Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthetic Peptides
2.2. Strains and Culture Media
2.3. Degradation of Gluten
2.4. Degradation of Mixed Gliadins in Solution
2.5. Degradation of Gliadin in Gel (Gliadin Zymography)
2.6. Degradation of Immunotoxic Peptides
2.7. Growth and Gluten Degradation
2.8. Complete Genome Sequencing of B. cereus
2.9. RP-HPLC
2.10. Mass Spectrometry
2.11. Statistical Analysis
3. Results
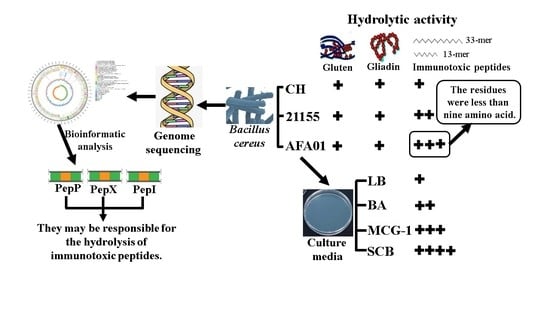
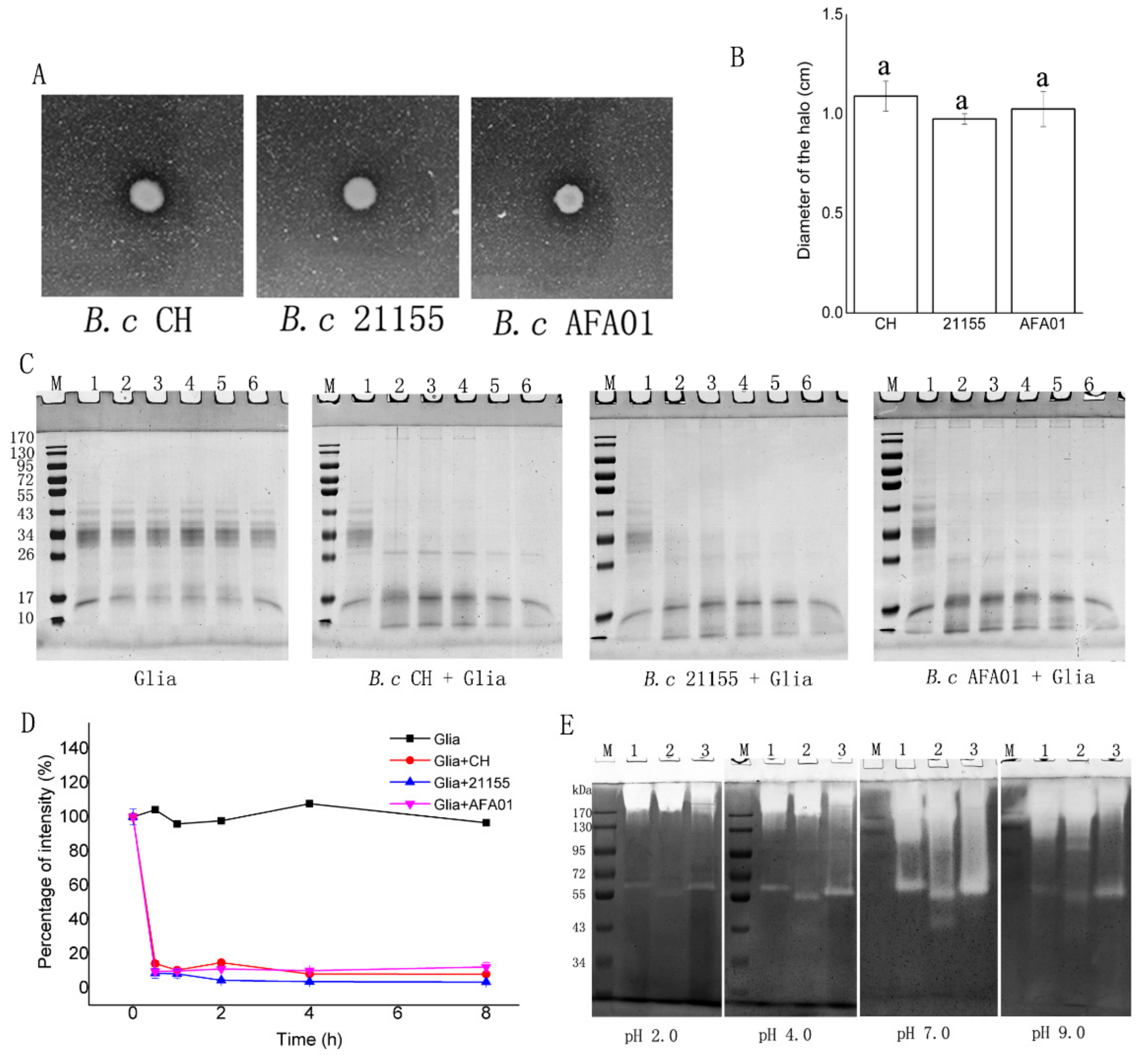
3.1. Degradation of Gluten and Gliadin by B. cereus
3.2. Peptidasic Activity against Immunotoxic Peptides
3.3. Influence of Media Composition on Protease Activity
3.4. Whole Genome Sequencing and Bioinformatic Analysis of B. cereus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barak, S.; Mudgil, D.; Khatkar, B.S. Biochemical and Functional Properties of Wheat Gliadins: A Review. Crit. Rev. Food Sci. Nutr. 2015, 55, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Hujoel, I.A.; Reilly, N.R.; Rubio-Tapia, A. Celiac Disease: Clinical Features and Diagnosis. Gastroenterol. Clin. N. Am. 2019, 48, 19–37. [Google Scholar] [CrossRef]
- Sollid, L.M.; Lundin, K.E.A. The Autoimmune Diseases, Sixth Ed.; Academic Press: Salt Lake City, UT, USA, 2020; pp. 849–869. [Google Scholar]
- Lebwohl, B.; Sanders, D.S.; Green, P.H.R. Coeliac disease. Lancet 2018, 391, 70–81. [Google Scholar] [CrossRef]
- Serena, G.; Kelly, C.P.; Fasano, A. Nondietary Therapies for Celiac Disease. Gastroenterol. Clin. N. Am. 2019, 48, 145–163. [Google Scholar] [CrossRef]
- Makharia, G.K.D. Current and Emerging Therapy for Celiac Disease. Front. Med. 2014, 1. [Google Scholar] [CrossRef] [Green Version]
- Rizzello, C.G.; De Angelis, M.; Di Cagno, R.; Camarca, A.; Silano, M.; Losito, I.; De Vincenzi, M.; De Bari, M.D.; Palmisano, F.; Maurano, F. Highly efficient gluten degradation by lactobacilli and fungal proteases during food processing: New perspectives for celiac disease. Appl. Environ. Microb. 2007, 73, 4499–4507. [Google Scholar] [CrossRef] [Green Version]
- Montserrat, V.; Bruins, M.J.; Edens, L.; Koning, F. Influence of dietary components on Aspergillus niger prolyl endoprotease mediated gluten degradation. Food Chem. 2015, 174, 440–445. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.A.; Kelly, C.P.; Green, P.H.; Marcantonio, A.; Wu, T.-T.; Mäki, M.; Adelman, D.C.; Ansari, S.; Ayub, K.; Basile, A.; et al. No Difference between Latiglutenase and Placebo in Reducing Villous Atrophy or Improving Symptoms in Patients with Symptomatic Celiac Disease. Gastroenterology 2017, 152, 787–798.