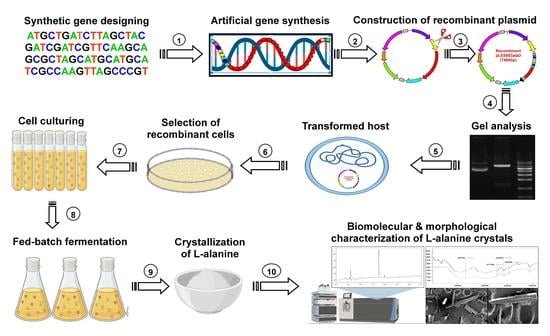
Metabolic Engineering of Pediococcus acidilactici BD16 for Heterologous Expression of Synthetic alaD Gene Cassette and L-Alanine Production in the Recombinant Strain Using Fed-Batch Fermentation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Growth Conditions
2.2. Method of Culturing of Host Strain under Microaerophilic Conditions
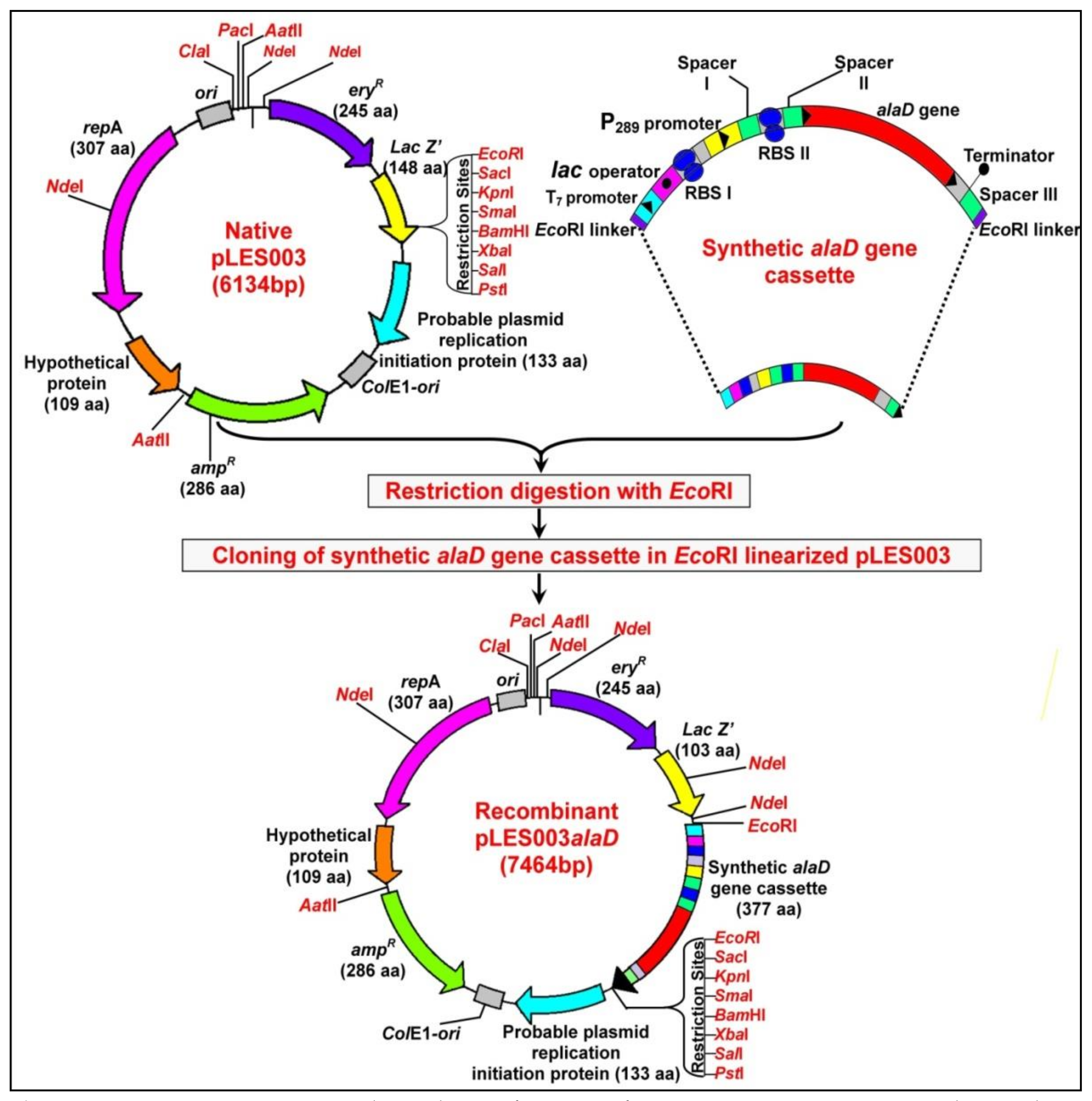
2.3. Designing of Synthetic Alanine Dehydrogenase (alaD) Gene Cassette
2.4. Subcloning of Synthetic alaD Gene Cassette and Construction of Recombinant Plasmid pLES003alaD
2.5. Preparation of the Competent P. acidilactici BD16 and Its Transformation by CaCl2-Heat Shock Method
2.6. Determination of Transformation Efficiency, Plasmid Segregational Stability and Copy Number of the Recombinant pLES003alaD Vector
2.7. Fed-Batch Fermentation for Production of L-Alanine
2.8. Quantitative Estimation of L-Alanine Production in Recombinant P. acidilactici BD16 (alaD+)
2.9. Extraction of L-Alanine from Recombinant Culture Broth by Crystallization
2.10. Analytical and Morphological Characterization of L-Alanine Crystals
2.11. Enantiomeric Purity of L-Alanine Produced by Recombinant P. acidilactici BD16 (alaD+)
2.12. Statistical Analysis
3. Results
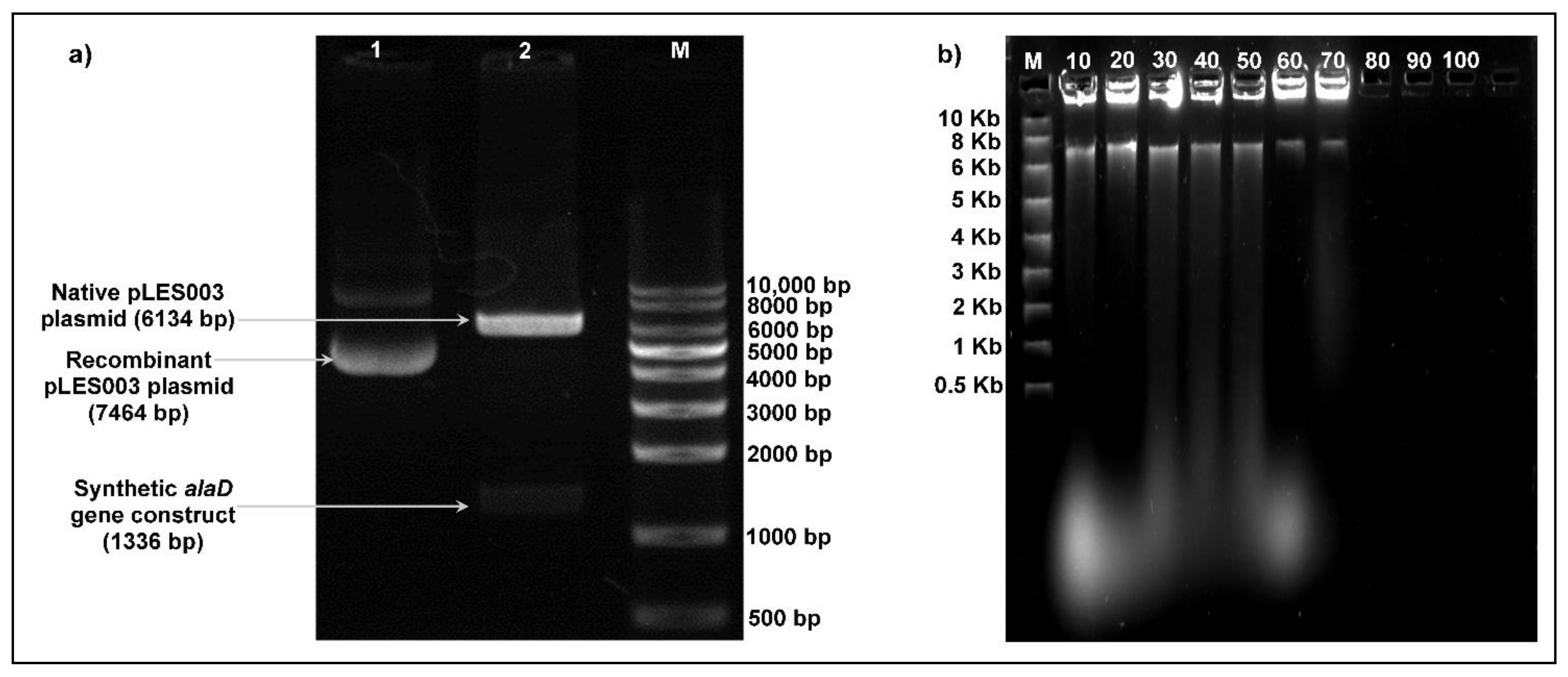
3.1. Designing of Synthetic alaD Gene Construct and Construction of Recombinant pLES003alaD Vector
3.2. Transformation Efficiency, Plasmid Copy Number and Segregational Stability of Recombinant pLES003alaD Vector
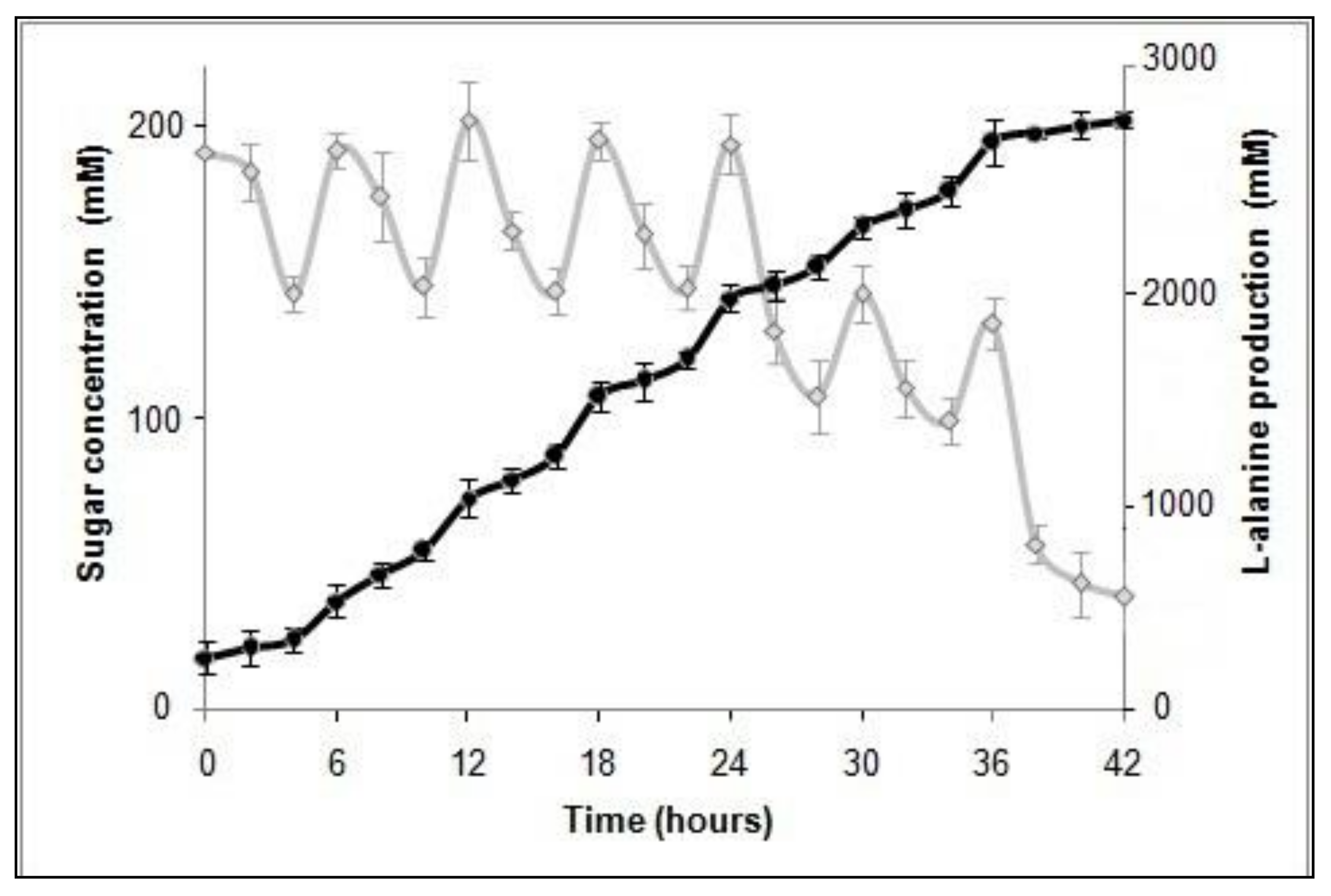
3.3. L-Alanine Production in Recombinant P. acidilactici BD16 (alaD+) Using Fed-Batch Fermentation
3.4. Crystallization and Enantiomeric Purity of Heterologously Produced L-Alanine
3.5. Morphological and Analytical Characterization of L-Alanine Crystals
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- D’Este, M.; Alvarado-Morales, M.; Angelidaki, I. Amino acids production focusing on fermentation technologies—A review. Biotechnol. Adv. 2018, 36, 14–25. [Google Scholar] [CrossRef] [Green Version]
