Enzymatic Production of Biologically Active 3-Methoxycinnamoylated Lysophosphatidylcholine via Regioselctive Lipase-Catalyzed Acidolysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Acidolysis Reaction of Egg-Yolk Phosphatidylcholine with 3-Methoxycinnamic Acid
2.3. Design of Experiment
2.4. Acidolysis of Egg-Yolk Phosphatidylcholine with 3-methoxycinnamic Acid Catalyzed by Novozym 435 in an Increased Scale
2.5. Methods of Analysis
2.5.1. Thin-Layer Chromatography (TLC)
2.5.2. Gas Chromatography (GC)
2.5.3. High-Performance Liquid Chromatography (HPLC)
3. Results and Discussion
3.1. Screening of Commercial Immobilized Lipases
3.2. Effect of Organic Solvents on Enzyme Activity
3.3. Model Fitting
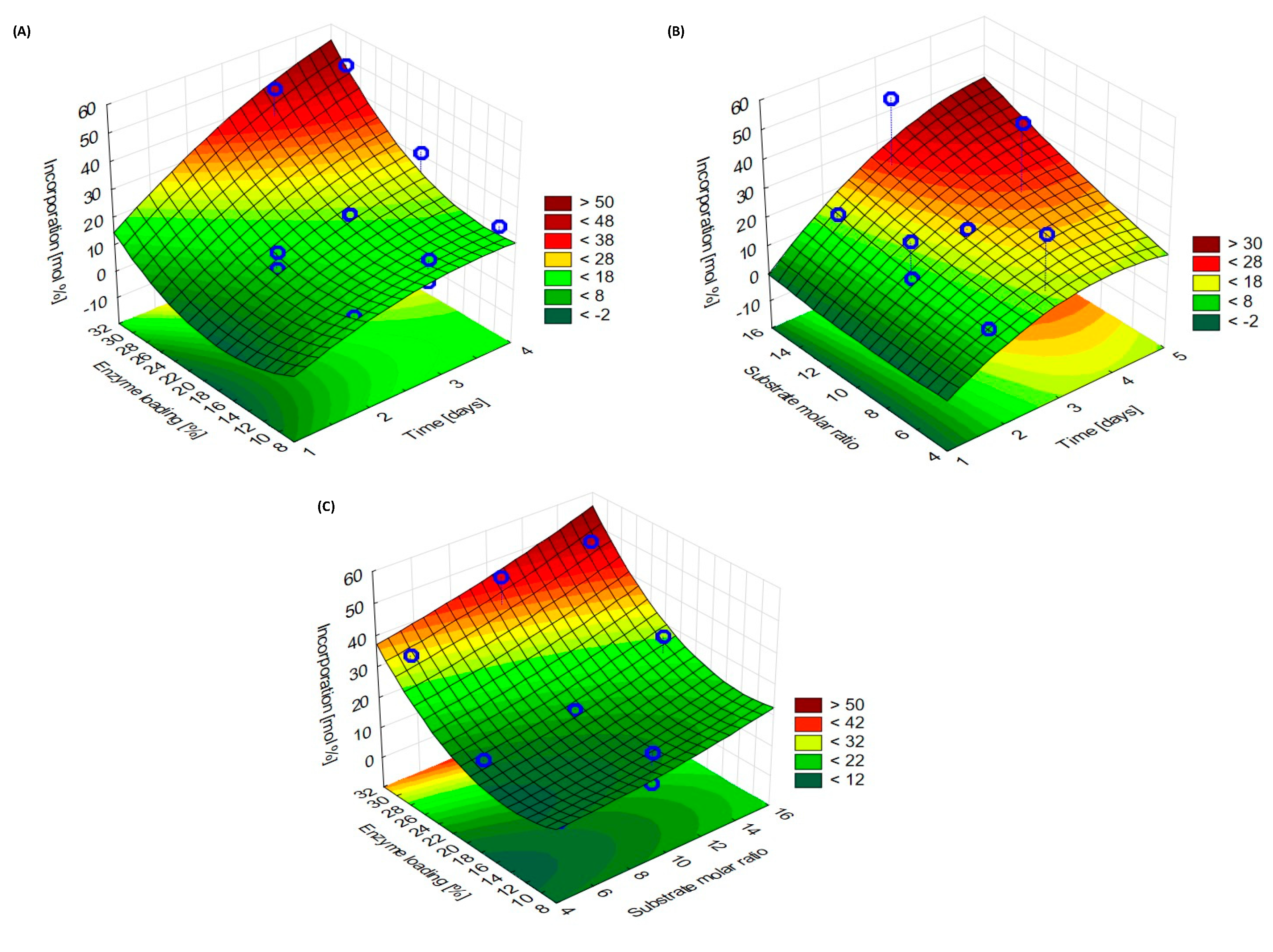
3.4. Response Surface Metodology Analysis and Interpretation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ang, X.; Chen, H.; Xiang, J.-Q.; Wei, F.; Quek, S.Y. Preparation and functionality of lipase-catalysed structured phospholipid—A review. Trends Food Sci. Technol. 2019, 88, 373–383. [Google Scholar] [CrossRef]
- Kim, B.H.; Akoh, C.C. Recent Research Trends on the Enzymatic Synthesis of Structured Lipids. J. Food Sci. 2015, 80, C1713–C1724. [Google Scholar] [CrossRef] [PubMed]
- Mnasri, T.; Herault, J.; Gauvry, L.; Loiseau, C.; Poisson, L.; Ergan, F.; Pencreac’h, G. Lipase-catalyzed production of lysophospholipids. OCL 2017, 24, D405. [Google Scholar] [CrossRef][Green Version]
- Villeneuve, P. Lipases in lipophilization reactions. Biotechnol. Adv. 2007, 25, 515–536. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Dhar, K.; Kanwar, S.S.; Arora, P.K. Lipase catalysis in organic solvents: Advantages and applications. Biol. Proced. Online 2016, 18, 2. [Google Scholar] [CrossRef]
