Novel Latex Microsphere Immunochromatographic Assay for Rapid Detection of Cadmium Ion in Asparagus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
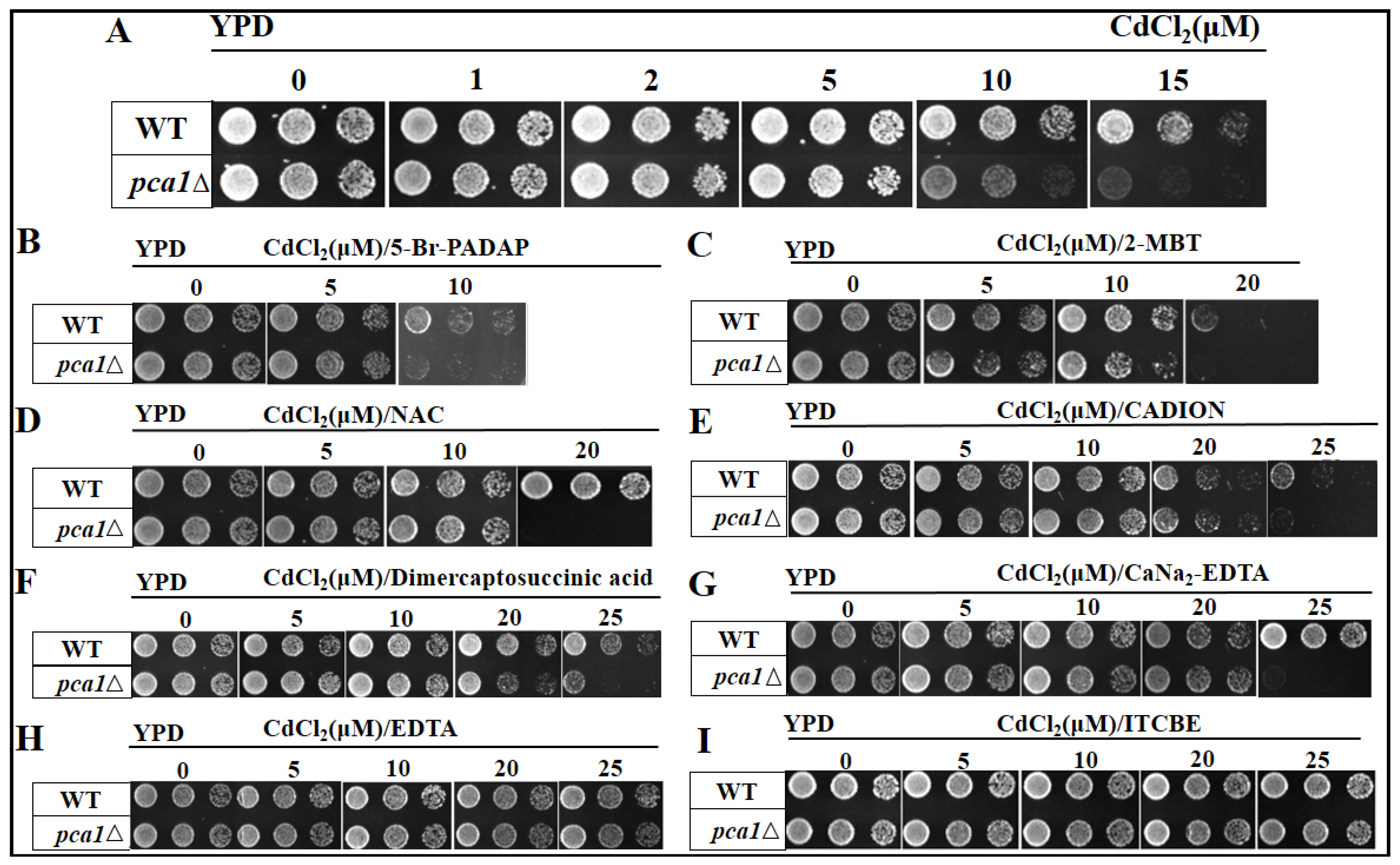
2.2. Screening of Chelators for Cd2+
2.3. Synthesis of Complete Antigen for Cd2+
2.3.1. Synthesis of Cd2+ Immunogen
Carrier Protein BSA and Method 1
BSA and Method 2
KLH and Method 1
KLH and Method 2
2.3.2. Synthesis of Coating Antigen
2.4. Preparation and Performance Evaluation of mAb
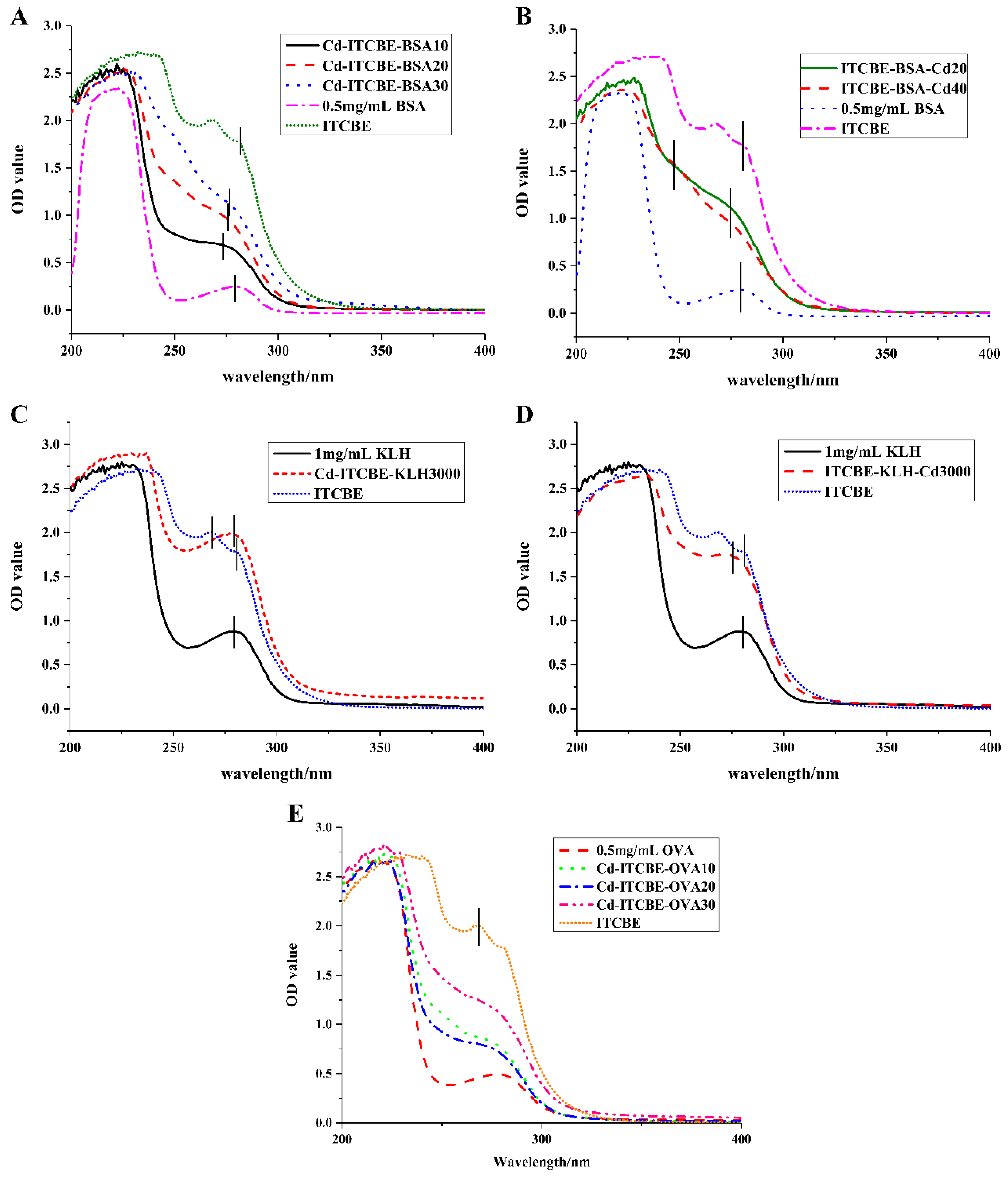
2.5. Identification of the Complete Antigen for Cd2+
2.6. Cross-Reactivity of mAb for Cd2+
2.7. Preparation of Latex Microsphere Immunochromatographic Test Strip for Cd2+
2.8. Sensitivity Evaluation of Test Strips
2.9. Specificity Evaluation of Test Strips
2.10. Sample Treatment and Determination
2.11. Statistical Analysis
3. Results and Discussion
3.1. Screening of Chelators for Cd2+
3.2. Synthesis and Identification of Complete Antigens for Cd2+
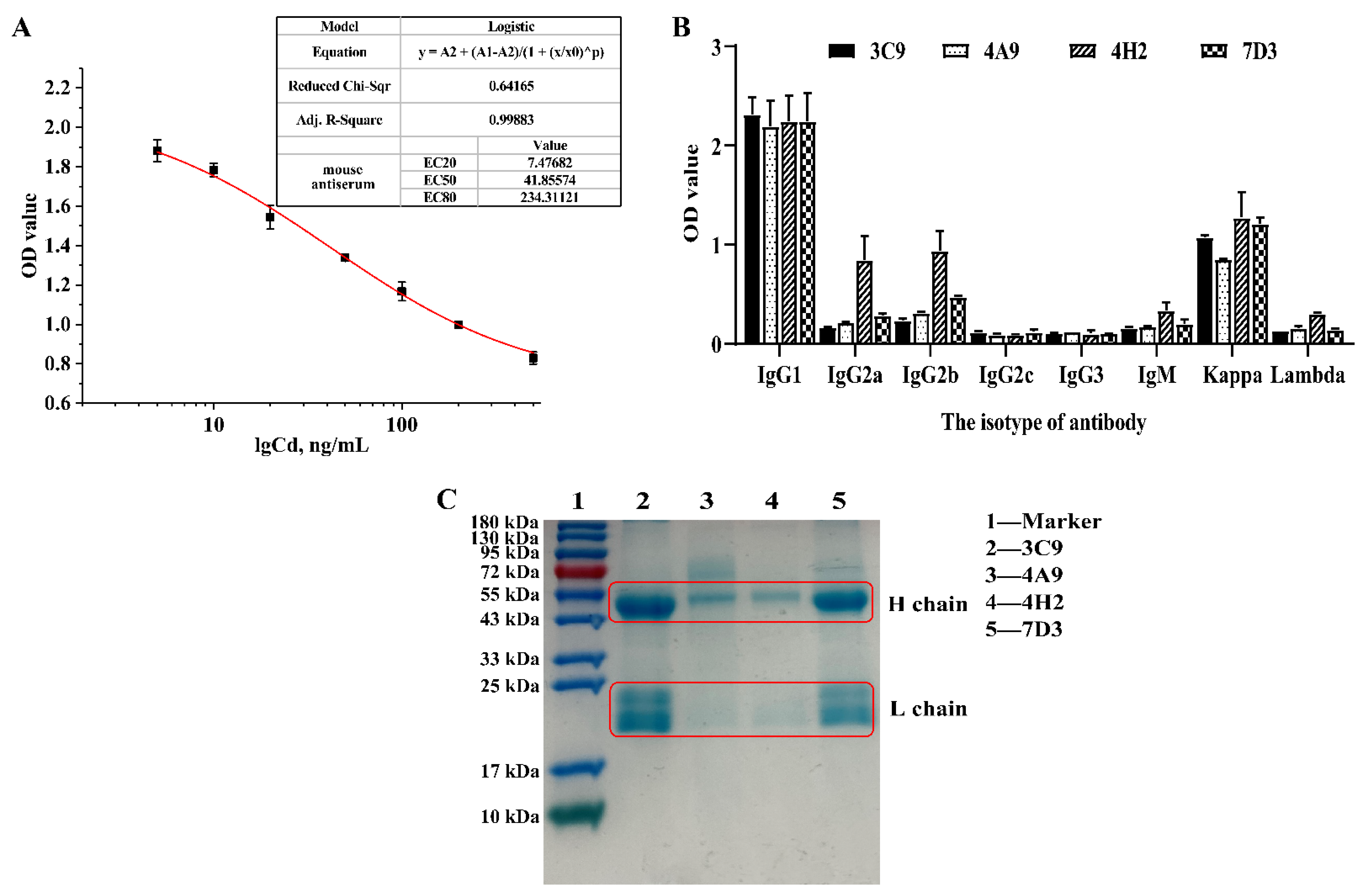
3.3. Animal Immunity and Screening of Positive Cells
3.4. Characterization of Specific mAbs against Cd2+
3.5. Preparation of Latex Microsphere Immunochromatographic Test Strips for Detection of Cd2+
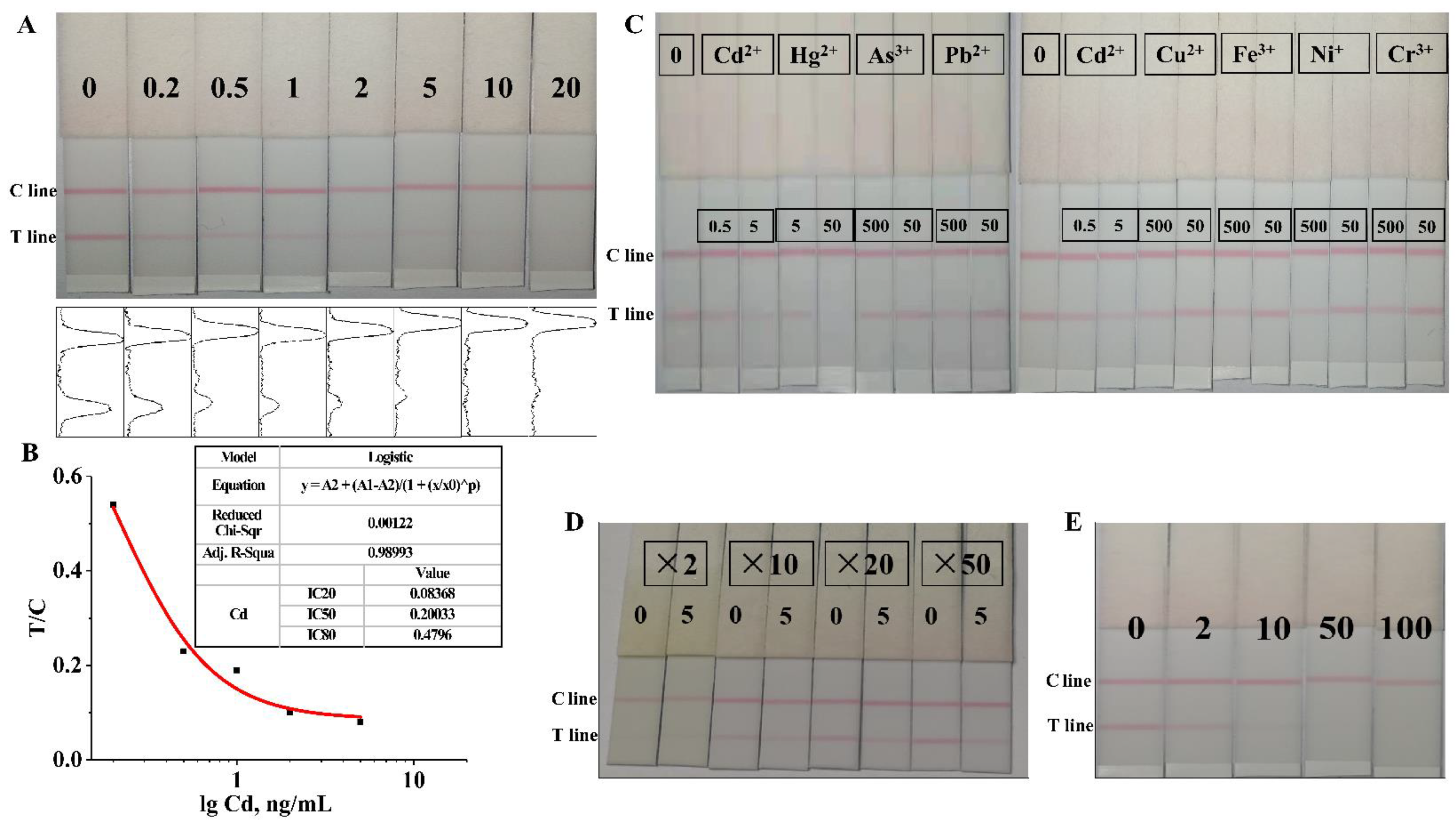
3.6. Sensitivity Evaluation of Latex Microsphere Immunochromatographic Test Strips
3.7. Specificity Evaluation of Latex Microsphere Immunochromatographic Test Strips
3.8. Detection of Cd2+ in Asparagus Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Zhou, Q.; Ren, B.; Luo, J.; Yuan, J.; Ding, X.; Bian, H.; Yao, X. Trends and health risks of dissolved heavy metal pollution in global river and lake water from 1970 to 2017. In Reviews of Environmental Contamination and Toxicology; de Voogt, P., Ed.; Springer International Publishing: Cham, Switzerland, 2020; Volume 251, pp. 1–24. [Google Scholar]
- Komarnicki, G.J. Lead and cadmium in indoor air and the urban environment. Environ. Pollut. 2005, 136, 47–61. [Google Scholar] [CrossRef] [PubMed]