- Hols, P.; Kleerebezem, M.; Schanck, A.N.; Ferain, T.; Hugenholtz, J.; Delcour, J.; De Vos, W.M. Conversion of Lactococcus lactis from homolactic to homoalanine fermentation through metabolic engineering. Nat. Biotechnol. 1999, 17, 588–592. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Smith, G.M.; Eiteman, M.A.; Altman, E. Aerobic production of alanine by Escherichia coli aceF ldhA mutants expressing the Bacillus sphaericus alaD gene. Appl. Microbiol. Biotechnol. 2004, 65, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Brennan, L.; Shine, A.; Hewage, C.; Malthouse, J.; Brindle, K.M.; McClenaghan, N.; Flatt, P.R.; Newsholme, P. A Nuclear Magnetic Resonance-Based Demonstration of Substantial Oxidative L-Alanine Metabolism and L-Alanine-Enhanced Glucose Metabolism in a Clonal Pancreatic -Cell Line: Metabolism of L-Alanine Is Important to the Regulation of Insulin Secretion. Diabetes 2002, 51, 1714–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tessem, M.-B.; Swanson, M.G.; Keshari, K.R.; Albers, M.J.; Joun, D.; Tabatabai, Z.L.; Simko, J.P.; Shinohara, K.; Nelson, S.J.; Vigneron, D.B.; et al. Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using1H HR-MAS spectroscopy of biopsy tissues. Magn. Reson. Med. 2008, 60, 510–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braverman, E.R.; Pfeiffer, C.E.; Blum, K.; Smayda, R. Alanine: The hypoglycemia helper. In The Healing Nutrients within: Facts, Findings and New Research on Amino Acids; Accessible Publishing Systems Pty Ltd.: Philadelphia, PA, USA, 2009. [Google Scholar]
- Leuchtenberger, W.; Huthmacher, K.; Drauz, K. Biotechnological production of amino acids and derivatives: Current status and prospects. Appl. Microbiol. Biotechnol. 2005, 69, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dave, U.C.; Kadeppagari, R.-K. Alanine dehydrogenase and its applications—A review. Crit. Rev. Biotechnol. 2019, 39, 648–664. [Google Scholar] [CrossRef] [PubMed]
- Mallakpour, S.; Dinari, M. Progress in Synthetic Polymers Based on Natural Amino Acids. J. Macromol. Sci. Part A 2011, 48, 644–679. [Google Scholar] [CrossRef]
