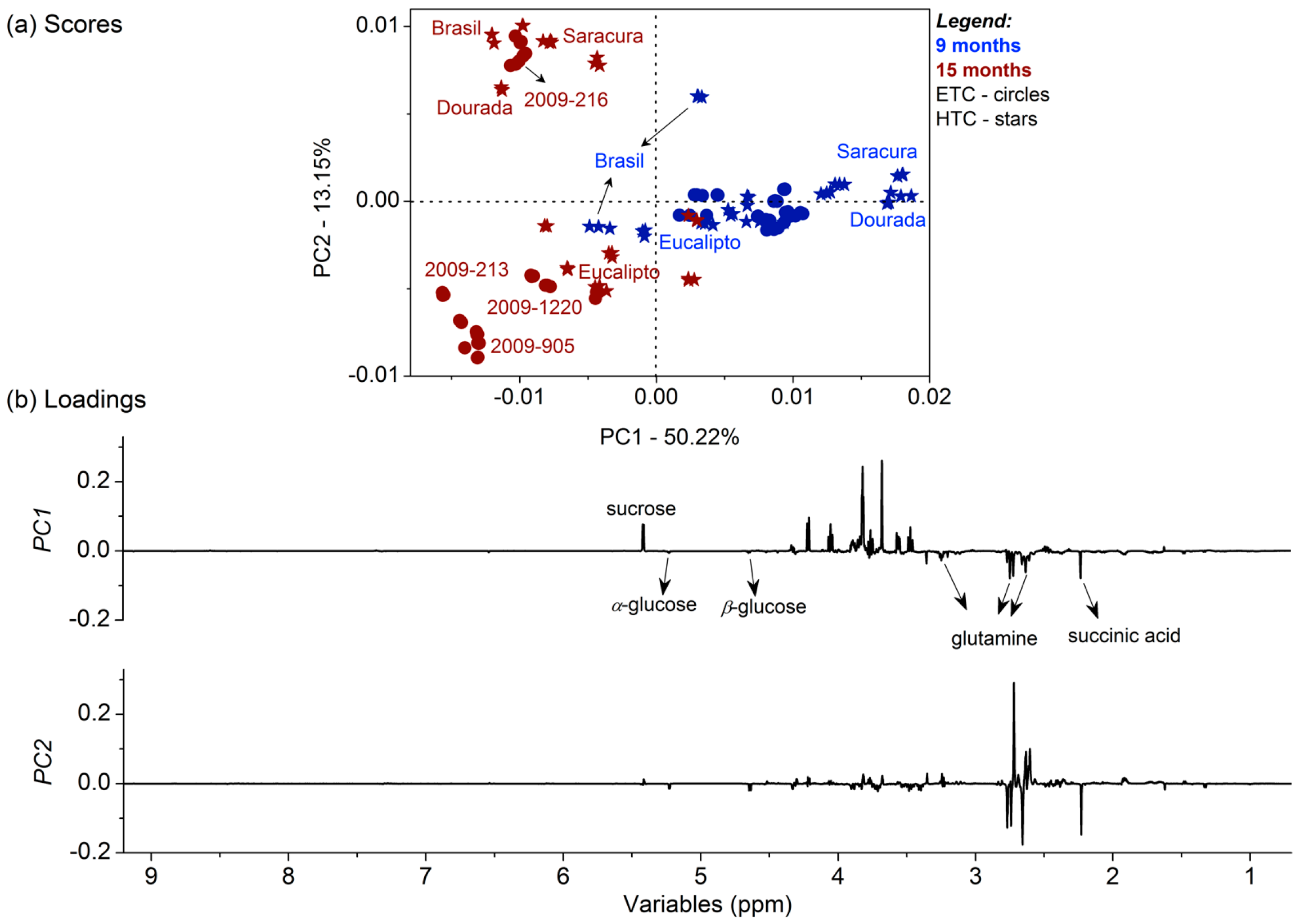
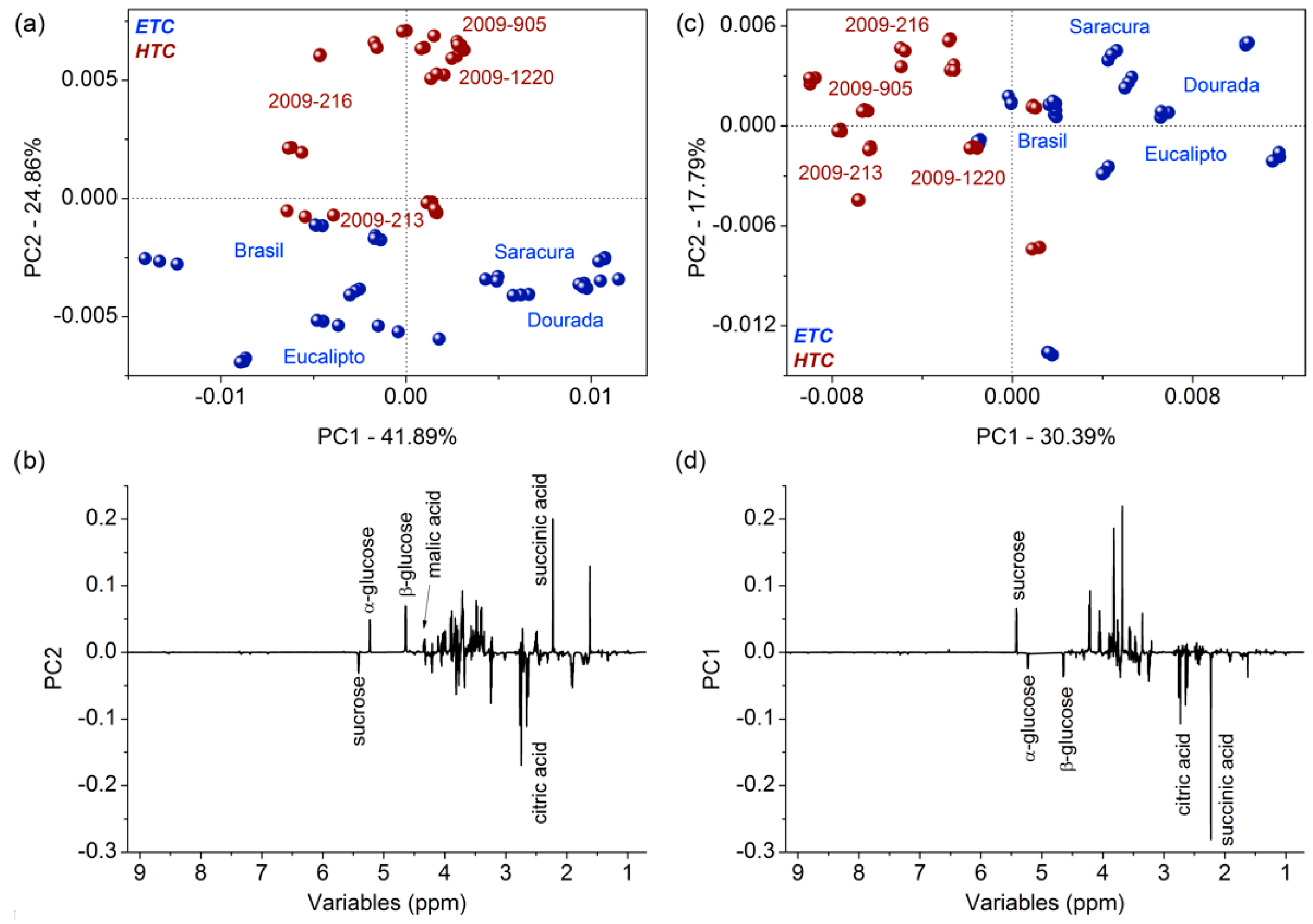
2.5. Chemometric Analysis of the 1H NMR Dataset
2.6. Metabolomic Pathway Analysis
2.7. Determination of Starch (Fresh and Dry Weight) and Cooking Time
3. Results and Discussion
3.1. Metabolomic Fingerprinting
3.2. Relative Quantitative 1H-13 HSQC NMR Analysis of the Lignin in Cassava Genotypes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siebers, T.; Catarino, B.; Agusti, J. Identification and expression analyses of new potential regulators of xylem development and cambium activity in cassava (Manihot esculenta). Planta 2017, 245, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Bayoumi, S.A.L.; Rowan, M.G.; Beeching, J.R.; Blagbrough, I.S. Constituents and secondary metabolite natural products in fresh and deteriorated cassava roots. Phytochemistry 2010, 71, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Sornyotha, S.; Kyu, K.L.; Ratanakhanokchai, K. Purification and detection of linamarin from cassava root cortex by high performance liquid chromatography. Food Chem. 2007, 104, 1750–1754. [Google Scholar] [CrossRef]
- Bredeson, J.V.; Lyons, J.B.; Prochnik, S.E.; Wu, G.A.; Ha, C.M.; Edsinger-Gonzales, E.; Grimwood, J.; Schmutz, J.; Rabbi, I.Y.; Egesi, C.; et al. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat. Biotechnol. 2016, 34, 562. [Google Scholar] [CrossRef] [Green Version]
- Zainuddin, I.M.; Fathoni, A.; Sudarmonowati, E.; Beeching, J.R.; Gruissem, W.; Vanderschuren, H. Cassava post-harvest physiological deterioration: From triggers to symptoms. Postharvest Biol. Technol. 2018, 142, 115–123. [Google Scholar] [CrossRef]
- EL-Sharkawy, M.A. Cassava biology and physiology. Plant Mol. Biol. 2003, 53, 621–641. [Google Scholar] [CrossRef]
- Bellotti, A.C.; Arias, B. Host plant resistance to whiteflies with emphasis on cassava as a case study. Crop Prot. 2001, 20, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Lima, A.C.S.; Rocha Viana, J.D.; Sousa Sabino, L.B.; Silva, L.M.R.; Silva, N.K.V.; Sousa, P.H.M. Processing of three different cooking methods of cassava: Effects on in vitro bioaccessibility of phenolic compounds and antioxidant activity. LWT-Food Sci. Technol. 2017, 76, 253–258. [Google Scholar] [CrossRef]