- López Marzo, A.M.; Pons, J.; Blake, D.A.; Merkoçi, A. All-integrated and highly sensitive paper based device with sample treatment platform for Cd2+ immunodetection in drinking/tap waters. Anal. Chem. 2013, 85, 3532–3538. [Google Scholar] [CrossRef] [PubMed]
- Kjellström, T.; Nordberg, G.F. A kinetic model of cadmium metabolism in the human being. Environ. Res. 1978, 16, 248–269. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Y.; Fu, H.; Cui, Z.; Shi, L.; Wang, L.; Liu, Z. Health risk of heavy metals in food crops grown on reclaimed tidal flat soil in the Pearl River Estuary, China. J. Hazard. Mater. 2012, 227–228, 148–154. [Google Scholar] [CrossRef]
- Filipič, M. Mechanisms of cadmium induced genomic instability. Mutat. Res. 2012, 733, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Bertin, G.; Averbeck, D. Cadmium: Cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie 2006, 88, 1549–1559. [Google Scholar] [CrossRef]
- Templeton, D.M.; Liu, Y. Multiple roles of cadmium in cell death and survival. Chem. Biol. Interact. 2010, 188, 267–275. [Google Scholar] [CrossRef]
- Rizwan, M.; Ali, S.; Zia Ur Rehman, M.; Rinklebe, J.; Tsang, D.C.W.; Bashir, A.; Maqbool, A.; Tack, F.M.G.; Ok, Y.S. Cadmium phytoremediation potential of Brassica crop species: A review. Sci. Total Environ. 2018, 631–632, 1175–1191. [Google Scholar] [CrossRef]
- Nawrot, T.; Plusquin, M.; Hogervorst, J.; Roels, H.A.; Celis, H.; Thijs, L.; Vangronsveld, J.; Van Hecke, E.; Staessen, J.A. Environmental exposure to cadmium and risk of cancer: A prospective population-based study. Lancet Oncol. 2006, 7, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Arai, H.; Nakamura, K.; Yoshida, Y.; Tawarada, K. Development of quantitative immunochromatographic kit and visual screening immunoassay strip for cadmium. Anal. Bioanal. Chem. 2016, 408, 6063–6068. [Google Scholar] [CrossRef]
- Igweze, Z.N.; Ekhator, O.C.; Orisakwe, O.E. Lead and cadmium in infant milk and cereal based formulae marketed in Nigeria: A probabilistic non-carcinogenic human health risk assessment. Rocz. Panstw. Zakl. Hig. 2020, 71, 303–311. [Google Scholar]