- Ortiz, C.; Ferreira, M.L.; Barbosa, O.; dos Santos, J.C.S.; Rodrigues, R.C.; Berenguer-Murcia, A.; Briand, L.E.; Fernandez-Lafuente, R. Novozymes 435: The “perfect” lipase immobilized biocatalyst? Catal. Sci. Technol. 2019, 9, 2380–2420. [Google Scholar] [CrossRef]
- Baddo, A.; Serban, S. Industrial application of immobilized enzymes—A review. Mol. Catal. 2019, 479, 110607. [Google Scholar]
- Jaeger, K.E.; Dijkstra, B.W.; Reetz, M.T. Bacterial Biocatalysts: Molecular Biology, three-dimensional structures, and biotechnological applications of lipases. Ann. Rev. Microbiol. 1999, 53, 315–351. [Google Scholar] [CrossRef]
- Zarevúcka, M.; Zdenék, W. Plant products for pharmacology: Application of enzymes in their transformations. Int. J. Mol. Sci. 2008, 9, 2447–2473. [Google Scholar] [CrossRef] [PubMed]
- Drzazga, A.; Okulus, M.; Rychlicka, M.; Biegała, A.; Gliszczyńska, A.; Gendaszewska-Darmach, E. Lysophosphatidylcholine Containing Anisic Acid Is Able to Stimulate Insulin Secretion Targeting G Protein Coupled Receptors. Nutrients 2020, 12, 1173. [Google Scholar] [CrossRef]
- Małodobra-Mazur, M.; Lewoń, D.; Cierzniak, A.; Okulus, M.; Gliszczyńska, A. Phospholipid derivatives of cinnamic acid restore insulin sensitivity in insulin resistance in 3T3-L1 adipocytes. Nutrients 2021, 13, 3619. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, M.; Świtalska, M.; Wietrzyk, J.; Maciejewska, G.; Gliszczyńska, A. Synthesis, Characterization, and In Vitro Cancer Cell Growth Inhibition Evaluation of Novel Phosphatidylcholines with Anisic and Veratric Acids. Molecules 2018, 23, 2022. [Google Scholar] [CrossRef]
