A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food-Pathogen Salmonella enterica
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Selection
2.2. Medium and Reagents
2.3. Antibacterial Activity Assay
2.4. Inhibition of Biofilm Formation
2.5. Antimicrobial Resistance Profile
2.6. Cytotoxicity Assays on Mammalian Cells
2.7. Statistical Analysis
3. Results and Discussion
3.1. Antimicrobial Activity of 1018-K6 on Planktonic Salmonella enterica Cells and Salmonella Resistance Profile
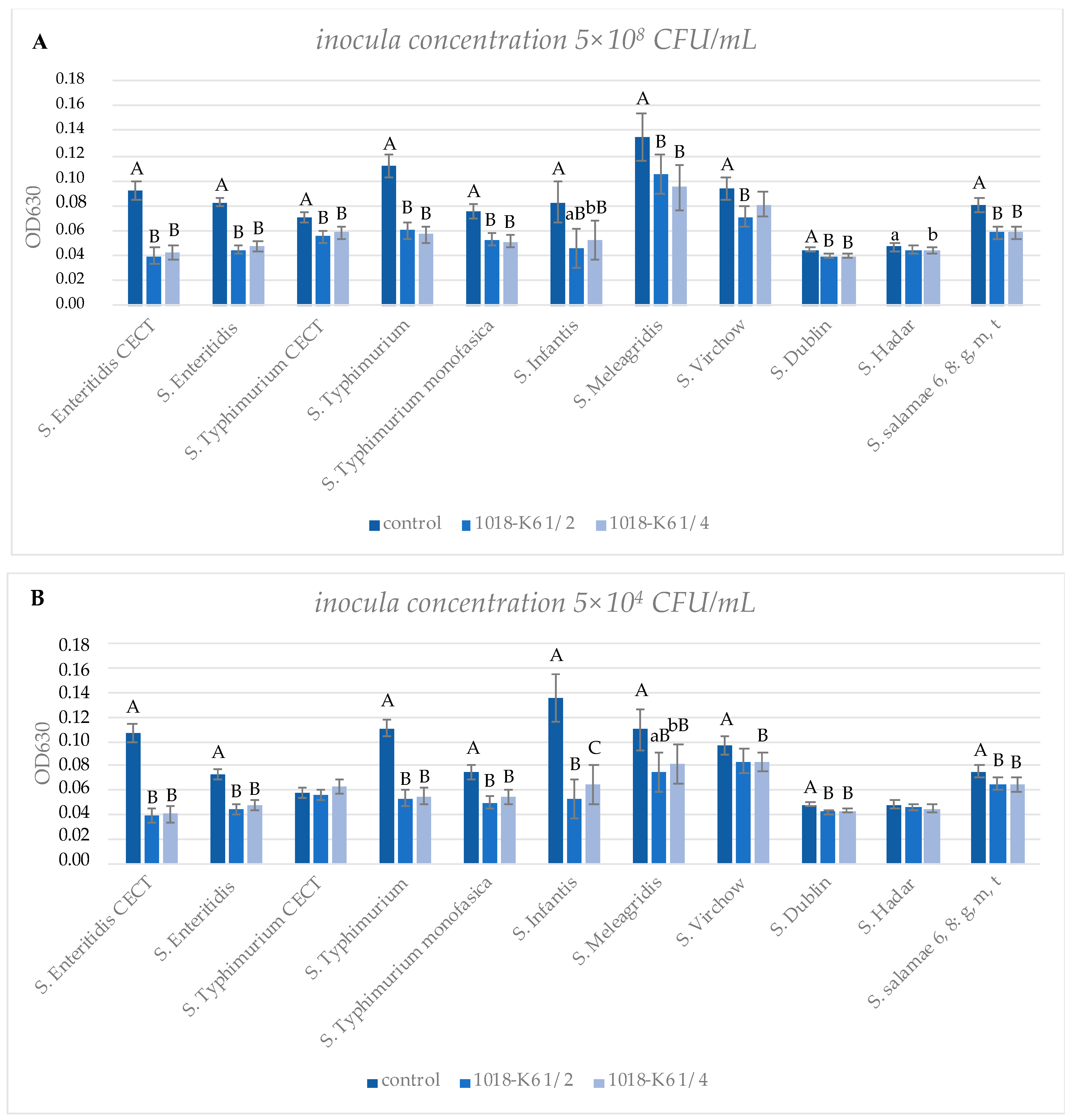
3.2. Evaluation of the Activity of 1018-K6 against Biofilm Formation of S. enterica
3.3. Evaluation of Cytotoxic Effects of 1018-K6 on Mammalian Fibroblast Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2019 Zoonoses Report. EFSA J. 2021, 19, 6406. [Google Scholar] [CrossRef]
- Parry, C.M.; Threlfall, E.J. Antimicrobial resistance in typhoidal and nontyphoidal salmonellae. Curr. Opin. Infect. Dis. 2008, 21, 531–538. [Google Scholar] [CrossRef]
- Anjum, M.F.; Choudhary, S.; Morrison, V.; Snow, L.C.; Mafura, M.; Slickers, P.; Ehricht, R.; Woodward, M.J. Identifying an-timicrobial resistance genes of human clinical relevance within Salmonella isolated from food animals in Great Britain. J. Antimicrob. Chemother. 2011, 66, 550–559. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hur, J.; Jawale, C.; Lee, J.H. Antimicrobial resistance of Salmonella isolated from food animals: A review. Food Res. Int. 2012, 45, 819–830. [Google Scholar] [CrossRef]
- European Food Safety Authority, and European Centre for Disease Prevention and Control. The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2018/2019. EFSA J. 2021, 19, e06490. [Google Scholar] [CrossRef]
- Pateiro, M.; Munekata, P.E.S.; Sant’Ana, A.S.; Domínguez, R.; Rodríguez-Lázaro, D.; Lorenzo, J.M. Application of essential oils as antimicrobial agents against spoilage and pathogenic microorganisms in meat products. Int. J. Food Microbiol. 2021, 337, 108966. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Duguid, J.P.; Anderson, E.S.; Campbell, I. Fimbriae and adhesive properties in salmonellae. J. Pathol. Bacteriol. 1966, 92, 107–137. [Google Scholar] [CrossRef]