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koga, S.; Rieder, A.; Ballance, S.; Uhlen, A.K.; Veiseth-Kent, E. Gluten-Degrading Proteases in Wheat Infected by Fusarium graminearum-Protease Identification and Effects on Gluten and Dough Properties. J. Agric. Food Chem. 2019, 67, 11025–11034. [Google Scholar] [CrossRef] [PubMed]
- Aamot, H.U.; Lyse, E.; Koga, S.; Nielsen, K.A.; Böcker, U.; Brodal, G.; Dill-Macky, R.; Uhlen, A.K.; Hofgaard, I.S. Microdochium majus and other fungal pathogens associated with reduced gluten quality in wheat grain. Int. J. Food Microbiol. 2020, 331, 108712. [Google Scholar] [CrossRef]
- Rashmi, B.S.; Gayathri, D.; Vasudha, M.; Prashantkumar, C.S.; Swamy, C.T.; Sunil, K.S.; Somaraja, P.K.; Prakash, P. Gluten hydrolyzing activity of Bacillus spp isolated from sourdough. Microb. Cell Factor. 2020, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jayawardana, I.A.; Boland, M.J.; Higgs, K.; Zou, M.; Loo, T.; Mcnabb, W.C.; Montoya, C.A. The kiwifruit enzyme actinidin enhances the hydrolysis of gluten proteins during simulated gas-trointestinal digestion. Food Chem. 2020, 341, 128239. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Lee, C.C.; Hsu, J.H.; Leu, W.M.; Meng, M. Efficient Hydrolysis of Gluten-Derived Celiac Disease-Triggering Immunogenic Peptides by a Bacterial Serine Protease from Burkholderia gladioli. Biomolecules 2021, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.J.; Wei, G. Experimental Strategy to Discover Microbes with Gluten-degrading Enzyme Activities. In Sensing Technologies for Global Health, Military Medicine, and Environmental Monitoring IV.; International Society for Optics and Photonics: Bellingham, WA, USA, 2014; Volume 9112, p. 91120D. [Google Scholar]
- Wood, E.J. Molecular Cloning. A Laboratory Manual. Biochem. Educ. 1983, 11, 82. [Google Scholar] [CrossRef]
- Caminero, A.; Herran, A.R.; Nistal, E.; Perez-Andres, J.; Vaquero, L.; Vivas, S.; Ruiz de Morales, J.M.; Albillos, S.M.; Casqueiro, J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: Isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol. Ecol. 2014, 88, 309–319. [Google Scholar] [CrossRef]
- Doddapaneni, K.K.; Tatineni, R.; Vellanki, R.N.; Rachcha, S.; Anabrolu, N.; Narakuti, V.; Mangamoori, L.N. Purification and characterization of a solvent and detergent-stable novel protease from Bacillus cereus. Microbiol. Res. 2009, 164, 383–390. [Google Scholar] [CrossRef]
