Response of Lactiplantibacillus plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of LAB Strains
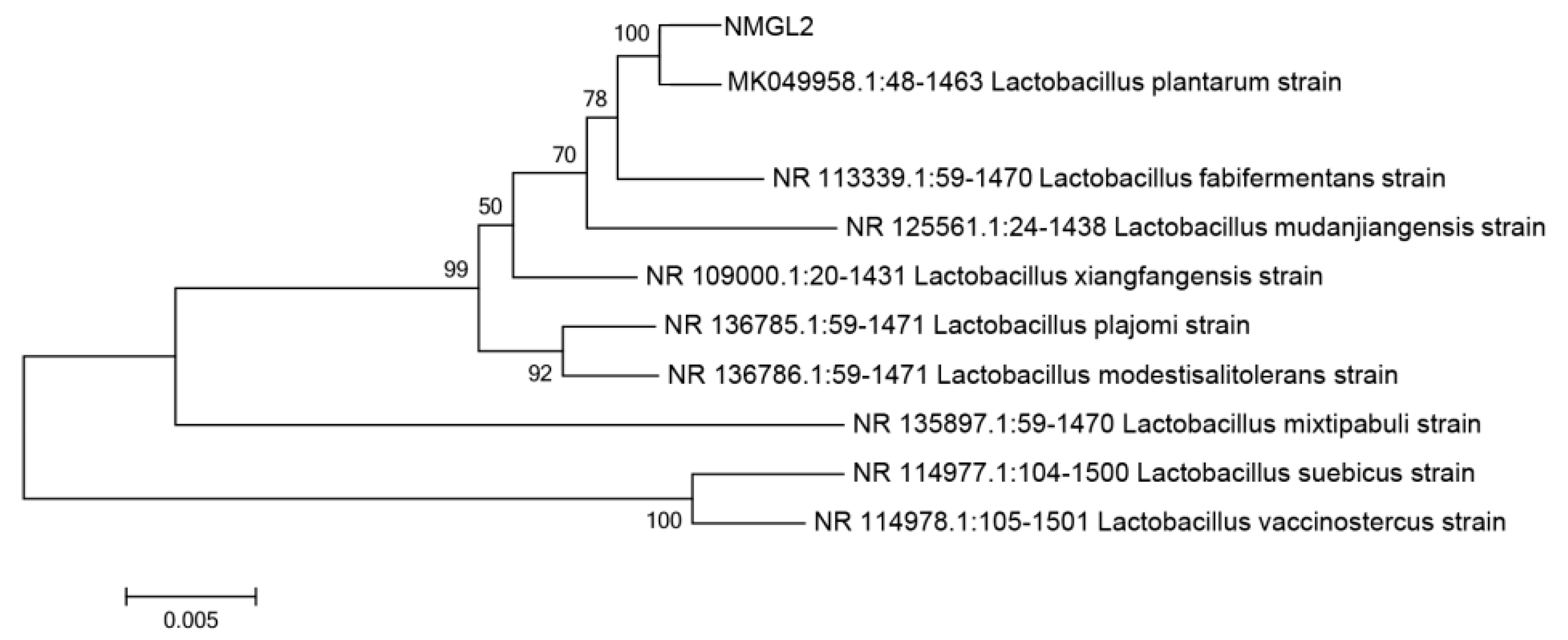
2.2. Identification of LAB Strains
2.3. Tolerance to Low Temperature
2.4. Tolerance to Low pH
2.5. Scanning Electron Microscopy
2.6. Preparation of Fermented Milk
2.7. Protein Extraction and Digestion
2.8. Liquid Chromatography-Mass Spectrometry Analysis
2.9. Data Analysis for LC-MS
2.10. Quantitative Data Analysis
2.11. Bioinformatics Analysis
3. Results and Discussion
3.1. Isolation and Identification of Lactic Acid Bacteria
3.2. Tolerance to Low-Temperature and Low pH
3.3. Morphological Change of Lpb. plantarum NMGL2 at Low Temperature and Low pH
3.4. Viability of Lpb. plantarum NMGL2 in Fermented Milk at Low Temperature
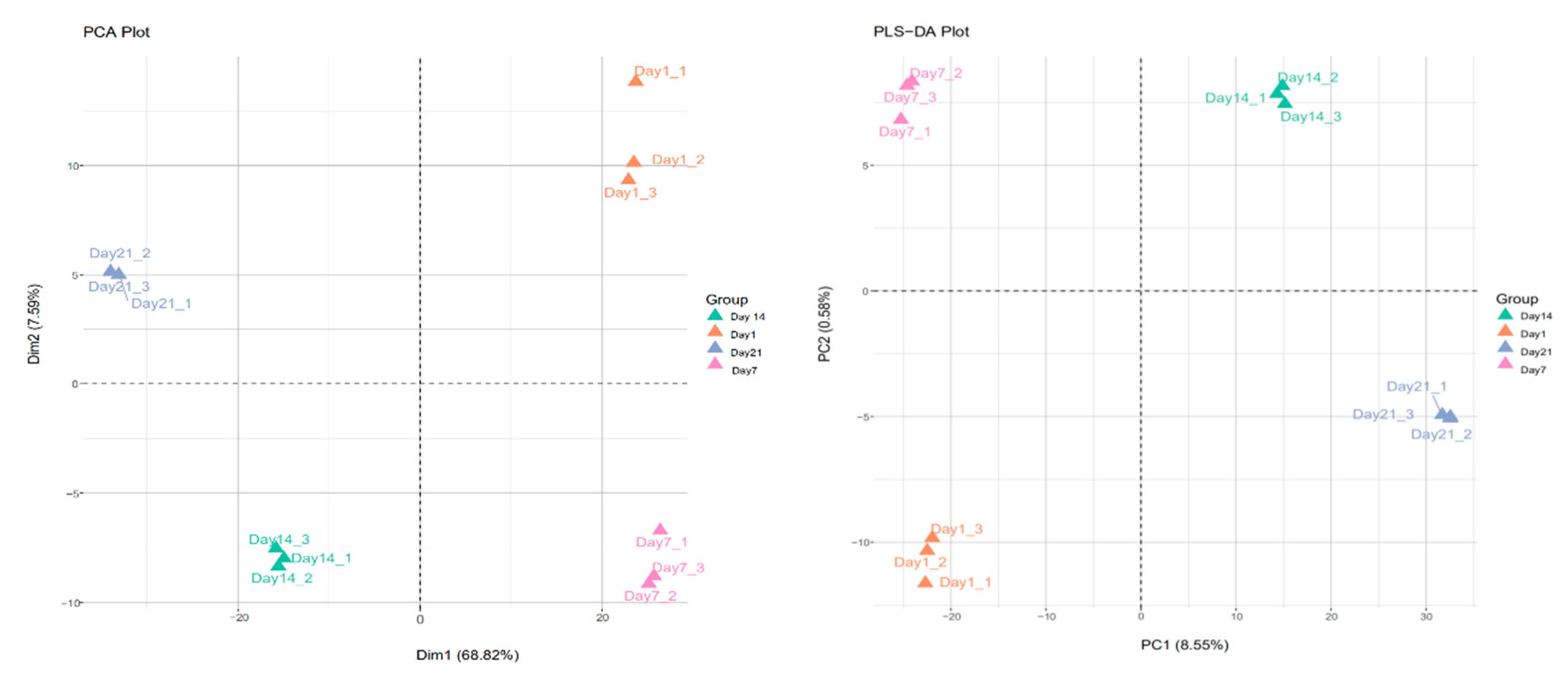
3.5. Proteomic Data Analysis by PCA and PLS-DA
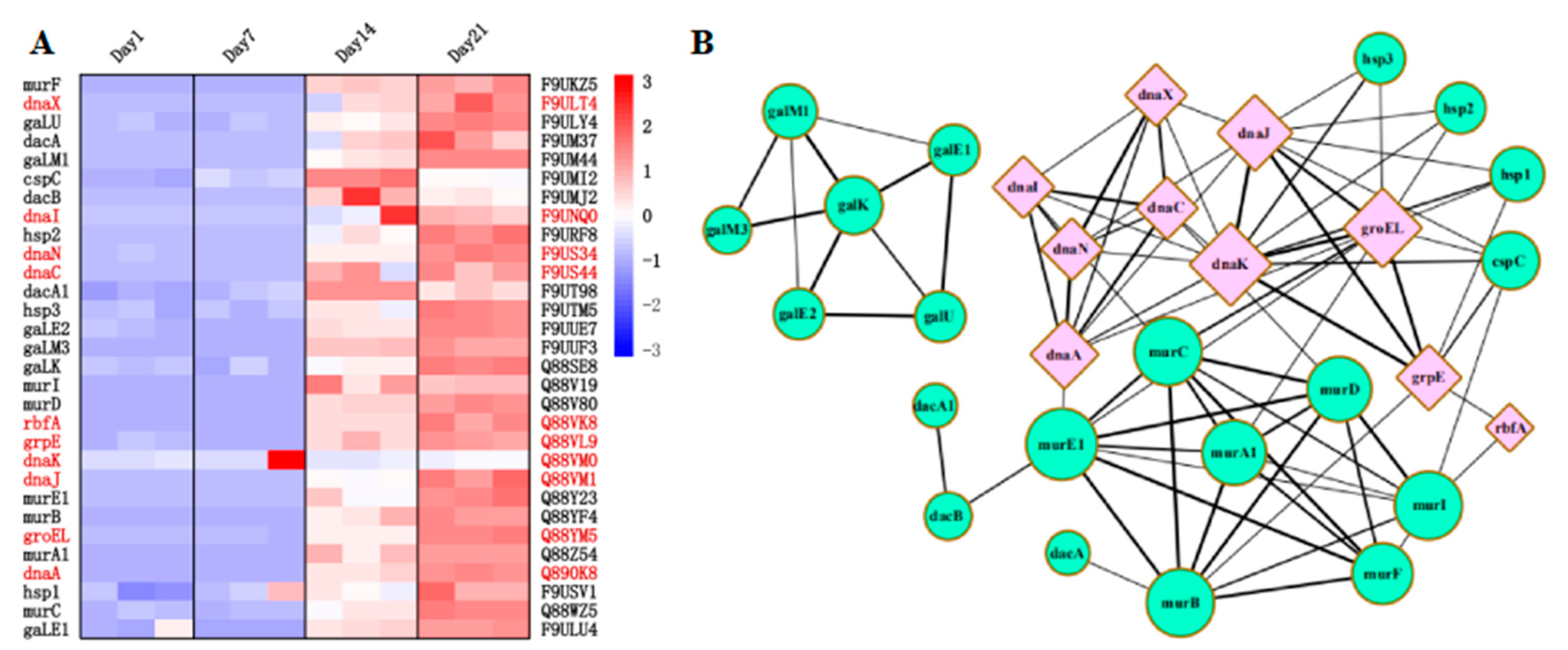
3.6. Differentially Expressed Proteins
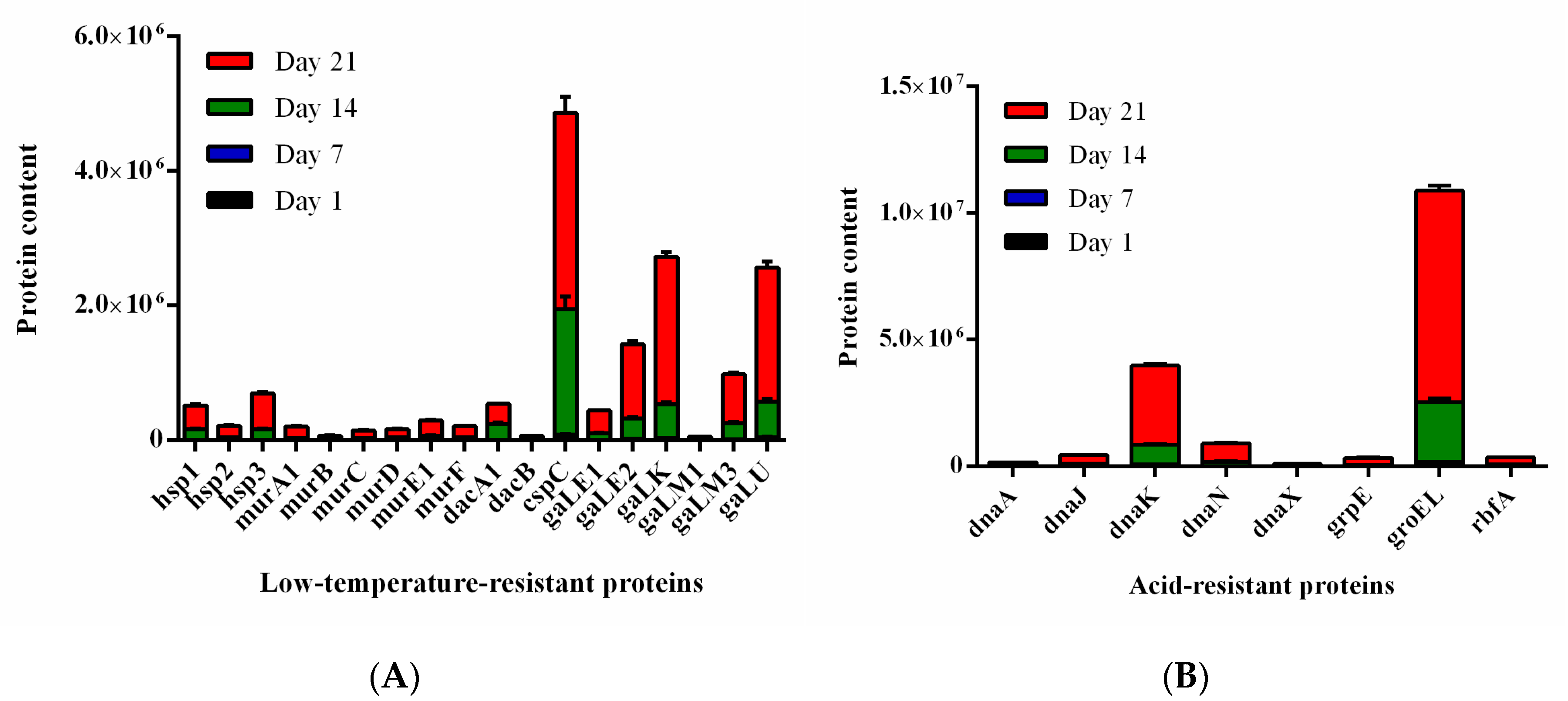
3.7. Screening of Low Temperature and Acid-Resistant Proteins
3.8. Parallel Reaction-Monitoring (PRM) Validation
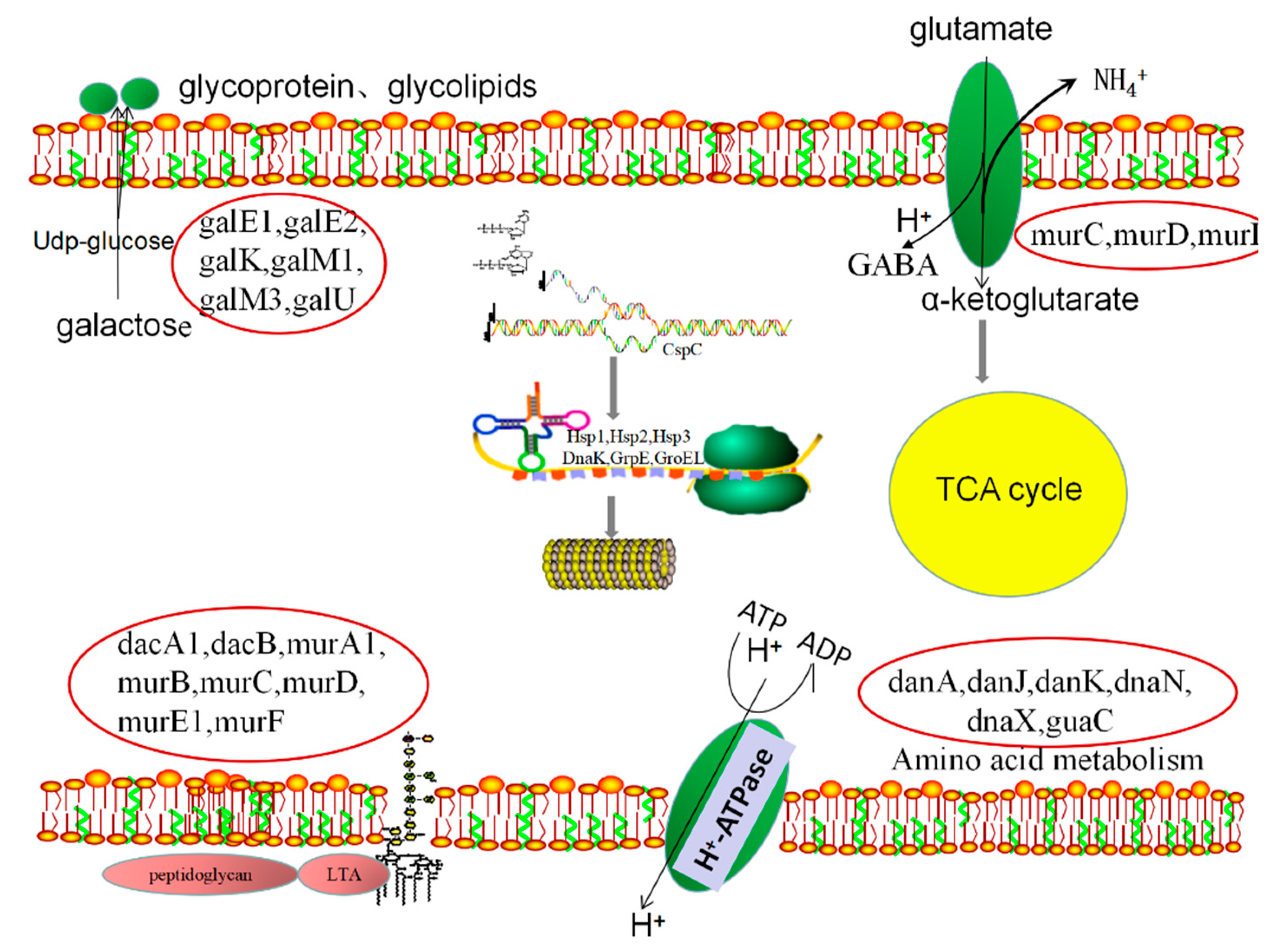
3.9. Metabolic Pathway Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, A.; Lee, Y.J.; Yoo, H.J.; Kim, M.; Chang, Y.; Lee, D.S.; Lee, J.H. Consumption of Dairy Yogurt Containing Lactobacillus paracasei ssp paracasei, Bifidobacterium animalis ssp lactis and Heat-Treated Lactobacillus plantarum Improves Immune Function Including Natural Killer Cell Activity. Nutrients 2017, 9, 558. [Google Scholar] [CrossRef]
- Pei, R.; Martin, D.A.; Dimarco, D.M.; Bolling, B.W. Evidence for the effects of yogurt on gut health and obesity. Crit. Rev. Food Sci. 2017, 57, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Mirghafourvand, M.; Rad, A.H.; Alizadeh, S.M.C.; Fardiazar, Z.; Shokri, K. The Effect of Probiotic Yogurt on Constipation in Pregnant Women: A Randomized Controlled Clinical Trial. Iran. Red Crescent Soc. 2016, 18, e39870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamani, B.; Golkar, H.R.; Farshbaf, S.; Emadi-Baygi, M.; Tajabadi-Ebrahimi, M.; Jafari, P.; Akhavan, R.