- Shibatani, T.; Kakimoto, T.; Chibata, I. Stimulation of L-asparate beta-decarboxylase formation by L-glutamate in Pseudomonas dacunhae and Improved production of L-alanine. Appl. Environ. Microbiol. 1979, 38, 359–364. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Jantama, K.; Moore, J.C.; Shanmugam, K.T.; Ingram, L.O. Production of l-alanine by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2007, 77, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, S.-I.; Katsumata, R. l-Alanine fermentation by an alanine racemase-deficient mutant of the dl-alanine hyperproducing bacterium Arthrobacter oxydans HAP-1. J. Ferment. Bioeng. 1998, 86, 385–390. [Google Scholar] [CrossRef]
- Ohashima, T.; Soda, K. Purification and properties of alanine dehydrogenase from Bacillus sphaericus. Eur. J. Biochem. 1979, 100, 29–39. [Google Scholar] [CrossRef] [Green Version]
- Örlygsson, J.; Anderson, R.; Svensson, B.H. Alanine as an end product during fermentation of monosaccharides by Clostridium strain P2. Antonie van Leeuwenhoek 1995, 68, 273–280. [Google Scholar] [CrossRef]
- Jojima, T.; Fujii, M.; Mori, E.; Inui, M.; Yukawa, H. Engineering of sugar metabolism of Corynebacterium glutamicum for production of amino acid l-alanine under oxygen deprivation. Appl. Microbiol. Biotechnol. 2010, 87, 159–165. [Google Scholar] [CrossRef]
- Yamamoto, S.; Gunji, W.; Suzuki, H.; Toda, H.; Suda, M.; Jojima, T.; Inui, M.; Yukawa, H. Overexpression of Genes Encoding Glycolytic Enzymes in Corynebacterium glutamicum Enhances Glucose Metabolism and Alanine Production under Oxygen Deprivation Conditions. Appl. Environ. Microbiol. 2012, 78, 4447–4457. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Deng, C.; Cui, W.-J.; Liu, Z.-M.; Zhou, Z.-M. Efficient L-Alanine Production by a Thermo-Regulated Switch in Escherichia coli. Appl. Biochem. Biotechnol. 2015, 178, 324–337. [Google Scholar] [CrossRef]