- Gutiérrez, S.; Pérez-Andrés, J.; Blanco, H.M.; Ferrero, M.A.; Vaquero, L.; Vivas, S.; Casqueiro, J.; Rodríguez-Aparicio, L.B. The human digestive tract has proteases capable of gluten hydrolysis. Mol. Metab. 2017, 6, 693–702. [Google Scholar] [CrossRef]
- Fernandez-Feo, M.; Wei, G.; Blumenkranz, G.; Dewhirst, F.E.; Schuppan, D.; Oppenheim, F.G.; Helmerhorst, E.J. The cultivable human oral gluten-degrading microbiome and its potential implications in coeliac disease and gluten sensitivity. Clin. Microbiol. Infect. 2013, 19, 384–394. [Google Scholar] [CrossRef] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Kooy-Winkelaar, Y.M.C.; Cordfunke, R.A.; Dragan, I.; Thompson, A.; Drijfhout, J.W.; Van Veelen, P.A.; Chen, H.; Koning, F. Abrogation of Immunogenic Properties of Gliadin Peptides through Transamidation by Microbial Transglutaminase Is Acyl-Acceptor Dependent. J. Agric. Food Chem. 2017, 65, 7542–7552. [Google Scholar] [CrossRef]
- Wieser, H. Chemistry of gluten proteins. Food Microbiol. 2007, 24, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Sieiro, P.; Redruello, B.; Ladero, V.; Cañedo, E.; Martin, M.C.; Fernández, M.; Alvarez, M.A. Solubilization of gliadins for use as a source of nitrogen in the selection of bacteria with gliadinase activity. Food Chem. 2015, 168, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagh, S.K.; Gadge, P.P.; Padul, M.V. Significant Hydrolysis of Wheat Gliadin by Bacillus tequilensis (10bT/HQ223107): A Pilot Study. Probiotics Antimicrob. Proteins 2017, 10, 662–667. [Google Scholar] [CrossRef]
- Wei, G.; Tian, N.; Valery, A.C.; Zhong, Y.; Schuppan, D.; Helmerhorst, E.J. Identification of Pseudolysin (lasB) as an Aciduric Glu-ten-Degrading Enzyme with High Therapeutic Potential for Celiac Disease. Am. J. Gastroenterol. 2015, 110, 899–908. [Google Scholar] [CrossRef] [Green Version]
- Saleem, M.; Rehman, A.; Yasmin, R.; Munir, B. Biochemical analysis and investigation on the prospective applications of alka-line protease from a Bacillus cereus strain. Mol. Biol. Rep. 2012, 39, 6399–6408. [Google Scholar] [CrossRef]
- Tian, N.; Faller, L.; Leffler, D.A.; Kelly, C.P.; Hansen, J.; Bosch, J.A.; Wei, G.; Paster, B.J.; Schuppan, D.; Helmerhorst, E.J. Salivary Gluten Degradation and Oral Microbial Profiles in Healthy Individuals and Celiac Disease Patients. Appl. Environ. Microbiol. 2017, 83, e03330-16. [Google Scholar] [CrossRef] [Green Version]