- Meharg, A.A.; Norton, G.J.; Deacon, C.; Williams, P.; Islam, M.R. Variation in rice cadmium related to human exposure. Environ. Sci. Technol. 2013, 47, 5613–5618. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, A.; Shah, M.H. Disparities of selected metal levels in the blood and scalp hair of ischemia heart disease patients and healthy subjects. Biol. Trace. Elem. Res. 2017, 180, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Chen, L.; Hu, K.; Yang, S.; Wen, X. Non-aqueous phase cold vapor generation and determination of trace cadmium by atomic fluorescence spectrometry. Spectrochim. Acta A. Mol. Biomol. Spectrosc. 2018, 203, 522–527. [Google Scholar] [CrossRef]
- Smirnova, S.V.; Ilin, D.V.; Pletnev, I.V. Extraction and ICP-OES determination of heavy metals using tetrabutylammonium bromide aqueous biphasic system and oleophilic collector. Talanta 2021, 221, 121485. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.S.; Ogawa, S.; Imura, Y.; Suzuki, M.; Yoshimura, E. Post-column detection of cadmium chelators by high-performance liquid chromatography using 5, 10, 15, 20-tetraphenyl-21H, 23H-porphinetetrasulfonic acid. J. Chromatogr. B. Analyt. Technol. Biomed. Life. Sci. 2020, 1141, 122025. [Google Scholar] [CrossRef]
- Tefera, W.; Liu, T.; Lu, L.; Ge, J.; Webb, S.M.; Seifu, W.; Tian, S. Micro-XRF mapping and quantitative assessment of Cd in rice (Oryza sativa L.) roots. Ecotoxicol. Environ. Saf. 2020, 193, 110245. [Google Scholar] [CrossRef]
- Wang, W.; Kong, W.; Shen, T.; Man, Z.; Zhu, W.; He, Y.; Liu, F.; Liu, Y. Application of laser-induced breakdown spectroscopy in detection of cadmium content in rice stems. Front. Plant. Sci. 2020, 11, 599616. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Zhao, Q.; Iqbal, M.N.; Dong, M.; Li, X.; Lin, M.; Wang, R.; Lei, F.; He, C.; Wang, S. Development of immunoassay methods based on monoclonal antibody and its application in the determination of cadmium ion. J. Hazard. Mater. 2021, 411, 124992. [Google Scholar] [CrossRef]