- Czarnecka, M.; Świtalska, M.; Wietrzyk, J.; Maciejewska, G.; Gliszczyńska, A. Synthesis and biological evaluation of phosphatidylcholines with cinnamic and 3-methoxycinnamic acids with potent antiproliferative activity. RSC Adv. 2018, 8, 35744–35752. [Google Scholar] [CrossRef]
- Palko-Łabuz, A.; Gliszczyńska, A.; Skonieczna, M.; Poła, A.; Wesołowska, O.; Środa-Pomianek, K. Conjugation with phospholipids as a modification increasing anticancer activity of phenolic acids in metastatic melanoma—In vitro and in silico studies. Int. J. Mol. Sci. 2021, 22, 8397. [Google Scholar] [CrossRef] [PubMed]
- Balakrishna, M.; Kaki, S.S.; Karuna, M.S.L.; Sarada, S.; Kumar, C.G.; Prasad, R.B.N. Synthesis and in vitro antioxidant and antimicrobial studies of novel structured phosphatidylcholines with phenolic acids. Food Chem. 2016, 221, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Anakanbil, S.; Perez, B.; Banerjee, C.; Guo, Z. New phenophospholipids equipped with multi-functionalities: Regiospecific synthesis and characterization. J. Colloid Interface Sci. 2018, 523, 169–178. [Google Scholar] [CrossRef]
- Yang, H.; Mu, Y.; Chen, H.; Xiu, Z.; Yang, T. Enzymatic synthesis of feruloylated lysophospholipid in a selected organic solvent medium. Food Chem. 2013, 141, 3317–3322. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Espinoza, M.-C.; Villeneuve, P. Phenolic acids enzymatic lipophilization. J. Agric. Food Chem. 2005, 53, 2779–2787. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, L.; wang, J.; Hu, Y.; Chen, S. Enzymatic Synthesis of Functional Structured Lipids from Glycerol and Naturally Phenolic Antioxidants. In Glycerine Production and Transformation—An Innovative Platform for Sustainable Biorefinery and Energy; Frediani, M., Bartoli, M., Rosi, L., Eds.; IntechOpen: London, UK, 2019; pp. 1–22. [Google Scholar]