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z. Clinical and metabolic response to probiotic supplementation in patients with rheumatoid arthritis: A randomized, double-blind, placebo-controlled trial. Int. J. Rheum. Dis. 2016, 19, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Akkasheh, G.; Kashani-Poor, Z.; Tajabadi-Ebrahimi, M.; Jafari, P.; Akbari, H.; Taghizadeh, M.; Memarzadeh, M.R.; Asemi, Z.; Esmaillzadeh, A. Clinical and metabolic response to probiotic administration in patients with major depressive disorder: A randomized, double-blind, placebo-controlled trial. Nutrition 2016, 32, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Passariello, A.; Agricole, P.; Malfertheiner, P. A critical appraisal of probiotics (as drugs or food supplements) in gastrointestinal diseases. Curr. Med. Res. Opin. 2014, 30, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Bae, D.; Lim, K.; Griffiths, M.W.; Oh, S. Cold stress improves the ability of Lactobacillus plantarum L67 to survive freezing. Int. J. Food Microbiol. 2014, 191, 135–143. [Google Scholar] [CrossRef]
- Chen, M.; Tang, H.; Chiang, M. Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 2017, 66, 20–27. [Google Scholar] [CrossRef]
- Cavicchioli, R.; Siddiqui, K.S.; Andrews, D.; Sowers, K.R. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 2002, 13, 253–261. [Google Scholar] [CrossRef]
- Berger, F.; Morellet, N.; Menu, F.; Potier, P. Cold shock and cold acclimation proteins in the psychrotrophic bacterium Arthrobacter globiformis SI55. J. Bacteriol. 1996, 178, 2999–3007. [Google Scholar] [CrossRef] [Green Version]
- Tribelli, P.M.; Lopez, N.I. Reporting Key Features in Cold-Adapted Bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Dhaulaniya, A.S.; Balan, B.; Kumar, M.; Agrawal, P.K.; Singh, D.K. Cold survival strategies for bacteria, recent advancement and potential industrial applications. Arch. Microbiol. 2019, 201, 1–16. [Google Scholar] [CrossRef]