- Uhlenbusch, I.; Sahm, H.; Sprenger, G.A. Expression of an L-alanine dehydrogenase gene in Zymomonas mobilis and excretion of L-alanine. Appl. Environ. Microbiol. 1991, 57, 1360–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, H.C.; Shin, H.S. Fed-Batch Cultures; Cambridge University Press (CUP): Cambridge, UK, 2013. [Google Scholar]
- Poontawee, R.; Limtong, S. Feeding Strategies of Two-Stage Fed-Batch Cultivation Processes for Microbial Lipid Production from Sugarcane Top Hydrolysate and Crude Glycerol by the Oleaginous Red Yeast Rhodosporidiobolusfluvialis. Microorganisms 2020, 8, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keasling, J.D. Manufacturing Molecules Through Metabolic Engineering. Science 2010, 330, 1355–1358. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Keasling, J.D. Synergies between synthetic biology and metabolic engineering. Nat. Biotechnol. 2011, 29, 693–695. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, R.S.; Gupta, G.; Ahmad, T.; Kaur, B. Metabolic Engineering of Enzyme-Regulated Bioprocesses Advances in Enzyme Technology; Elsevier: Amsterdam, The Netherlands, 2019; pp. 293–323. [Google Scholar]
- Sharma, A.; Gupta, G.; Ahmad, T.; Kaur, B.; Hakeem, K.R. Tailoring cellular metabolism in lactic acid bacteria through metabolic engineering. J. Microbiol. Methods 2020, 170, 105862. [Google Scholar] [CrossRef]
- Kaur, B.; Chakraborty, D.; Kumar, B. Metabolic engineering of Pediococcus acidilactici BD16 for production of vanillin through ferulic acid catabolic pathway and process optimization using response surface methodology. Appl. Microbiol. Biotechnol. 2014, 98, 8539–8551. [Google Scholar] [CrossRef]
- Kaur, B.; Kumar, B.; Kaur, G.; Chakraborty, D.; Kaur, K. Application of recombinant Pediococcus acidilactici BD16 (fcs + /ech + ) in malolactic fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 3015–3028. [Google Scholar] [CrossRef] [PubMed]
- Kaur, T.; Balgir, P.P.; Kaur, B. Correction to: Construction of a shuttle expression vector for lactic acid bacteria. J. Genet. Eng. Biotechnol. 2020, 18, 1. [Google Scholar] [CrossRef]