- Shan, L.; Molberg, Ø.; Parrot, I.; Hausch, F.; Filiz, F.; Gray, G.M.; Sollid, L.M.; Khosla, C. Structural basis for gluten intolerance in celiac sprue. Science 2002, 297, 2275–2279. [Google Scholar] [CrossRef] [Green Version]
- Wieser, H.; Koehler, P. Detoxification of Gluten by Means of Enzymatic Treatment. J. AOAC Int. 2012, 95, 356–363. [Google Scholar] [CrossRef]
- Haddar, A.; Fakhfakh-Zouari, N.; Hmidet, N.; Frikha, F.; Nasri, M.; Kamoun, A.S. Low-cost fermentation medium for alkaline protease production by Bacillus mojavensis A21 using hulled grain of wheat and sardinella peptone. J. Biosci. Bioeng. 2010, 110, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Berthold-Pluta, A.; Pluta, A.; Garbowska, M.; Stefańska, I. Prevalence and Toxicity Characterization of Bacillus cereus in Food Products from Poland. Foods 2019, 8, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joo, H.-S.; Kumar, C.G.; Park, G.-C.; Kim, K.T.; Paik, S.R.; Chang, C.-S. Optimization of the production of an extracellular alkaline protease from Bacillus horikoshii. Process. Biochem. 2002, 38, 155–159. [Google Scholar] [CrossRef]
- Zervas, A.; Aggerbeck, M.R.; Allaga, H.; Güzel, M.; Hendriks, M.; Jonuškienė, I.; Kedves, O.; Kupeli, A.; Lamovšek, J.; Mülner, P.; et al. Identification and Characterization of 33 Bacillus cereus sensu lato Isolates from Agricultural Fields from Eleven Widely Distributed Countries by Whole Genome Sequencing. Microorganisms 2020, 8, 2028. [Google Scholar] [CrossRef] [PubMed]
- Amador, M.D.L.M.; Arévalo-Rodríguez, M.; Durán, E.M.; Reyes, J.C.M.; Martín, C.S. A new microbial gluten-degrading prolyl endopeptidase: Potential application in celiac disease to reduce gluten immunogenic peptides. PLoS ONE 2019, 14, e0218346. [Google Scholar] [CrossRef] [Green Version]
- Francavilla, R.; De Angelis, M.; Rizzello, C.G.; Cavallo, N.; Dal Bello, F.; Gobbetti, M. Selected Probiotic Lactobacilli Have the Capacity to Hydrolyze Gluten Peptides during Simulated Gastrointestinal Digestion. Appl. Environ. Microbiol. 2017, 83, e00376-17. [Google Scholar] [CrossRef] [Green Version]
- De Angelis, M.; Cassone, A.; Rizzello, C.G.; Gagliardi, F.; Minervini, F.; Calasso, M.; Di Cagno, R.; Francavilla, R.; Gobbetti, M. Mechanism of degradation of immunogenic gluten epitopes from Triticum turgidum L. var. durum by sourdough lactobacilli and fungal proteases. Appl. Environ. Microbiol. 2010, 76, 508–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Medium | Composition |
|---|---|
| BA | Peptone 10 g·L−1, Casein peptone 10 g·L−1, Yeast extract 2.0 g·L−1, Glucose 1.0 g·L−1, Sodium chloride 5.0 g·L−1, Agar 13 g·L−1, pH 7.0 ± 0.2 |