- Vrancken, G.; Rimaux, T.; Weckx, S.; De Vuyst, L.; Leroy, F. Environmental pH determines citrulline and ornithine release through the arginine deiminase pathway in Lactobacillus fermentum IMDO 130101. Int. J. Food Microbiol. 2009, 135, 216–222. [Google Scholar] [CrossRef]
- Desmond, C.; Fitzgerald, G.F.; Stanton, C.; Ross, R.P. Improved stress tolerance of GroESL-overproducing Lactococcus lactis and orobiotic Lactobacillus paracasei NFBC 338. Appl. Environ. Microb. 2004, 70, 5929–5936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez Montoro, B.; Benomar, N.; Caballero Gomez, N.; Ennahar, S.; Horvatovich, P.; Knapp, C.W.; Galvez, A.; Abriouel, H. Proteomic analysis of Lactobacillus pentosus for the identification of potential markers involved in acid resistance and their influence on other probiotic features. Food Microbiol. 2018, 72, 31–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, H.; Liu, J.; Miao, S.; Zhao, Y.; Zhu, H.; Qiao, M.; Saris, P.E.J.; Qiao, J. Contribution of YthA, a PspC Family Transcriptional Regulator of Lactococcus lactis F44 Acid Tolerance and Nisin Yield: A Transcriptomic Approach. Appl. Environ. Microb. 2018, 84, e02483-17. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.; Zhang, W.; Sun, T.; Wu, J.; Yue, X.; Meng, H.; Zhang, H. Proteomic analysis of responses of a new probiotic bacterium Lactobacillus casei Zhang to low acid stress. Int. J. Food Microbiol. 2011, 147, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Rho, B.; Pi, K.; Kim, H.; Choi, Y. Proteomic analysis of protein expression in Lactobacillus plantarum in response to alkaline stress. J. Biotechnol. 2011, 153, 1–7. [Google Scholar] [CrossRef]
- Koponen, J.; Laakso, K.; Koskenniemi, K.; Kankainen, M.; Savijoki, K.; Nyman, T.A.; de Vos, W.M.; Tynkkynen, S.; Kalkkinen, N.; Varmanen, P. Effect of acid stress on protein expression and phosphorylation in Lactobacillus rhamnosus GG. J. Proteom. 2012, 75, 1357–1374. [Google Scholar] [CrossRef]