- Sharma, A.; Noda, M.; Sugiyama, M.; Ahmad, A.; Kaur, B. Production of Functional Buttermilk and Soymilk Using Pediococcus acidilactici BD16 (alaD+). Molecules 2021, 26, 4671. [Google Scholar] [CrossRef]
- Wada, T.; Noda, M.; Kashiwabara, F.; Jeon, H.J.; Shirakawa, A.; Yabu, H.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Characterization of four plasmids harboured in a Lactobacillus brevis strain encoding a novel bacteriocin, brevicin 925A, and construction of a shuttle vector for lactic acid bacteria and Escherichia coli. Microbiology 2009, 155, 1726–1737. [Google Scholar] [CrossRef] [Green Version]
- Shah, S.; Rathod, I.; Kanakia, D. Colorimetry method for estimation of glycine, alanine and isoleucine. Indian J. Pharm. Sci. 2007, 69, 462. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugars. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Seethalakshmi, K.; Perumal, S.; Selvarajan, P. Studies on growth, morphology, spectral and me-chanical properties of some doped l-alanine family of single crystals. Int. J. Curr. Res. Rev. 2012, 4, 53–61. [Google Scholar]
- Sherovski, P.; Stefova, M.; Ristovska, N. Simultaneous RP-HPLC-DAD determination of dansyl amino acids in chemically treated human hair. Maced. J. Chem. Chem. Eng. 2018, 37. [Google Scholar] [CrossRef]
- Noda, M.; Miyauchi, R.; Danshiitsoodol, N.; Matoba, Y.; Kumagai, T.; Sugiyama, M. Expression of Genes Involved in Bacteriocin Production and Self-Resistance in Lactobacillus brevis 174A Is Mediated by Two Regulatory Proteins. Appl. Environ. Microbiol. 2018, 84, e02707-17. [Google Scholar] [CrossRef] [Green Version]
- Katsumata, R.; Hashimoto, S.I. Kyowa Hakko Kogyo Co Ltd. Process for Producing Alanine. U.S. Patent 5,559,016, 24 September 1996. [Google Scholar]
- Smith, G.M.; Lee, S.A.; Reilly, K.C.; Eiteman, M.A.; Altman, E. Fed-batch two-phase production of alanine by a metabolically engineered Escherichia coli. Biotechnol. Lett. 2006, 28, 1695–1700. [Google Scholar] [CrossRef]