- Rychlicka, M.; Niezgoda, N.; Gliszczyńska, A. Lipase-Catalyzed Acidolysis of Egg-Yolk Phosphatidylcholine with Citronellic Acid. New Insight into Synthesis of Isoprenoid-Phospholipids. Molecules 2018, 23, 314. [Google Scholar] [CrossRef] [PubMed]
- Rychlicka, M.; Maciejewska, G.; Niezgoda, N.; Gliszczyńska, A. Production of feruloylated lysophospholipids via a one-step enzymatic interesterification. Food Chem. 2020, 323, 126802. [Google Scholar] [CrossRef]
- Rychlicka, M.; Niezgoda, N.; Gliszczyńska, A. Development and Optimization of Lipase-Catalyzed Synthesis of Phospholipids Containing 3,4-Dimethoxycinnamic Acid by Response Surface Methodology. Catalysts 2020, 10, 588. [Google Scholar] [CrossRef]
- Laane, C.; Boeren, S.; Vos, K.; Veeger, C. Rules for optimization of biocatalysis in organic solvents. Biotechnol. Bioeng. 1987, 30, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Rychlicka, M.; Gliszczyńska, A. Interesterification of Egg-Yolk Phosphatidylcholine with p-Methoxycinnamic Acid Catalyzed by Immobilized Lipase B from Candida Antarctica. Catalysts 2020, 10, 1181. [Google Scholar] [CrossRef]
- Okulus, M.; Gliszczyńska, A. Enzymatic synthesis of O-methylated phenophospholipids by lipase-catalyzed acidolysis of egg-yolk phosphatidylcholine with anisic and veratric acids. Catalysts 2020, 10, 538. [Google Scholar] [CrossRef]