- Ngeve, J.M. Cassava root yields and culinary qualities as affected by harvest age and test environment. J. Sci. Food Agric. 2003, 83, 249–257. [Google Scholar] [CrossRef]
- Butarelo, S.S.; Beleia, A.; Fonseca, I.C.d.B.; Ito, K.C. Hidratação de tecidos de raízes de mandioca (Manihot esculenta Crantz.) e gelatinização do amido durante a cocção. Food Sci. Technol. 2004, 24, 311–315. [Google Scholar] [CrossRef] [Green Version]
- Filho, E.G.A.; Braga, L.N.; Silva, L.M.A.; Miranda, F.R.; Silva, E.O.; Canuto, K.M.; Miranda, M.R.; de Brito, E.S.; Zocolo, G.J. Physiological changes for drought resistance in different species of Phyllanthus. Sci. Rep. 2018, 8, 15141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S. NMR metabolomics: A look ahead. J. Magn. Reson. 2019, 306, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR Spectroscopy for Metabolomics Research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inkrod, C.; Raita, M.; Champreda, V.; Laosiripojana, N. Characteristics of Lignin Extracted from Different Lignocellulosic Materials via Organosolv Fractionation. BioEnergy Res. 2018, 11, 277–290. [Google Scholar] [CrossRef]
- do Prado Apparecido, R.; Barros Lopes, T.I.; Braz Alcantara, G. NMR-based foodomics of common tubers and roots. J. Pharm. Biomed. Anal. 2022, 209, 114527. [Google Scholar] [CrossRef]
- Spraul, M.; Schutz, B.; Humpfer, E.; Mortter, M.; Schafer, H.; Koswigb, S.; Rinke, P. Mixture Analysis by NMR as Applied to Fruit Juice Quality Control. Magn. Reson. Chem. 2009, 47, S130–S137. [Google Scholar] [CrossRef]
- Freitas, J.V.B.; Alves Filho, E.G.; Silva, L.M.A.; Zocolo, G.J.; de Brito, E.S.; Gramosa, N.V. Chemometric analysis of NMR and GC datasets for chemotype characterization of essential oils from different species of Ocimum. Talanta 2018, 180, 329–336. [Google Scholar] [CrossRef]
- Mansfield, S.D.; Kim, H.; Lu, F.; Ralph, J. Whole plant cell wall characterization using solution-state 2D NMR. Nat. Protoc. 2012, 7, 1579. [Google Scholar] [CrossRef]
- Pinheiro, F.G.C.; Soares, A.K.L.; Santaella, S.T.; e Silva, L.M.A.; Canuto, K.M.; Caceres, C.A.; de Freitas Rosa, M.; de Andrade Feitosa, J.P.; Leitao, R.C. Optimization of the acetosolv extraction of lignin from sugarcane bagasse for phenolic resin production. Ind. Crops Prod. 2017, 96, 80–90. [Google Scholar] [CrossRef]