- Sanchez, B.; Champomier-Verges, M.; Collado, M.D.C.; Anglade, P.; Baraige, F.; Sanz, Y.; Reyes-Gavilan, C.G.D.L.; Margolles, A.; Zagorec, M. Low-pH adaptation and the acid tolerance response of Bifidobactetium longum biotype longum. Appl. Environ. Microb. 2007, 73, 6450–6459. [Google Scholar] [CrossRef] [Green Version]
- Chongde, W.; Juan, Z.; Wei, C.; Miao, W.; Guocheng, D.; Jian, C. A combined physiological and proteomic approach to reveal lactic-acid-induced alterations in Lactobacillus casei Zhang and its mutant with enhanced lactic acid tolerance. Appl. Microbiol. Biotechnol. 2012, 93, 707–722. [Google Scholar]
- Behera, S.S.; Ray, R.C.; Zdolec, N. Lactobacillus plantarum with Functional Properties: An Approach to Increase Safety and Shelf-Life of Fermented Foods. Biomed. Res. Int. 2018, 2018, 9361614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.L.; Martoni, C.J.; Parent, M.; Prakash, S. Cholesterol-lowering efficacy of a microencapsulated bile salt hydrolase-active Lactobacillus reuteri NCIMB 30242 yoghurt formulation in hypercholesterolaemic adults. Brit. J. Nutr. 2012, 107, 1505–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Jin, M.; Meng, J.; Gao, S.; Lu, R. Exopolysaccharide from Lactobacillus planterum LP6: Antioxidation and the effect on oxidative stress. Carbohyd. Polym. 2013, 98, 1147–1152. [Google Scholar] [CrossRef]
- Belkacem-Hanfi, N.; Fhoula, I.; Semmar, N.; Guesmi, A.; Perraud-Gaime, I.; Ouzari, H.; Boudabous, A.; Roussos, S. Lactic acid bacteria against post-harvest moulds and ochratoxin A isolated from stored wheat. Biol. Control. 2014, 76, 52–59. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, W.; Guo, X.; Guo, T.; Zheng, Y.; Wang, Y.; Hao, Y.; Yang, Z. Survival and Effect of Exopolysaccharide-Producing Lactobacillus plantarum YW11 on the Physicochemical Properties of Ice Cream. Pol. J. Food Nutr. Sci. 2017, 67, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Na, L.; Song, M.; Likang, Q. Screening and application of lactic acid bacteria and yeasts with L-lactic acid-producing and antioxidant capacity in traditional fermented rice acid. Food Sci. Nutr. 2020, 8, 6095–6111. [Google Scholar]