- Han, G.; Poornachary, S.K.; Chow, P.S.; Tan, R.B.H. Understanding Growth Morphology Changes of γ-Glycine anddl-Alanine Polar Crystals in Pure Aqueous Solutions. Cryst. Growth Des. 2010, 10, 4883–4889. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of Infrared Spectra, A Practical Approach Encyclopedia of Analytical Chemistry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Raj, C.J.; Das, S.J. Growth and characterization of nonlinear optical active l-alanine formate crystal by modified Sankaranarayanan–Ramasamy (SR) method. J. Cryst. Growth 2007, 304, 191–195. [Google Scholar] [CrossRef]
- Thilak, T.; Ahamed, M.B.; Marudhu, G.; Vinitha, G. Effect of KDP on the growth, thermal and optical properties of l-alanine single crystals. Arab. J. Chem. 2016, 9, 676–680. [Google Scholar] [CrossRef] [Green Version]
- Perry, J.J. Production of L-alanine by biotin-deficient Micrococcus sodonensis. J. Bacteriol. 1967, 94, 1249–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasserman, S.A.; Walsh, C.T.; Botstein, D. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 1983, 153, 1439–1450. [Google Scholar] [CrossRef] [Green Version]

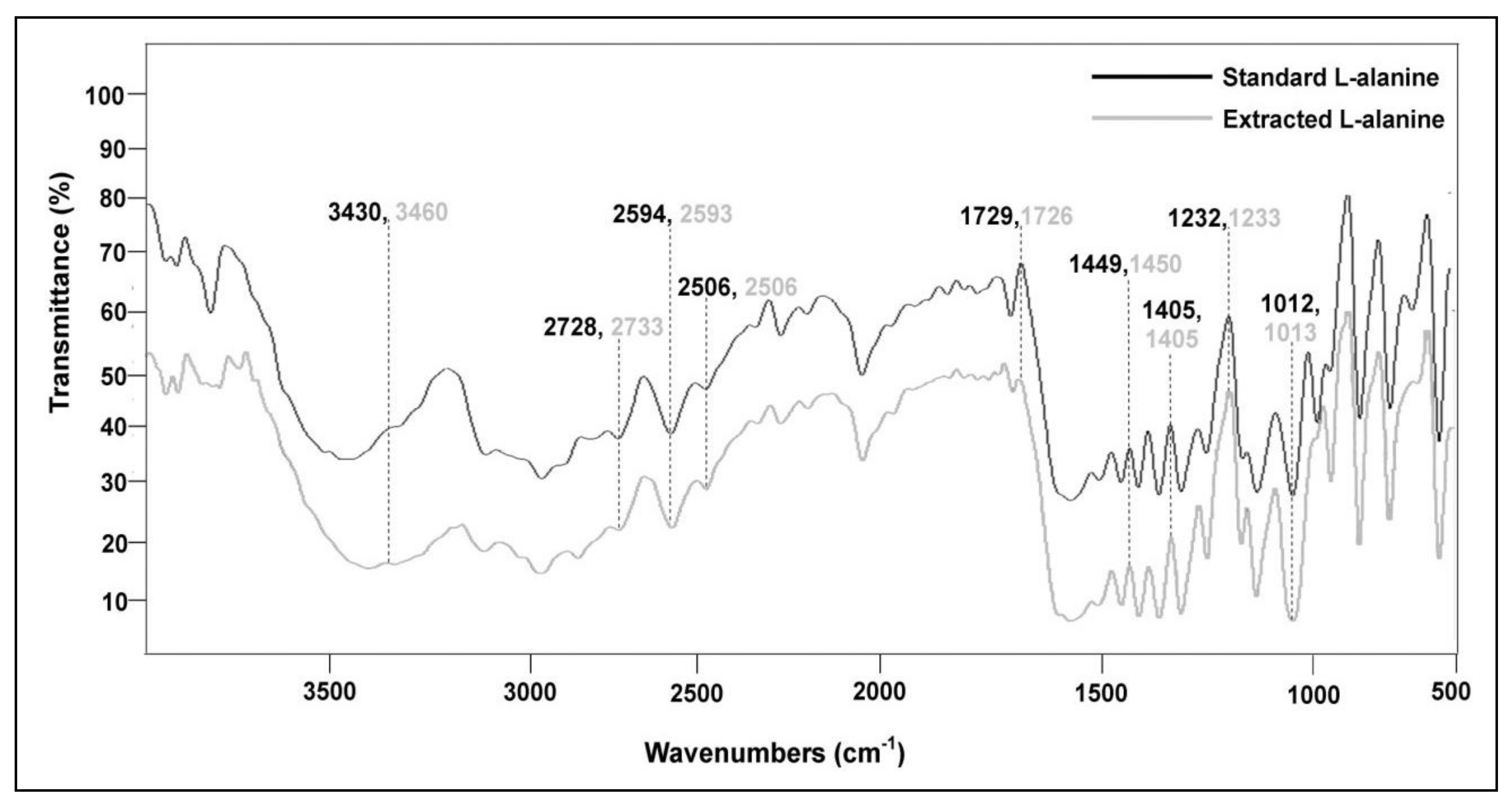
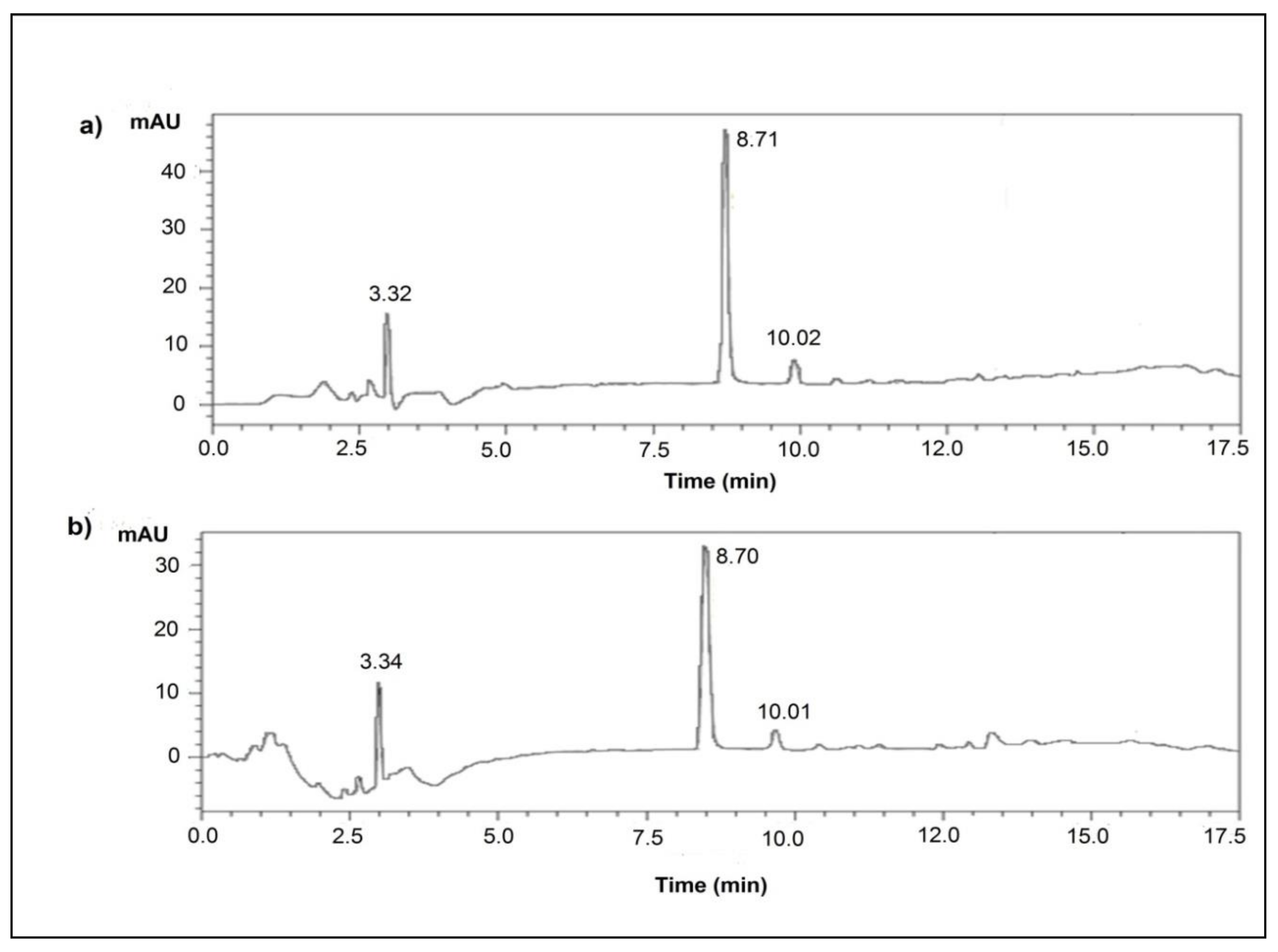
| Microorganism | Genetic Modifications | Growth and Fermentation Conditions | Fermentation Process | Time (h) | L-Alanine Production in g/L (mM) | Enantiomeric Purity (%) | Reference |
|---|---|---|---|---|---|---|---|
| Zymomonas mobilis CP4(pZY73) (Gram –ve) | Bacillus sphaericus IFO3525 alaD | Mineral salts medium containing 280 mM glucose, 85 mM ammonium sulfate; under anaerobic conditions | Batch | 26 | 7.5 (84 mM) | Not reported | [18] |
| Corynebacterium glutamicum AL107 (pOBP107) (Gram +ve) | Arthrobacter oxydans HAP-1 alaD | Corn steep liquor medium containing 1110 mM glucose, 44.8 mM DL-alanine; limited oxygen conditions | Batch | 70 | 71 (797 mM) | >99 | [35] |
| E coli AL1 (pOBP1) (Gram –ve) | A. oxydans HAP-1 alaD | Mineral salt medium containing 110 mM glucose; limited oxygen conditions | Batch | 40 | 8 (90 mM) | Not reported | [35] |
| Arthrobacter oxydans DAN 75 (Gram +ve) | Alanine racemacedeficient | Mineral salt medium containing 832.6 mM glucose, 2.24 mM D-alanine; shaking conditions | Two-stage fed-batch | 120 | 77 (864 mM) | 98 | [12] |