- Bai, Y.-Y.; Xiao, L.-P.; Shi, Z.-J.; Sun, R.-C. Structural variation of bamboo lignin before and after ethanol organosolv pretreatment. Int. J. Mol. Sci. 2013, 14, 21394–21413. [Google Scholar] [CrossRef]
- Sousa, S.; Magalhães, A.; Ferreira, M.M.C. Optimized bucketing for NMR spectra: Three case studies. Chemom. Intell. Lab. Syst. 2013, 122, 93–102. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Lever, J.; Krzywinski, M.; Altman, N. Correction: Corrigendum: Classification evaluation. Nat. Methods 2016, 13, 890. [Google Scholar] [CrossRef] [Green Version]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res 2009, 37, W652–W660. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wu, J.e.; Li, Z.; Liu, Q.; Zhao, X.; Yang, H. Metabolomic analysis of energy regulated germination and sprouting of organic mung bean (Vigna radiata) using NMR spectroscopy. Food Chem. 2019, 286, 87–97. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, J.e.; Lim, Z.Y.; Aggarwal, A.; Yang, H.; Wang, S. Evaluation of the metabolic response of Escherichia coli to electrolysed water by 1H NMR spectroscopy. LWT-Food Sci. Technol. 2017, 79, 428–436. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, J.E.; Lim, Z.Y.; Lai, S.; Lee, N.; Yang, H. Metabolite profiling of Listeria innocua for unravelling the inactivation mechanism of electrolysed water by nuclear magnetic resonance spectroscopy. Int. J. Food Microbiol. 2018, 271, 24–32. [Google Scholar] [CrossRef]
- Holm, J.; Björck, I.; Drews, A.; Asp, N.-G. A Rapid Method for the Analysis of Starch. Starch-Stärke 1986, 38, 224–226. [Google Scholar] [CrossRef]
- Alves Filho, E.G.; Sartori, L.; Silva, L.; Silva, B.F.; Fadini, P.S.; Soong, R.; Simpson, A.; Ferreira, A.G. Non-targeted analyses of organic compounds in urban wastewater. Magn. Reson. Chem. 2015, 53, 704–710. [Google Scholar] [CrossRef]
- Alves Filho, E.G.; Sartori, L.; Silva, L.; Venâncio, T.; Carneiro, R.L.; Ferreira, A.G. 1H qNMR and Chemometric Analyses of Urban Wastewater. J. Braz. Chem. Soc. 2015, 26, 1257–1264. [Google Scholar] [CrossRef]