- Abe, K.; Sakurai, Y.; Okuyama, A.; Sasaki, K.; Tawarada, K. Simplified method for determining cadmium concentrations in rice foliage and soil by using a biosensor kit with immunochromatography. J. Sci. Food Agric. 2009, 89, 1097–1100. [Google Scholar] [CrossRef]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten years of lateral flow immunoassay technique applications: Trends, challenges and future perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Xing, C.R.; Kuang, H.; Hao, C.L.; Liu, L.Q.; Wang, L.B.; Xu, C.L. A silver enhanced and sensitive strip sensor for Cadmium detection. Food. Agric. Immunol. 2014, 25, 287–300. [Google Scholar] [CrossRef]
- Xiao, M.; Fu, Q.Q.; Shen, H.C.; Chen, Y.; Xiao, W.; Yan, D.G.; Tang, X.J.; Zhong, Z.Y.; Tang, Y. A turn-on competitive immunochromatographic strips integrated with quantum dots and gold nano-stars for cadmium ion detection. Talanta 2018, 178, 644–649. [Google Scholar] [CrossRef]
- Fu, Q.; Liu, H.L.; Wu, Z.; Liu, A.; Yao, C.; Li, X.; Xiao, W.; Yu, S.; Luo, Z.; Tang, Y. Rough surface Au@Ag core-shell nanoparticles to fabricating high sensitivity SERS immunochromatographic sensors. J. Nanobiotechnol. 2015, 13, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Chen, X.; Liu, Z.; Wang, J.; Hua, Q.; Liang, J.; Shen, X.; Xu, Z.; Lei, H.; Sun, Y. Latex microsphere immunochromatography for quantitative detection of dexamethasone in milk and pork. Food. Chem. 2021, 345, 128607. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.; Li, Q.; Wang, Y.; Wang, Y.; Li, G.; Shi, J.; Ding, P.; Guo, J.; Deng, R.; et al. A strip test for the optical determination of influenza virus H3 subtype using gold nanoparticle coated polystyrene latex microspheres. Mikrochim. Acta 2020, 187, 306. [Google Scholar] [CrossRef]
- Yu, L.L.; Ding, J.Z.; Wen, L.Y.; Lou, D.; Yan, X.L.; Lin, L.J.; Lu, S.H.; Lin, D.D.; Zhou, X.N. Development of a rapid dipstick with latex immunochromatographic assay (DLIA) for diagnosis of schistosomiasis japonica. Parasites Vectors 2011, 4, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.W.; Xu, Y.; Liu, S.J.; He, J.F.; Long, F.Y. Concentration and potential health risk of heavy metals in market vegetables in Chongqing, China. Ecotoxicol. Environ. Saf. 2011, 74, 1664–1669. [Google Scholar] [CrossRef]