- Sun, S.; Zhu, S.; Bi, Y. Solvent-free enzymatic synthesis of feruloylated structured lipids by the transesterification of ethyl ferulate with castor oil. Food Chem. 2014, 158, 292–295. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Vikbjerg, A.F.; Xu, X. Enzymatic modification of phospholipids for functional applications and human nutrition. Biotechnol. Adv. 2005, 23, 203–259. [Google Scholar] [CrossRef]
- Virto, C.; Adlercreutz, P. Lysophosphatidylcholine synthesis with Candida antarctica lipase B (Novozym 435). Enzym. Microb. Technol. 2000, 26, 630–635. [Google Scholar] [CrossRef]
- Katsoura, M.H.; Polydera, A.C.; Tsironis, L.D.; Petraki, M.P.; Rajacic, S.K.; Tselepis, A.D.; Stamatis, H. Efficient enzymatic preparation of hydroxycinnamates in ionic liquids enhances their antioxidant effect on lipoproteins oxidative modification. New Biotechnol. 2009, 26, 83–91. [Google Scholar] [CrossRef]
- Choo, W.-S.; Birch, E.J.; Stewart, I. Radical scavenging activity of Lipophilized products from transesterification of flaxseed oil with cinnamic acid or ferulic acid. Lipids 2009, 44, 807–815. [Google Scholar] [CrossRef]
- Guyot, B.; Bosquette, B.; Pina, M.; Graille, J. Esterification of phenolic acids from green coffee with an immobilized lipase from Candida antarctica in solvent-free medium. Biotechnol. Lett. 1997, 19, 529–532. [Google Scholar] [CrossRef]
- Safari, M.; Karboune, S.; St-Louis, R.; Kermasha, S. Enzymatic synthesis of structured phenolic lipids by incorporation of selected phenolic acids into triolein. Biocatal. Biotransform. 2006, 24, 272–279. [Google Scholar] [CrossRef]
- Karboune, S.; St-Louis, R.; Kermasha, S. Enzymatic synthesis of structured phenolic lipids by acidolysis of flaxseed oil with selected phenolic acids. J. Mol. Catal. B Enzym. 2008, 52–53, 96–105. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipases in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef]