- Alves Filho, E.G.; Silva, L.M.; Teofilo, E.M.; Larsen, F.H.; de Brito, E.S. 1H NMR spectra dataset and solid-state NMR data of cowpea (Vigna unguiculata). Data Brief 2017, 11, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Balayssac, S.; Trefi, S.; Gilard, V.; Malet-Martino, M.; Martino, R.; Delsuc, M.-A. 2D and 3D DOSY 1H NMR, a useful tool for analysis of complex mixtures: Application to herbal drugs or dietary supplements for erectile dysfunction. J. Pharm. Biomed. Anal. 2009, 50, 602–612. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.L.; Cai, Y.; Davies, A.P.; Lewis, J. 1H and 13C NMR assignments of some green tea polyphenols. Magn. Reson. Chem. 1996, 34, 887–890. [Google Scholar] [CrossRef]
- Nord, L.I.; Vaag, P.; Duus, J.Ø. Quantification of organic and amino acids in beer by 1H NMR spectroscopy. Anal. Chem. 2004, 76, 4790–4798. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef]
- Ye, Y.; Yang, R.; Lou, Y.; Chen, J.; Yan, X.; Tang, H. Effects of food processing on the nutrient composition of Pyropia yezoensis products revealed by NMR-based metabolomic analysis. J. Food Nutr. Res. 2014, 2, 749–756. [Google Scholar] [CrossRef] [Green Version]
- Izquierdo-García, J.L.; Villa, P.; Kyriazis, A.; del Puerto-Nevado, L.; Pérez-Rial, S.; Rodriguez, I.; Hernandez, N.; Ruiz-Cabello, J. Descriptive review of current NMR-based metabolomic data analysis packages. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 59, 263–270. [Google Scholar] [CrossRef]
- Vega-Mas, I.; Cukier, C.; Coleto, I.; González-Murua, C.; Limami, A.M.; González-Moro, M.B.; Marino, D. Isotopic labelling reveals the efficient adaptation of wheat root TCA cycle flux modes to match carbon demand under ammonium nutrition. Sci. Rep. 2019, 9, 8925. [Google Scholar] [CrossRef] [Green Version]
- van der Merwe, M.J.; Osorio, S.; Araujo, W.L.; Balbo, I.; Nunes-Nesi, A.; Maximova, E.; Carrari, F.; Bunik, V.I.; Persson, S.; Fernie, A.R. Tricarboxylic Acid Cycle Activity Regulates Tomato Root Growth via Effects on Secondary Cell Wall Production. Plant Physiol. 2010, 153, 611–621. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Song, X.; Bartels, D. Enzymes and Metabolites in Carbohydrate Metabolism of Desiccation Tolerant Plants. Proteomes 2016, 4, 40. [Google Scholar] [CrossRef] [Green Version]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and Raffinose Constitute a Novel Function to Protect Plants from