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999, 285, 901–906. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, E.; Inouhe, M.; Joho, M.; Tohoyama, H. The cadmium-resistant gene, CAD2, which is a mutated putative copper-transporter gene (PCA1), controls the intracellular cadmium-level in the yeast S. cerevisiae. Curr. Genet. 2000, 37, 79–86. [Google Scholar] [CrossRef]
- Adle, D.J.; Sinani, D.; Kim, H.; Lee, J. A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 2007, 282, 947–955. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.J.; Zhai, Y.F.; Tang, Y.; Wang, H.; Liu, B.; Guo, C.W. A competitive indirect enzyme-linked immunoassay for lead ion measurement using mAbs against the lead-DTPA complex. Environ. Pollut. 2010, 158, 1376–1380. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, H.; Pschenitza, M.; Niessner, R.; Li, Y.; Knopp, D.; Deng, A. Highly sensitive and specific determination of mercury(II) ion in water, food and cosmetic samples with an ELISA based on a novel monoclonal antibody. Anal. Bioanal. Chem. 2012, 403, 2519–2528. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Kim, Y.S.; Kumar, V. Heavy metal toxicity: An update of chelating therapeutic strategies. J. Trace Elem. Med. Biol. 2019, 54, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Kong, T.; Hao, X.Q.; Li, X.B.; Liu, G.W.; Zhang, Z.G.; Yang, Z.J.; Wang, Z.; Tang, J.; Yang, W.; Sun, J. Preparation of novel monoclonal antibodies against chelated cadmium ions. Biol. Trace Elem. Res. 2013, 152, 117–124. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, L.; Lou, Y.; Yu, H.; Li, X.; Blake, D.A.; Liu, F. Preparation of specific monoclonal antibodies (MAbs) against heavy metals: MAbs that recognize chelated cadmium ions. J. Agric. Food. Chem. 2007, 55, 7648–7653. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, V.H.; Ullah, I.; Dunwell, J.M.; Tibbett, M. Bioremediation potential of Cd by transgenic yeast expressing a metallothionein gene from Populus trichocarpa. Ecotoxicol. Environ. Saf. 2020, 202, 110917. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Otani, M.; Uraguchi, S.; Akihiro, T.; Fujiwara, T. Rice ABCG43 is Cd inducible and confers Cd tolerance on yeast. Biosci. Biotechnol. Biochem. 