| GA | Wheat gluten 23 g·L−1, Sodium chloride 5.0 g·L−1, Glucose 1.0 g·L−1, Sodium succinate 0.5 g·L−1, Soluble starch 1.0 g·L−1, Sodium pyruvate 1.0 g·L−1, Soluble pyrophosphate 0.25 g·L−1, L-Arginine 1.0 g·L−1, L-Cysteine 0.5 g·L−1, Haemin0.01 g·L−1, Vitamin K 0.001 g·L−1, Sodium bicarbonate 0.4 g·L−1, Agar 12 g·L−1 |
| LB | Tryptone 10 g·L−1, Yeast extract 5.0 g·L−1, Sodium chloride 10 g·L−1 |
| MCG-1 | Glucose 20 g·L−1, Gluten 30 g·L−1, CaCl2 0.05 g·L−1, ZnSO4 0.07 g·L−1, L-cysteine 0.05 g·L−1, Tween 80 0.1%, 60 mM Phosphate Buffer (pH 6.5), Agar 16 g·L−1 |
| SCB | Starch 10 g·L−1, Casein 3.0 g·L−1, KNO3 2.0 g·L−1, NaCl 2.0 g·L−1, K2HPO4 2.0 g·L−1, MgSO4 0.05 g·L−1, CaCl2 0.02 g·L−1, FeSO4 0.01 g·L−1, pH 7.2 |
| The Number of Genes | ||||
|---|---|---|---|---|
| KO ID | KO Description | CH | 21155 | AFA01 |
| K01297 | muramoyltetrapeptide carboxypeptidase [EC:3.4.17.13] | 1 | 1 | 1 |
| K07258 | serine-type D-Ala-D-Ala carboxypeptidase [EC:3.4.16.4] | 4 | 4 | 4 |
| K01299 | carboxypeptidase Taq [EC:3.4.17.19] | 1 | 1 | 1 |
| K08602 | oligoendopeptidase F [EC:3.4.24.-] | 3 | 3 | 3 |
| K03798 | cell division protease FtsH [EC:3.4.24.-] | 1 | 1 | 1 |
| K01265 | methionyl aminopeptidase [EC:3.4.11.18] | 3 | 3 | 3 |
| K19689 | aminopeptidase [EC:3.4.11.-] | 3 | 3 | 3 |
| K01258 | tripeptide aminopeptidase [EC:3.4.11.4] | 2 | 2 | 2 |
| K01255 | leucyl aminopeptidase [EC:3.4.11.1] | 1 | 1 | 1 |
| K03100, K12380 | signal peptidase I [EC:3.4.21.89] | 7 | 7 | 7 |
| K03101 | signal peptidase II [EC:3.4.23.36] | 1 | 1 | 1 |
| K08600 | sortase B [EC:3.4.22.71] | 1 | 1 | 1 |
| K02236 | leader peptidase (prepilin peptidase)/N-methyltransferase [EC:3.4.23.43 2.1.1.-] | 1 | 1 | 1 |
| K05995 | dipeptidase E [EC:3.4.13.21] | 1 | 1 | 1 |
| K01270 | dipeptidase D [EC:3.4.13.-] | 1 | 1 | 1 |
| K01273 | membrane dipeptidase [EC:3.4.13.19] | 1 | 1 | 1 |
| K08651 | thermitase [EC:3.4.21.66] | 1 | 1 | 1 |
| K17733 | peptidoglycan LD-endopeptidase CwlK [EC:3.4.-.-] | 1 | 1 | 1 |
| K01419 | ATP-dependent HslUV protease, peptidase subunit HslV [EC:3.4.25.2] | 1 | 1 | 1 |
| K01338 K04076 | ATP-dependent Lon protease [EC:3.4.21.53] | 1 | 1 | 1 |
| K01358 | ATP-dependent Clp protease, protease subunit [EC:3.4.21.92] | 2 | 2 | 2 |
| K20742 | gamma-D-glutamyl-L-lysine dipeptidyl-peptidase [EC:3.4.14.13] | 1 | 1 | 1 |
| K01304 | pyroglutamyl-peptidase [EC:3.4.19.3] | 1 | 1 | 1 |
| K08777 | neutral peptidase B [EC:3.4.24.-] | 1 | 1 | 1 |
| K21472 | peptidoglycan LD-endopeptidase LytH [EC:3.4.-.-] | 1 | 1 | 1 |