- Fu, B.; Vasudevan, P.T. Effect of Solvent−Co-solvent Mixtures on Lipase-Catalyzed Transesterification of Canola Oil. Energy Fuels 2010, 24, 4646–4651. [Google Scholar] [CrossRef]
- Ferreira, S.L.C.; Bruns, R.E.; Ferreira, H.S.; Matos, G.D.; David, J.M.; Brand, G.C.; Silva, E.G.P.; Reis, P.S.; Souza, A.S.; Santos, W.N.L. Box-Behnken design: An alternative for the optimization of analytical methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef]
- Amin, N.A.S.; Anggoro, D.D. Optimization of direct conversion of methane to liquid fuels over Cu loaded W/ZSM-5 catalyst. Fuel 2004, 83, 487–494. [Google Scholar] [CrossRef]
- Soo, E.L.; Salleh, A.B.; Basri, M.; Rahman, R.N.Z.A.; Kamaruddin, K. Response surface methodological study on lipase-catalyzed synthesis of amino acid surfactants. Process. Biochem. 2004, 39, 1511–1518. [Google Scholar] [CrossRef]
- Li, L.; Du, W.; Liu, D.; Wang, L.; Li, Z. Lipase-catalyzed transesterification of rapeseed oils from biodiesel production with a novel organic solvent as the reaction medium. J. Mol. Catal. B Enzym 2006, 43, 58–62. [Google Scholar] [CrossRef]
- Yahya, A.R.M.; Anderson, W.A.; Moo-Young, M. Ester synthesis in lipase-catalyzed reactions. Enzym. Microb. Technol. 1998, 23, 438–450. [Google Scholar] [CrossRef]
- Shieh, C.J.; Akoh, C.C.; Koehler, P.E. Four-factor response surface optimization of the enzymatic modification of triolein to structured lipids. J. Am. Oil Chem. Soc. 1995, 72, 619–623. [Google Scholar] [CrossRef]