2011, 75, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Chen, Q.A.; Zhang, Y.; Wang, R.; Jin, N.; Pang, J.; Wang, S. Development of ELISA and colloidal gold immunoassay for tetrodotoxin detetcion based on monoclonal antibody. Biosens. Bioelectron. 2015, 71, 256–260. [Google Scholar] [CrossRef]
- Loomans, E.E.; Roelen, A.J.; Van Damme, H.S.; Bloemers, H.P.; Gribnau, T.C.; Schielen, W.J. Assessment of the functional affinity constant of monoclonal antibodies using an improved enzyme-linked immunosorbent assay. J. Immunol. Methods. 1995, 184, 207–217. [Google Scholar] [CrossRef]
- Blake, D.A.; Chakrabarti, P.; Khosraviani, M.; Hatcher, F.M.; Westhoff, C.M.; Goebel, P.; Wylie, D.E.; Blake, R.C. Metal binding properties of a monoclonal antibody directed toward metal-chelate complexes. J. Biol. Chem. 1996, 271, 27677–27685. [Google Scholar] [CrossRef] [Green Version]
- Love, R.A.; Villafranca, J.E.; Aust, R.M.; Nakamura, K.K.; Jue, R.A.; Major, J.G., Jr.; Radhakrishnan, R.; Butler, W.F. How the anti-(metal chelate) antibody CHA255 is specific for the metal ion of its antigen: X-ray structures for two Fab’/hapten complexes with different metals in the chelate. Biochemistry 1993, 32, 10950–10959. [Google Scholar] [CrossRef]
- Jones, R.M.; Yu, H.; Delehanty, J.B.; Blake, D.A. Monoclonal antibodies that recognize minimal differences in the three-dimensional structures of metal-chelate complexes. Bioconjug. Chem. 2002, 13, 408–415. [Google Scholar] [CrossRef]
- Pento, J.T. Monoclonal antibodies for the treatment of cancer. Anticancer Res. 2017, 37, 5935–5939. [Google Scholar]
- Rudenko, N.; Fursova, K.; Shepelyakovskaya, A.; Karatovskaya, A.; Brovko, F. Antibodies as Biosensors’ Key Components: State-of-the-Art in Russia 2020–2021. Sensors 2021, 21, 7614. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, L.; Liu, L.; Kuang, H.; Xu, C. Colloidal gold-based immunochromatographic strip assay for the rapid detection of three natural estrogens in milk. Food Chem. 2018, 259, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Z.; Zong, S.; Huang, Z.; Zhang, P.; Cui, Y. A SERS-based immunoassay with highly increased sensitivity using gold/silver core-shell nanorods. Biosens. Bioelectron. 