| K21471 | peptidoglycan DL-endopeptidase CwlO [EC:3.4.-.-] | 1 | 1 | 1 |
| K11749 | regulator of sigma E protease [EC:3.4.24.-] | 1 | 1 | 1 |
| K06383 | stage II sporulation protein GA [EC:3.4.23.-] | 1 | 1 | 1 |
| K06402 | stage IV sporulation protein FB [EC:3.4.24.-] | 1 | 1 | 1 |
| K06399 | stage IV sporulation protein B [EC:3.4.21.116] | 1 | 1 | 1 |
| K06012 | spore protease [EC:3.4.24.78] | 1 | 1 | 1 |
| K14647 | minor extracellular serine protease Vpr [EC:3.4.21.-] | 1 | 1 | 1 |
| K03797 | carboxyl-terminal processing protease [EC:3.4.21.102] | 1 | 1 | 1 |
| K08303 | putative protease [EC:3.4.-.-] | 2 | 2 | 2 |
| K19701 | aminopeptidase YwaD [EC:3.4.11.6 3.4.11.10] | 1 | 1 | 1 |
| K01271 | Xaa-Pro dipeptidase [EC:3.4.13.9] | 2 | 2 | 2 |
| K01262 | Xaa-Pro aminopeptidase [EC:3.4.11.9] | 1 | 1 | 1 |
| K01281 | X-Pro dipeptidyl-peptidase [EC:3.4.14.11] | 1 | 1 | 1 |
| K01286 | D-alanyl-D-alanine carboxypeptidase [EC:3.4.16.4] | 12 | 12 | 10 |
| K07260 | zinc D-Ala-D-Ala carboxypeptidase [EC:3.4.17.14] | 7 | 5 | 3 |
| K19702 | aminopeptidase S [EC:3.4.11.24] | 1 | 0 | 0 |
| K01387 | microbial collagenase [EC:3.4.24.3] | 5 | 4 | 5 |
| K01400 | bacillolysin [EC:3.4.24.28] | 1 | 1 | 2 |
| K09607 | immune inhibitor A [EC:3.4.24.-] | 4 | 3 | 4 |
| K07284 | sortase A [EC:3.4.22.70] | 2 | 4 | 2 |
| K13275 | major intracellular serine protease [EC:3.4.21.-] | 3 | 1 | 1 |
| K20486 | lantibiotic leader peptide-processing serine protease [EC:3.4.21.-] | 2 | 0 | 0 |
| K01356 | repressor LexA [EC:3.4.21.88] | 2 | 3 | 2 |
| K01259 | proline iminopeptidase [EC:3.4.11.5] | 2 | 3 | 4 |
| Total | 99 | 94 | 91 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Wu, Y.; Yuan, J.; Yuan, J.; Wang, Z.; Gao, J.; Chen, H. Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides. Foods 2021, 10, 1725. https://doi.org/10.3390/foods10081725
Lu J, Wu Y, Yuan J, Yuan J, Wang Z, Gao J, Chen H. Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides. Foods. 2021; 10(8):1725. https://doi.org/10.3390/foods10081725
Chicago/Turabian StyleLu, Jun, Yong Wu, Juanli Yuan, Jin Yuan, Zhongliang Wang, Jinyan Gao, and Hongbing Chen. 2021. "Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides" Foods 10, no. 8: 1725. https://doi.org/10.3390/foods10081725
APA StyleLu, J., Wu, Y., Yuan, J., Yuan, J., Wang, Z., Gao, J., & Chen, H. (2021). Characterization of Bacillus cereus AFA01 Capable of Degrading Gluten and Celiac-Immunotoxic Peptides. Foods, 10(8), 1725. https://doi.org/10.3390/foods10081725