- Tratsiak, K.; Prudnikova, T.; Drienovska, I.; Damborsky, J.; Brynda, J.; Pachl, P.; Kuty, M.; Chaloupkova, R.; Rezacova, P.; Smatanova, I.K. Crystal structure of the cold-adapted haloalkane dehalogenase DpcA from Psychrobacter cryohalolentis K5. Acta Cryst. Sect. F-Struct. Biol. Commun. 2019, 75, 324–331. [Google Scholar] [CrossRef]
- Pinto, A.; Barbosa, J.; Albano, H.; Isidro, J.; Teixeira, P. Screening of Bacteriocinogenic Lactic Acid Bacteria and Their Characterization as Potential Probiotics. Microorganisms 2020, 8, 393. [Google Scholar] [CrossRef] [Green Version]
- Hamon, E.; Horvatovich, P.; Marchioni, E.; Aoude-Werner, D.; Ennahar, S. Investigation of potential markers of acid resistance in Lactobacillus plantarum by comparative proteomics. J. Appl. Microbiol. 2014, 116, 134–144. [Google Scholar] [CrossRef]
- Onyango, L.A.; Dunstan, R.H.; Gottfries, J.; von Eiff, C.; Roberts, T.K. Effect of Low Temperature on Growth and Ultra-Structure of Staphylococcus spp. PLoS ONE 2012, 7, e29031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hidalgo-Cantabrana, C.; Sanchez, B.; Milani, C.; Ventura, M.; Margolles, A.; Ruas-Madiedo, P. Genomic Overview and Biological Functions of Exopolysaccharide Biosynthesis in Bifidobacterium spp. Appl. Environ. Microb. 2014, 80, 9–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; He, M.; Wu, C. Cross protection of lactic acid bacteria during environmental stresses: Stress responses and underlying mechanisms. LWT-Food Sci. Technol. 2021, 144. [Google Scholar] [CrossRef]