| Lactococcus lactis NZ3950 (pNZ2650) (Gram +ve) | B. sphaericus IFO3525 alaD, ΔldhA | M17 rich medium containing 100 mM glucose, 2.25 mM D-alanine, 150 mM Ammonium sulfate; shaking conditions at 120rpm | Batch | 17 | 13 (146 mM) | 85–90 | [2] |
| L. lactis PH3950 (pNZ2650) (Gram +ve) | B. sphaericus IFO3525 alaD, ΔldhA, Δalr | M17 rich medium containing 100 mM glucose, 2.25 mM D-alanine; 150 mM Ammonium sulfate; shaking conditions at 120rpm | Batch | 17 | Not reported | >99 | [2] |
| E. coli ALS887 (pTrc99A-alaD) (Gram –ve) | B. sphaericus IFO3525 alaD, ΔldhA, ΔaceF | Medium containing 666 mM glucose, 1261 mM ammonium chloride; oxygen limited conditions | Two-stage, fed-batch | 27 | 32 (359 mM) | Not reported | [3] |
| E. coli ALS929 (pTrc99A-alaD) (Gram –ve) | B. sphaericus IFO3525 alaD, Δpfl, Δpps, ΔpoxB, ΔldhA, ΔaceF | Medium containing 999 mM glucose; oxygen limited conditions | Two-stage fed-batch | 23 h anaerobic phase (48 h total fermentation time) | 88 (988 mM) | Not reported | [36] |
| E. coliXZ132 (Gram –ve) | Geobacillus stearothermophilus alaD, Δpfl, ΔackA, ΔadhE, ΔldhA, ΔmgsA, ΔdadX | Low salt medium containing 666 mM glucose; anaerobic conditions | Batch | 48 | 114 (1279 mM) | >99 | [11] |
| C. glutamicum (Gram +ve) | Lysinibacillus sphaericus alaD, gapA, ΔldhA, Δppc, Δalr | Minimal salts medium containing 888 mM glucose, 52.97 mM ammonium sulfate; Limited oxygen conditions | Fed-batch | 32 | 98 (1097 mM) | 99.5% | [15] |
| C. glutamicum GLY3/pCRD500 (Gram +ve) | L. sphaericus alaD, ΔldhA, Δppc; stepwise over-expression and chromosomal integration of four glycolytic enzymes encoded by gapA, pyk, pfk, gpi genes | BT medium containing 1600 mM glucose; with agitation but without aeration. | Two stage fed-batch | 48 | 216 (2430 mM) | - | [16] |
| C. glutamicum GLY3/pCRD914 (Gram +ve) | L. sphaericus alaD | BT medium containing 1600 mM glucose; with agitation but without aeration. | Two stage fed-batch | 72 | 275 (3090 mM) | - | [16] |