2012, 38, 94–99. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Tawarada, K.; Okuhata, H.; Miyasaka, H.; Okuyama, A.; Maruyama, Y.; Ohmura, N. Development of MAb-based immunochromatographic assay for cadmium from biological samples. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 2006, 1842–1845. [Google Scholar] [PubMed]
- Xu, L.; Xing, C.; Liu, L.; Song, S.; Kuang, H.; Xu, C. Quick, easy, cheap, effective, rugged and safe strategy for quantifying cadmium polluted rice. Food Agric. Immunol. 2016, 27, 783–795. [Google Scholar] [CrossRef]
- Liu, G.L.; Wang, J.F.; Li, Z.Y.; Liang, S.Z.; Wang, X.N. Immunoassay for cadmium detection and quantification. Biomed. Environ. Sci. 2009, 22, 188–193. [Google Scholar] [CrossRef]
- He, H.; Tang, B.; Sun, C.; Yang, S.G.; Zheng, W.J.; Huang, Z.C. Preparation of hapten-specific monoclonal antibody for cadmium and its ELISA application to aqueous samples. Front. Environ. Sci. Eng. China 2011, 5, 409–416. [Google Scholar] [CrossRef]
- Gao, W.; Nan, T.G.; Tan, G.Y.; Zhao, H.W.; Wang, B.M.; Li, Q.X.; Meng, F.Y. Development of a sensitive monoclonal antibody-based enzyme-linked immunosorbent assay for the analysis of cadmium ions in water, soil and rape samples. Food Agric. Immunol. 2012, 23, 27–39. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.M.; Wu, X.L.; Liu, L.Q.; Zhu, J.P.; Xu, L.G.; Kuang, H. Development of a fluorescent immunoassay strip for the rapid quantitative detection of cadmium in rice. Food Agric. Immunol. 2020, 31, 501–512. [Google Scholar] [CrossRef] [Green Version]


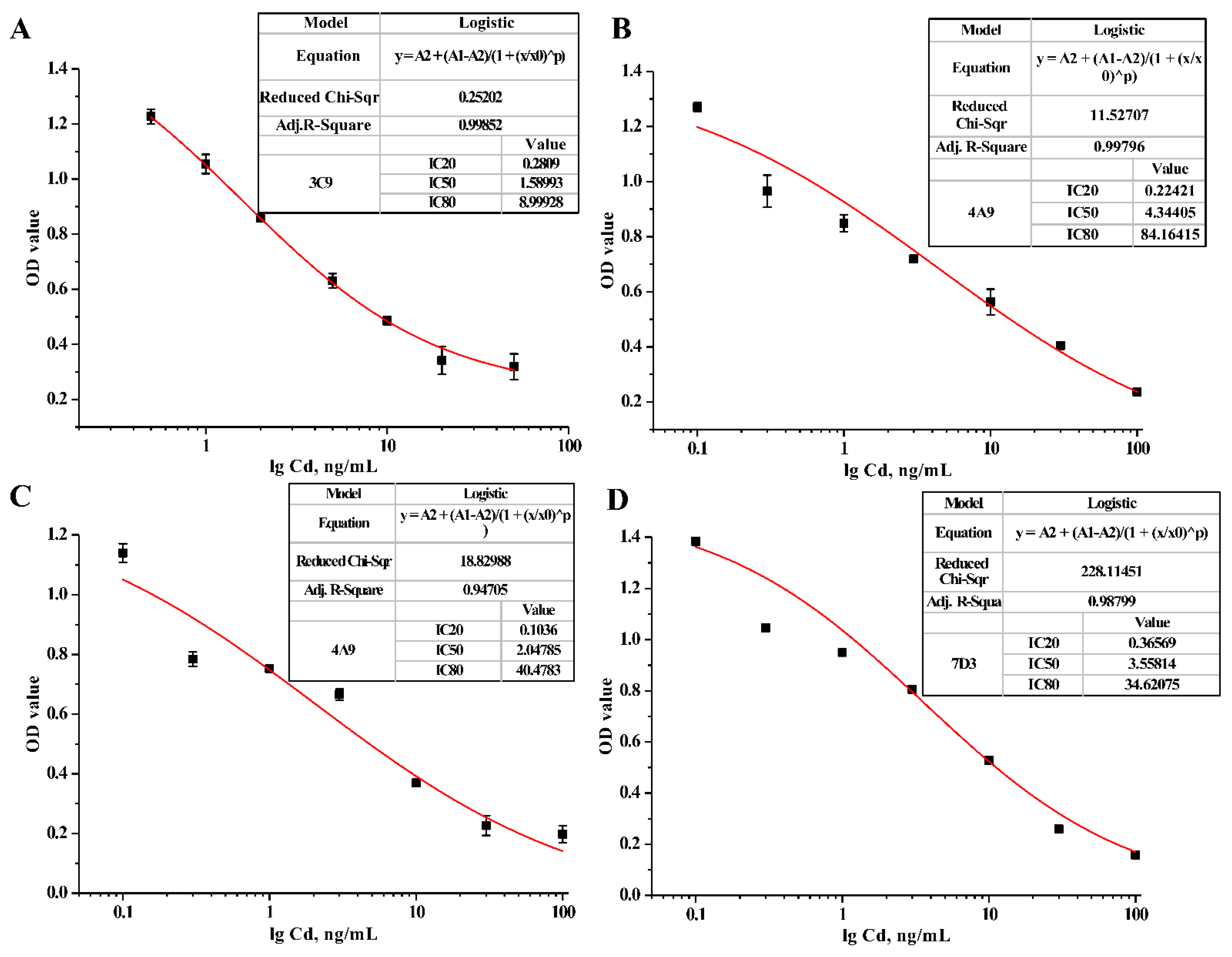

| Cell Strain | IC50 (ng/mL) | LOD (ng/mL) | Linear Range (ng/mL) |
|---|---|---|---|
| 3C9 | 1.59 | 0.13 | 0.28–9.00 |
| 4A9 | 4.34 | 0.04 | 0.22–84.16 |
| 4H2 | 2.05 | 0.02 | 0.10–40.48 |
| 7D3 | 3.56 | 0.10 | 0.37–34.62 |
| mAb Number | 3C9 | 4A9 | 4H2 | 7D3 | ||||
|---|---|---|---|---|---|---|---|---|
| EDTA-Meta Ions | IC50, ng/mL | CR% | IC50, ng/mL | CR% | IC50, ng/mL | CR% | IC50, ng/mL | CR% |