- Sanchez, B.; Champomier-Verges, M.; Stuer-Lauridsen, B.; Ruas-Madiedo, P.; Anglade, P.; Baraige, F.; de Los Reyes-Gavilan, C.G.; Johansen, E.; Zagorec, M.; Margolles, A. Adaptation and response of Bifidobacterium animalis subsp lactis to bile: A proteomic and physiological approach. Appl. Environ. Microb. 2007, 73, 6757–6767. [Google Scholar] [CrossRef] [Green Version]
- Smart, J.B.; Thomas, T.D. Effect of oxygen on lactose metabolism in lactic streptococci. Appl. Environ. Microb. 1987, 53, 533–541. [Google Scholar] [CrossRef] [Green Version]
- Polo, L.; Mañes-Lázaro, R.; Olmeda, I.; Cruz-Pio, L.E.; Medina, Á.; Ferrer, S.; Pardo, I. Influence of freezing temperatures prior to freeze-drying on viability of yeasts and lactic acid bacteria isolated from wine. J. Appl. Microbiol. 2017, 122, 1603–1614. [Google Scholar] [CrossRef] [Green Version]
- Graumann, P.; Marahiel, M.A. Some like it cold: Response of microorganisms to cold shock. Arch. Microbiol. 1996, 166, 293–300. [Google Scholar] [CrossRef]
- Ma, J.; Xu, C.; Liu, F.; Hou, J.; Shao, H.; Yu, W. Stress adaptation and cross-protection of Lactobacillus plantarum KLDS 1.0628. CyTA-J. Food 2021, 19, 72–80. [Google Scholar] [CrossRef]
- Matsui, T.; Ichikawa, H.; Fujita, T.; Takagi, T.; Osada-Oka, M.; Minamiyama, Y. Histidine deficiency attenuates cell viability in rat intestinal epithelial cells by apoptosis via mitochondrial dysfunction. J. Nutr. Intermed. Metab. 2017, 8, 21–28. [Google Scholar] [CrossRef]



| OD600 nm Value | OD600 nm Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Incubation Time | 24 h | 48 h | 72 h | Incubation Time | 24 h | 48 h | 72 h | ||||||
| Incubation Temperature | 4 °C | 10 °C | 4 °C | 10 °C | 4 °C | 10 °C | Incubation Temperature | 4 °C | 10 °C | 4 °C | 10 °C | 4 °C | 10 °C |
| Lpb. plantarum NMGL2 | 0.12 ± 0.02 | 0.20 ± 0.02 | 0.25 ± 0.03 | 0.90 ± 0.04 | 0.68 ± 0.04 | 1.11 ± 0.03 | Lpb. plantarum P4 | 0.12 ± 0.02 | 0.15 ± 0.03 | 0.22 ± 0.03 | 0.72 ± 0.04 | 0.33 ± 0.02 | 0.99 ± 0.04 |
| Lcb. casei JS2 | 0.12 ± 0.03 | 0.17 ± 0.01 | 0.22 ± 0.02 | 0.65 ± 0.03 | 0.46 ± 0.03 | 0.86 ± 0.03 | Lpb. plantarum PC10 | 0.12 ± 0.03 | 0.16 ± 0.04 | 0.18 ± 0.02 | 0.83 ± 0.03 | 0.31 ± 0.04 | 1.08 ± 0.03 |