- Harikrisna, S.; Sattur, A.P.; Karant, N.G. Lipase-catalyzed synthesis of isoamyl isobutyrate optimization using central composite rotatable design. Process. Biochem. 2001, 37, 9–16. [Google Scholar] [CrossRef]
- Manohar, B.; Divakar, S. Application of surface plots and statistical designs to selected lipase catalysed esterification reactions. Process. Biochem. 2004, 39, 847–853. [Google Scholar] [CrossRef]
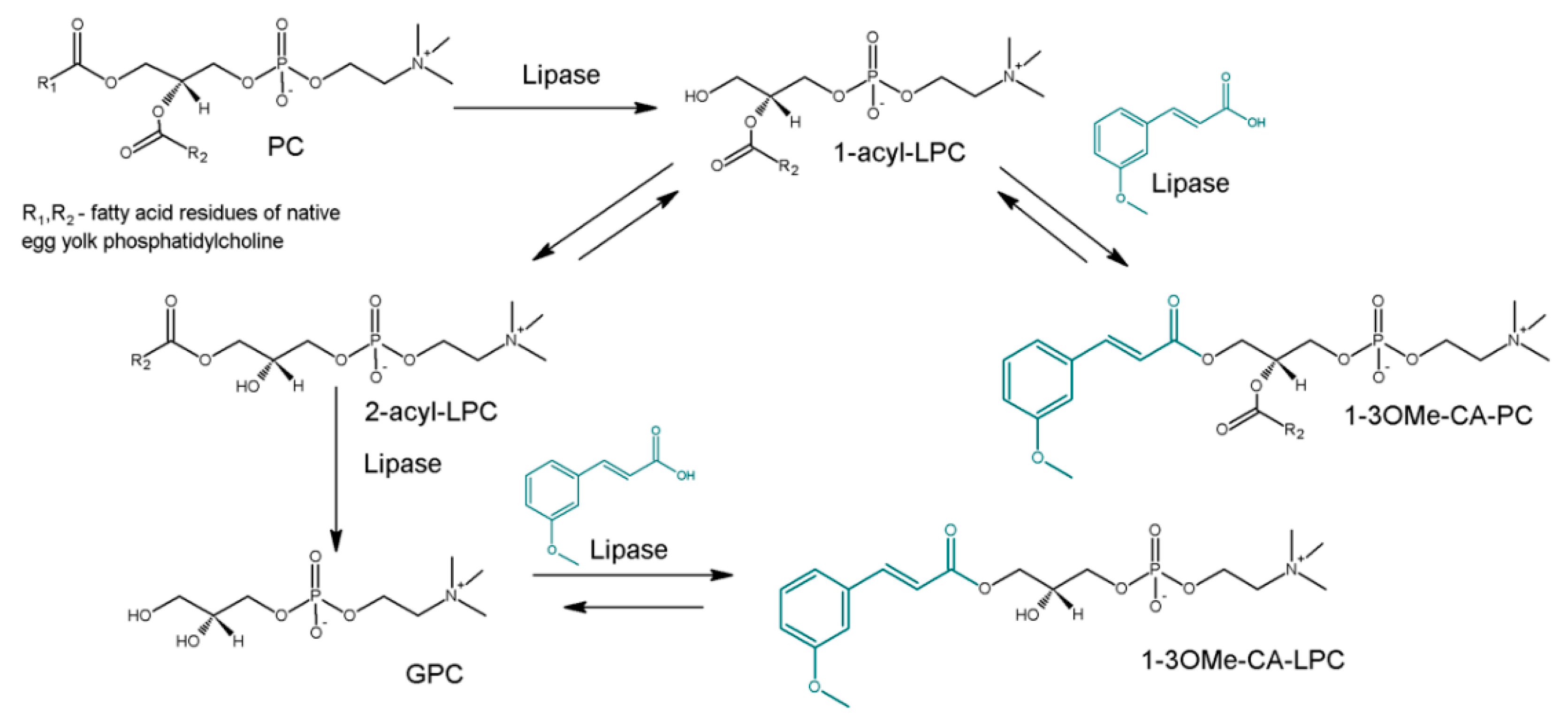
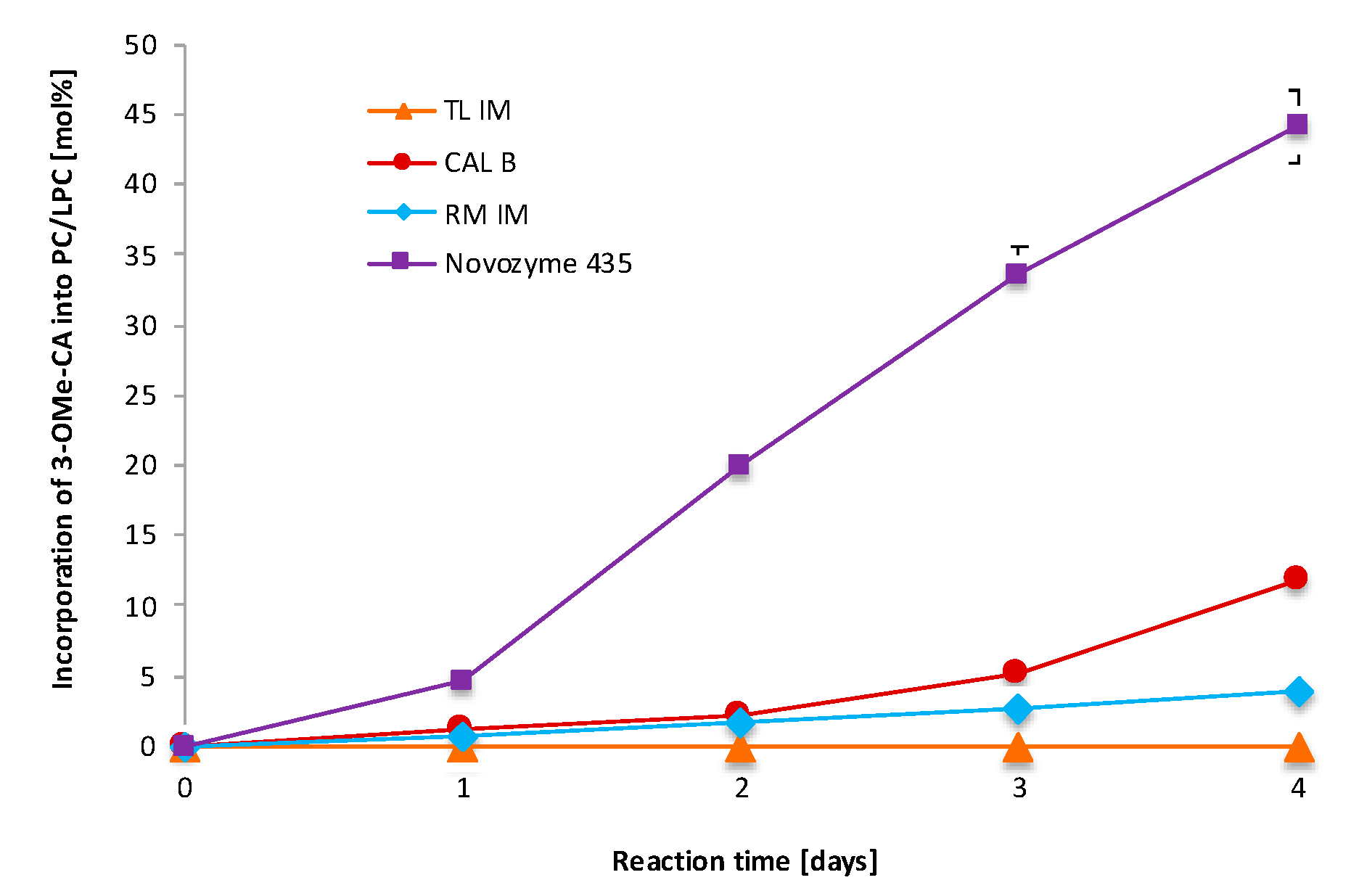
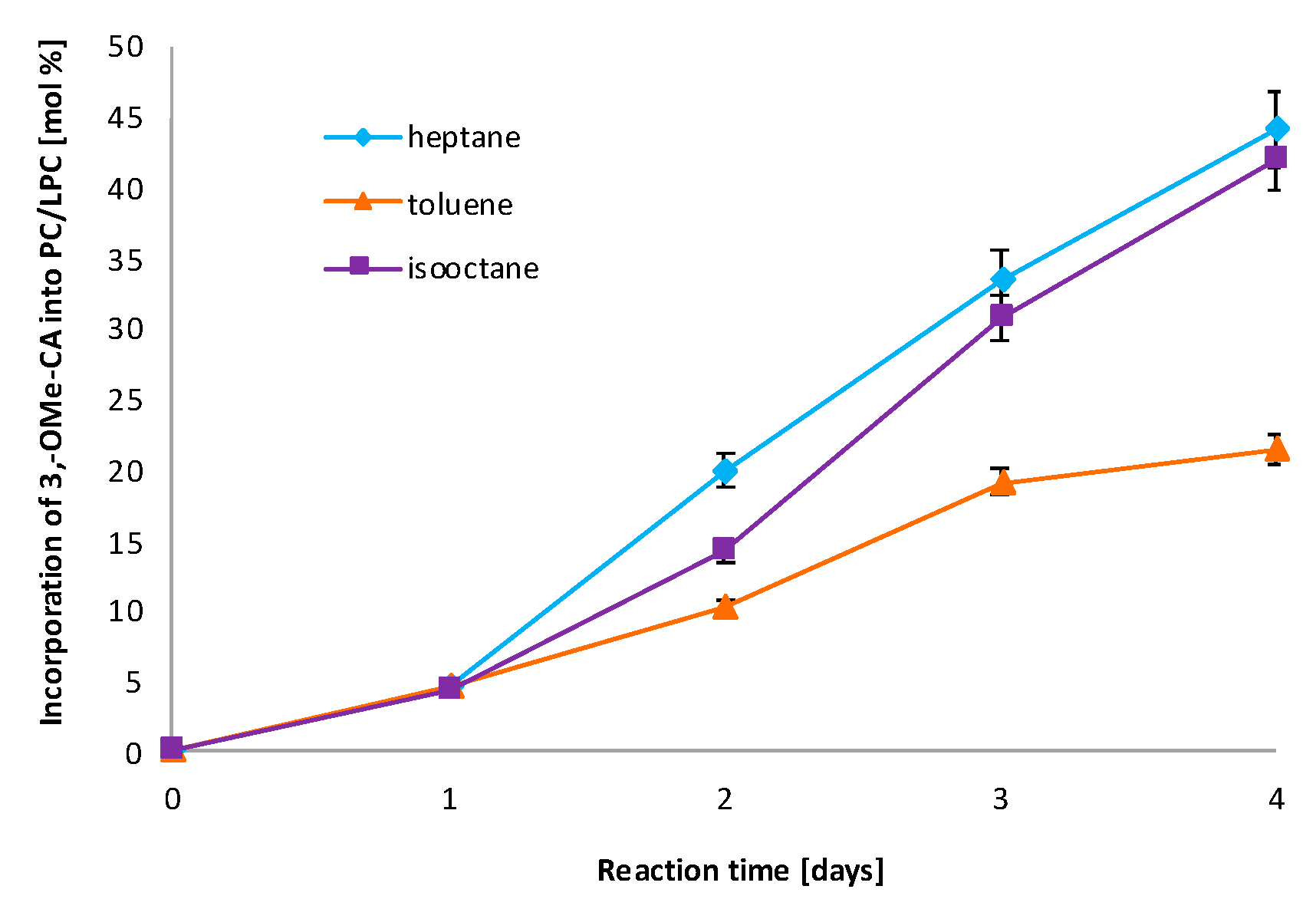
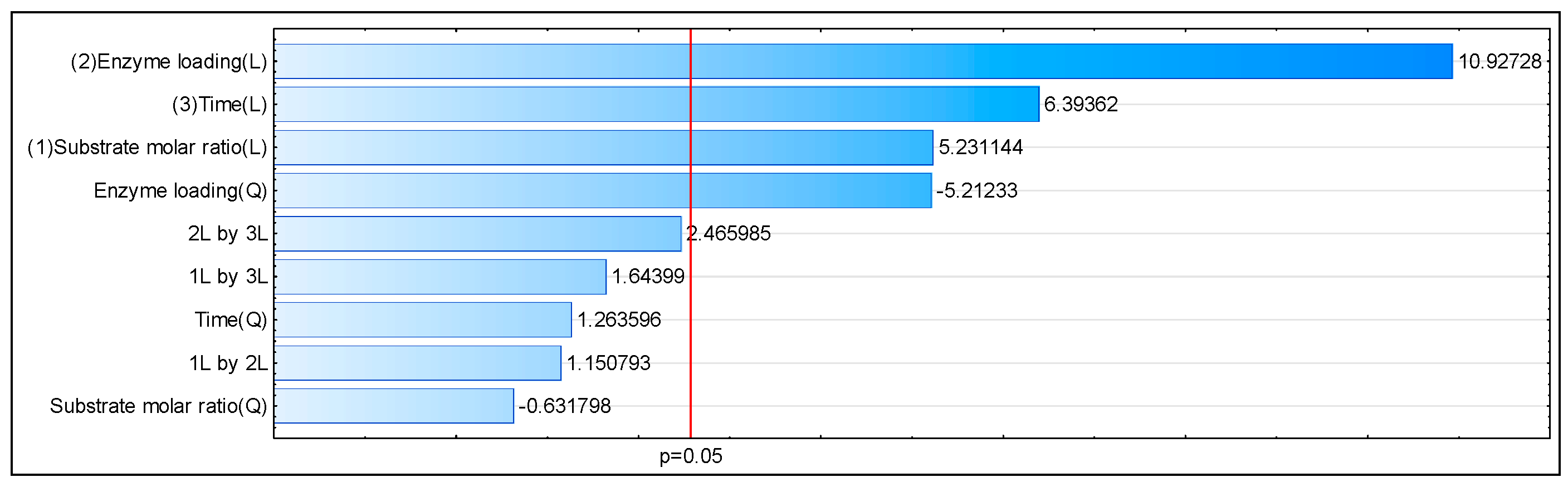

| Run | Substrate Molar Ratio PC/3-OMe-CA | Enzyme Loading [%] | Reaction Time [Days] | Incorporation of 3-OMe-CA into PC/LPC [mol%] a (Experimental) | Incorporation of 3-OMe-CA to PC/LPC [mol%] (Predicted) |
|---|---|---|---|---|---|
| 1 | 5 | 10 | 3 | 11 ± 0.4 | 13 |
| 2 | 15 | 10 | 3 | 19 ± 0.6 | 20 |
| 3 | 5 | 30 | 3 | 34 ± 1.2 | 33 |
| 4 | 15 | 30 | 3 | 49 ± 2.3 | 47 |
| 5 | 5 | 20 | 2 | 10 ± 0.6 | 8 |
| 6 | 15 | 20 | 2 | 16 ± 0.9 | 14 |
| 7 | 5 | 20 | 4 | 15 ± 0.7 | 17 |
| 8 | 15 | 20 | 4 | 31 ± 1.1 | 33 |
| 9 | 10 | 10 | 2 | 10 ± 0.5 | 10 |
| 10 | 10 | 30 | 2 | 23 ± 0.9 | 26 |
| 11 | 10 | 10 | 4 | 20 ± 0.7 | 17 |
| 12 | 10 | 30 | 4 | 48 ± 1.8 | 48 |
| 13 | 10 | 20 | 3 | 19 ± 0.7 | 19 |
| 14 | 10 | 20 | 3 | 19 ± 0.6 | 19 |
| 15 | 10 | 20 | 3 | 19 ± 0.2 | 19 |
| Evaluated Factors | Sum of Squares | Degrees of Freedom | Medium Square | F-Value | p-Value |
|---|---|---|---|---|---|
| (1) Substrate molar ratio (L) | 253.1250 | 1 | 253.125 | 27.3649 | 0.003379 |
| Substrate molar ratio (Q) | 3.692 | 1 | 3.692 | 0.3992 | 0.555275 |
| (2) Enzyme loading(L) | 1104.500 | 1 | 1104.500 | 119.4054 | 0.000112 |
| Enzyme loading (Q) | 251.308 | 1 | 251.308 | 27.1684 | 0.003432 |