| E. coli B0016-060BC (Gram –ve) | G. stearothermophilus alaD, ∆ack, ∆pta, ∆pflB, ∆adhE, ∆frdA, ∆ldhA ∆alr | M9-1 medium containing 2837 mM glucose; oxygen limited phase at 42 °C | Two stage fed-batch | 24 | 120.8 (1347 mM) | - | [17] |
| P. acidilacticiBD16 (Gram +ve) (Wild type) | No genetic manipulation | MRS medium containing 100 mM dextrose; under microaerophilic and stationary conditions | Batch | 24 | 0.42 (4.71 mM) | - | Present study |
| P. acidilactici BD16 (Gram +ve) (Codon optimized) | pLES003 containing synthetic alaD gene cassette | Minimal Salt Medium containing 1400 mM dextrose, 8.22 mM tri-ammonium citrate; oxygen limited conditions | Fed-batch | 42 | 217.54(2442mM) | 97% | Present study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, A.; Noda, M.; Sugiyama, M.; Kaur, B.; Ahmad, A. Metabolic Engineering of Pediococcus acidilactici BD16 for Heterologous Expression of Synthetic alaD Gene Cassette and L-Alanine Production in the Recombinant Strain Using Fed-Batch Fermentation. Foods 2021, 10, 1964. https://doi.org/10.3390/foods10081964
Sharma A, Noda M, Sugiyama M, Kaur B, Ahmad A. Metabolic Engineering of Pediococcus acidilactici BD16 for Heterologous Expression of Synthetic alaD Gene Cassette and L-Alanine Production in the Recombinant Strain Using Fed-Batch Fermentation. Foods. 2021; 10(8):1964. https://doi.org/10.3390/foods10081964
Chicago/Turabian StyleSharma, Anshula, Masafumi Noda, Masanori Sugiyama, Baljinder Kaur, and Ajaz Ahmad. 2021. "Metabolic Engineering of Pediococcus acidilactici BD16 for Heterologous Expression of Synthetic alaD Gene Cassette and L-Alanine Production in the Recombinant Strain Using Fed-Batch Fermentation" Foods 10, no. 8: 1964. https://doi.org/10.3390/foods10081964
APA StyleSharma, A., Noda, M., Sugiyama, M., Kaur, B., & Ahmad, A. (2021). Metabolic Engineering of Pediococcus acidilactici BD16 for Heterologous Expression of Synthetic alaD Gene Cassette and L-Alanine Production in the Recombinant Strain Using Fed-Batch Fermentation. Foods, 10(8), 1964. https://doi.org/10.3390/foods10081964