| Lpb. plantarum P5 | 0.12 ± 0.02 | 0.15 ± 0.02 | 0.22 ± 0.03 | 0.80 ± 0.04 | 0.45 ± 0.03 | 1.04 ± 0.02 | Lpb. plantarum P7 | 0.12 ± 0.02 | 0.16 ± 0.04 | 0.21 ± 0.04 | 0.33 ± 0.02 | 0.27 ± 0.01 | 0.45 ± 0.02 |
| Lab. brevis PC7 | 0.10 ± 0.02 | 0.13 ± 0.03 | 0.21 ± 0.01 | 0.64 ± 0.04 | 0.45 ± 0.02 | 0.87 ± 0.04 | Lab. brevis PC2 | 0.10 ± 0.02 | 0.13 ± 0.04 | 0.21 ± 0.04 | 0.41 ± 0.03 | 0.27 ± 0.03 | 0.59 ± 0.04 |
| Lpb. plantarum NMGL1 | 0.13 ± 0.04 | 0.16 ± 0.02 | 0.22 ± 0.02 | 0.37 ± 0.04 | 0.41 ± 0.02 | 0.91 ± 0.03 | Lab. brevis PC6 | 0.12 ± 0.01 | 0.10 ± 0.03 | 0.21 ± 0.02 | 0.37 ± 0.02 | 0.27 ± 0.03 | 0.48 ± 0.04 |
| Lcb. casei JS1 | 0.12 ± 0.02 | 0.16 ± 0.01 | 0.21 ± 0.04 | 0.66 ± 0.02 | 0.37 ± 0.02 | 0.84 ± 0.02 | Lpb. plantarum P3 | 0.11 ± 0.02 | 0.11 ± 0.03 | 0.20 ± 0.03 | 0.51 ± 0.04 | 0.23 ± 0.04 | 0.79 ± 0.03 |
| Lab. brevis PC5 | 0.11 ± 0.01 | 0.13 ± 0.04 | 0.21 ± 0.04 | 0.66 ± 0.02 | 0.36 ± 0.02 | 0.93 ± 0.03 | Lab. brevis PC3 | 0.10 ± 0.01 | 0.13 ± 0.02 | 0.20 ± 0.02 | 0.37 ± 0.04 | 0.22 ± 0.03 | 0.51 ± 0.03 |
| Lpb. pentosus PC11 | 0.12 ± 0.04 | 0.17 ± 0.03 | 0.23 ± 0.04 | 0.77 ± 0.01 | 0.35 ± 0.01 | 0.99 ± 0.02 | Lab. brevis PC8 | 0.10 ± 0.01 | 0.11 ± 0.04 | 0.20 ± 0.04 | 0.27 ± 0.03 | 0.19 ± 0.04 | 0.40 ± 0.04 |
| Lpb. plantarum P6 | 0.12 ± 0.01 | 0.15 ± 0.03 | 0.22 ± 0.02 | 0.73 ± 0.03 | 0.35 ± 0.01 | 0.94 ± 0.01 | Lab. brevis PC9 | 0.10 ± 0.03 | 0.13 ± 0.02 | 0.20 ± 0.01 | 0.37 ± 0.02 | 0.17 ± 0.02 | 0.51 ± 0.02 |
| Lpb. plantarum P1 | 0.12 ± 0.02 | 0.15 ± 0.01 | 0.22 ± 0.03 | 0.67 ± 0.03 | 0.34 ± 0.02 | 0.88 ± 0.01 | Lpb. plantarum P2 | 0.12 ± 0.01 | 0.13 ± 0.01 | 0.12 ± 0.02 | 0.14 ± 0.04 | 0.13 ± 0.02 | 0.14 ± 0.01 |
| Lpb. plantarum NMGL3 | 0.14 ± 0.01 | 0.17 ± 0.02 | 0.25 ± 0.01 | 0.70 ± 0.04 | 0.34 ± 0.02 | 0.97 ± 0.02 | |||||||
| OD600 nm Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | 6 | 4 | 3 | |||||||||
| Incubation temperatures (h) | 4 | 8 | 12 | 4 | 8 | 12 | 16 | 4 | 8 | 12 | 16 | 20 |