Oxidative Damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peshev, D.; Vergauwen, R.; Moglia, A.; Hideg, E.; Van den Ende, W. Towards understanding vacuolar antioxidant mechanisms: A role for fructans? J. Exp. Bot. 2013, 64, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, R.; Wolf, S. Phloem Transport: Cellular Pathways and Molecular Trafficking. Annu. Rev. Plant Biol. 2009, 60, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.S.; Liu, C.W.; Lin, Y.L.; Li, C.Y.; Wang, A.Z.; Chien, M.W.; Wang, C.S.; Lai, C.C. Mapping and comparative proteomic analysis of the starch biosynthetic pathway in rice by 2D PAGE/MS. Plant Mol. Biol. 2017, 95, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.J.C.B.; Souza, C.R.B.; Cascardo, J.C.M.; Junior, C.B.; Campos, L. Identification and characterization of a novel cassava (Manihot esculenta Crantz) clone with high free sugar content and novel starch. Plant Mol. Biol. 2004, 56, 643–659. [Google Scholar] [CrossRef] [PubMed]
- Denyer, K.A.Y.; Johnson, P.; Zeeman, S.; Smith, A.M. The control of amylose synthesis. J. Plant Physiol. 2001, 158, 479–487. [Google Scholar] [CrossRef]
- Raemakers, K.; Schreuder, M.; Suurs, L.; Furrer-Verhorst, H.; Vincken, J.-P.; de Vetten, N.; Jacobsen, E.; Visser, R.G.F. Improved Cassava Starch by Antisense Inhibition of Granule-bound Starch Synthase I. Mol. Breed. 2005, 16, 163–172. [Google Scholar] [CrossRef]
- Gutiérrez, A.; Rodríguez, I.M.; del Río, J.C. Chemical Characterization of Lignin and Lipid Fractions in Industrial Hemp Bast Fibers Used for Manufacturing High-Quality Paper Pulps. J. Agric. Food Chem. 2006, 54, 2138–2144. [Google Scholar] [CrossRef] [Green Version]
- Sette, M.; Wechselberger, R.; Crestini, C. Elucidation of Lignin Structure by Quantitative 2D NMR. Chem. Eur. J. 2011, 17, 9529–9535. [Google Scholar] [CrossRef]

| Cooking Characteristics | |||||||
|---|---|---|---|---|---|---|---|
| Model | 2 LV a (%) | Bias b | r2 cal c | RMSEC d | r2 val e | RMSECV f | RMSEC/RMSEV g |
| 9 months | 61.82 | −1.03 × 10−3 | 0.80 | 0.22 | 0.80 | 0.22 | 1 |
| 15 months | 92.54 | −4.29 × 10−3 | 0.92 | 0.14 | 0.91 | 0.15 | 0.93 |
| Cooking Time | |||||||
|---|---|---|---|---|---|---|---|
| Model | 5 LV a (%) | Bias b | r2 cal c | RMSEC d | r2 val e | RMSECV f | RMSEC/RMSEV g |