| EDTA-Cd | 1.61 ± 0.01 | 100 | 4.32 ± 0.03 | 100 | 2.01 ± 0.02 | 100 | 3.62 ± 0.03 | 100 |
| EDTA-Hg | >10 | <16% | >50 | <8% | >50 | <4% | >10 | <36% |
| EDTA-Pb | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| EDTA-As | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| EDTA-Cu | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| EDTA-Fe | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| EDTA-Ni | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| EDTA-Cr | >500 | <0.3% | >500 | <0.8% | >500 | <0.4% | >500 | <0.7% |
| Methods | Linear Detection Range (ng/mL) | IC50 (ng/mL) | Detection Limit (ng/mL) | Detection Time | Reference | |
|---|---|---|---|---|---|---|
| vLOD | LOQ | |||||
| ic-ELISA | - | 45.6 | - | 1.953 | >2 h | [51] |
| ic-ELISA | 0.1–1000 | - | - | 0.1 | >2 h | [52] |
| ic-ELISA | 0.2–40 | 2.59 | - | 0.08 | >3 h | [53] |
| Silver-enhanced ICA strip | 0.5–5 | - | 5 | 0.35 | >30 min | [23] |
| Competitive ICA strip | 0.25–8 | - | - | 0.18 | 1 h | [24] |
| Fuorescent immunoassay strip | 3.8–48.9 | - | - | 1.93 | 12 min | [54] |
| LMIA strip | 0.08–0.48 | 0.20 | 2.00 | 0.05 | 3–5 min | this work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, N.; Zhu, Q.; Zhu, J.; Jia, J.; Wei, X.; Wang, Y. Novel Latex Microsphere Immunochromatographic Assay for Rapid Detection of Cadmium Ion in Asparagus. Foods 2022, 11, 78. https://doi.org/10.3390/foods11010078
Xu N, Zhu Q, Zhu J, Jia J, Wei X, Wang Y. Novel Latex Microsphere Immunochromatographic Assay for Rapid Detection of Cadmium Ion in Asparagus. Foods. 2022; 11(1):78. https://doi.org/10.3390/foods11010078
Chicago/Turabian StyleXu, Naifeng, Qiaojuan Zhu, Jiangxiong Zhu, Jingze Jia, Xinlin Wei, and Yuanfeng Wang. 2022. "Novel Latex Microsphere Immunochromatographic Assay for Rapid Detection of Cadmium Ion in Asparagus" Foods 11, no. 1: 78. https://doi.org/10.3390/foods11010078
APA StyleXu, N., Zhu, Q., Zhu, J., Jia, J., Wei, X., & Wang, Y. (2022). Novel Latex Microsphere Immunochromatographic Assay for Rapid Detection of Cadmium Ion in Asparagus. Foods, 11(1), 78. https://doi.org/10.3390/foods11010078