| Lpb. plantarum NMGL2 | 0.86 ± 0.02 | 1.29 ± 0.01 | 1.44 ± 0.02 | 0.35 ± 0.01 | 0.67 ± 0.02 | 0.92 ± 0.01 | 1.22 ± 0.02 | 0.19 ± 0.01 | 0.25 ± 0.02 | 0.31 ± 0.02 | 0.34 ± 0.02 | 0.47 ± 0.02 |
| Lcb. casei JS2 | 0.74 ± 0.01 | 1.21 ± 0.02 | 1.39 ± 0.02 | 0.33 ± 0.02 | 0.62 ± 0.01 | 0.88 ± 0.02 | 1.24 ± 0.03 | 0.17 ± 0.02 | 0.18 ± 0.01 | 0.21 ± 0.01 | 0.45 ± 0.01 | 0.48 ± 0.03 |
| Lpb. plantarum P5 | 0.74 ± 0.02 | 1.15 ± 0.02 | 1.40 ± 0.01 | 0.31 ± 0.02 | 0.56 ± 0.01 | 0.76 ± 0.02 | 1.07 ± 0.02 | 0.16 ± 0.01 | 0.18 ± 0.02 | 0.19 ± 0.02 | 0.25 ± 0.01 | 0.27 ± 0.02 |
| Lab. brevis PC7 | 0.25 ± 0.03 | 0.50 ± 0.01 | 0.82 ± 0.02 | 0.12 ± 0.03 | 0.19 ± 0.02 | 0.30 ± 0.01 | 0.40 ± 0.01 | 0.10 ± 0.01 | 0.10 ± 0.02 | 0.10 ± 0.01 | 0.12 ± 0.02 | 0.11 ± 0.03 |
| Lpb. plantarum NMGL1 | 0.88 ± 0.02 | 1.33 ± 0.02 | 1.50 ± 0.03 | 0.41 ± 0.02 | 0.74 ± 0.02 | 0.94 ± 0.02 | 1.28 ± 0.02 | 0.18 ± 0.02 | 0.21 ± 0.01 | 0.25 ± 0.02 | 0.34 ± 0.01 | 0.38 ± 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yao, M.; Lai, T.; Zhao, H.; Wang, Y.; Yang, Z. Response of Lactiplantibacillus plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics. Foods 2021, 10, 1514. https://doi.org/10.3390/foods10071514
Zhang M, Yao M, Lai T, Zhao H, Wang Y, Yang Z. Response of Lactiplantibacillus plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics. Foods. 2021; 10(7):1514. https://doi.org/10.3390/foods10071514
Chicago/Turabian StyleZhang, Min, Mengke Yao, Tiantian Lai, Hua Zhao, Yihui Wang, and Zhennai Yang. 2021. "Response of Lactiplantibacillus plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics" Foods 10, no. 7: 1514. https://doi.org/10.3390/foods10071514
APA StyleZhang, M., Yao, M., Lai, T., Zhao, H., Wang, Y., & Yang, Z. (2021). Response of Lactiplantibacillus plantarum NMGL2 to Combinational Cold and Acid Stresses during Storage of Fermented Milk as Analyzed by Data-Independent Acquisition Proteomics. Foods, 10(7), 1514. https://doi.org/10.3390/foods10071514