| 9 months | 88.35 | −1.4 × 10−14 | 0.91 | 3.31 | 0.86 | 4.00 | 0.83 |
| 15 months | 74.17 | −7.1 × 10−15 | 0.91 | 3.42 | 0.88 | 4.10 | 0.83 |
| Percentage of starch at fresh root | |||||||
| Model | 8 LV a (%) | Bias b | r2 cal c | RMSEC d | r2 val e | RMSECV f | RMSEC/RMSEV g |
| 9 months | 86.74 | −3.5 × 10−15 | 0.96 | 0.70 | 0.88 | 1.27 | 0.55 |
| 15 months | 84.95 | 0 | 0.98 | 0.57 | 0.96 | 0.92 | 0.62 |
| Starch at dried base | |||||||
| Model | 8 LV a (%) | Bias b | r2 cal c | RMSEC d | r2 val e | RMSECV f | RMSEC/RMSEV g |
| 9 months | 87.33 | −2.8 × 10−14 | 0.96 | 1.17 | 0.88 | 2.06 | 0.57 |
| 15 months | 82.80 | −2.8 × 10−14 | 0.98 | 1.20 | 0.94 | 1.92 | 0.62 |
| Genotype | S/G |
|---|---|
| Saracura | 0.6255 |
| BRS Dourada | 0.8385 |
| Eucalipto | 0.8655 |
| BRS Brasil | 0.79321 |
| 2009.0213 | 0.8395 |
| 2009.0216 | 0.8513 |
| 2009.0905 | 0.7965 |
| 2009.1220 | 0.8320 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, L.M.A.; Alves Filho, E.G.; Martins, R.M.; Oliveira, W.J.D.J.; Vidal, C.S.; de Oliveira, L.A.; de Brito, E.S. NMR-Based Metabolomic Approach for Evaluation of the Harvesting Time and Cooking Characteristics of Different Cassava Genotypes. Foods 2022, 11, 1651. https://doi.org/10.3390/foods11111651
Silva LMA, Alves Filho EG, Martins RM, Oliveira WJDJ, Vidal CS, de Oliveira LA, de Brito ES. NMR-Based Metabolomic Approach for Evaluation of the Harvesting Time and Cooking Characteristics of Different Cassava Genotypes. Foods. 2022; 11(11):1651. https://doi.org/10.3390/foods11111651
Chicago/Turabian StyleSilva, Lorena Mara A., Elenilson G. Alves Filho, Robson M. Martins, Willyane J. D. J. Oliveira, Cristine S. Vidal, Luciana A. de Oliveira, and Edy S. de Brito. 2022. "NMR-Based Metabolomic Approach for Evaluation of the Harvesting Time and Cooking Characteristics of Different Cassava Genotypes" Foods 11, no. 11: 1651. https://doi.org/10.3390/foods11111651
APA StyleSilva, L. M. A., Alves Filho, E. G., Martins, R. M., Oliveira, W. J. D. J., Vidal, C. S., de Oliveira, L. A., & de Brito, E. S. (2022). NMR-Based Metabolomic Approach for Evaluation of the Harvesting Time and Cooking Characteristics of Different Cassava Genotypes. Foods, 11(11), 1651. https://doi.org/10.3390/foods11111651