| (3) Time of reaction (L) | 378.125 | 1 | 378.125 | 40.8784 | 0.001387 |
| Time of reaction (Q) | 14.769 | 1 | 14.769 | 1.5967 | 0.262086 |
| 1 by 2 | 12.250 | 1 | 12.250 | 1.3243 | 0.301861 |
| 1 by 3 | 25.000 | 1 | 25.000 | 2.7027 | 0.161099 |
| 2 by 3 | 56.250 | 1 | 56.250 | 6.0811 | 0.056810 |
| Error | 46.250 | 5 | 9.250 | ||
| Total error | 2153.733 | 14 | |||
| R2 = 0.98771 |
| Fatty and 3-OMe-CA Acids | Native PC | Modified PC |
| C16:0 (PA) | 36 | 10 |
| C16:1 (OPA) | 3 | 1 |
| C18:0 (SA) | 16 | 3 |
| C18:1 (OA) | 23 | 13 |
| C18:2 (LA) | 17 | 12 |
| C20:4 (AA) | 5 | 3 |
| 3-OMe-CA | - | 58 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Okulus, M.; Rychlicka, M.; Gliszczyńska, A. Enzymatic Production of Biologically Active 3-Methoxycinnamoylated Lysophosphatidylcholine via Regioselctive Lipase-Catalyzed Acidolysis. Foods 2022, 11, 7. https://doi.org/10.3390/foods11010007
Okulus M, Rychlicka M, Gliszczyńska A. Enzymatic Production of Biologically Active 3-Methoxycinnamoylated Lysophosphatidylcholine via Regioselctive Lipase-Catalyzed Acidolysis. Foods. 2022; 11(1):7. https://doi.org/10.3390/foods11010007
Chicago/Turabian StyleOkulus, Marta, Magdalena Rychlicka, and Anna Gliszczyńska. 2022. "Enzymatic Production of Biologically Active 3-Methoxycinnamoylated Lysophosphatidylcholine via Regioselctive Lipase-Catalyzed Acidolysis" Foods 11, no. 1: 7. https://doi.org/10.3390/foods11010007
APA StyleOkulus, M., Rychlicka, M., & Gliszczyńska, A. (2022). Enzymatic Production of Biologically Active 3-Methoxycinnamoylated Lysophosphatidylcholine via Regioselctive Lipase-Catalyzed Acidolysis. Foods, 11(1), 7. https://doi.org/10.3390/foods11010007