- Wang, H.; Ye, K.; Wei, X.; Cao, J.; Xu, X.; Zhou, G. Occurrence, antimicrobial resistance and biofilm formation of Salmonella isolates from a chicken slaughter plant in China. Food Control. 2013, 33, 378–384. [Google Scholar] [CrossRef]
- Schonewille, E.; Nesse, L.L.; Hauck, R.; Windhorst, D.; Hafez, H.M.; Vestby, L.K. Biofilm building capacity of Salmonella enterica strains from the poultry farm environment. FEMS Immunol. Med. Microbiol. 2012, 65, 360–365. [Google Scholar] [CrossRef]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Zharkova, M.S.; Orlov, D.S.; Golubeva, O.Y.; Chakchir, O.B.; Eliseev, I.E.; Grinchuk, T.M.; Shamova, O.V. Application of an-timicrobial peptides of the innate immune system in combination with conventional antibiotics—A novel way to combat antibiotic resistance? Front. Cell. Infect. Microbiol. 2019, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Mika, J.T.; Moiset, G.; Cirac, A.D.; Feliu, L.; Bardají, E.; Planas, M.; Sengupta, D.; Marrink, S.; Poolman, B. Structural basis for the enhanced activity of cyclic antimicrobial peptides: The case of BPC194. Biochim. Biophys. Acta Biomembr. 2011, 1808, 2197–2205. [Google Scholar] [CrossRef]
- Maróti, G.; Kereszt, A.; Kondorosi, É.; Mergaert, P. Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 2011, 162, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Genet. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nat. Cell Biol. 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Yeaman, M.R.; Yount, N.; Hauger, R.L.; Grigoriadis, D.E.; Dallman, M.F.; Plotsky, P.M.; Vale, W.W.; Dautzenberg, F.M. Mechanisms of Antimicrobial Peptide Action and Resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef]
- Starr, C.G.; Ghimire, J.; Guha, S.; Hoffmann, J.P.; Wang, Y.; Sun, L.; Landreneau, B.N.; Kolansky, Z.D.; Kilanowski-Doroh, I.M.; Sammarco, M.C.; et al. Synthetic molecular evolution of host cell-compatible, antimicrobial peptides effective against drug-resistant, biofilm-forming bacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 8437–8448. [Google Scholar] [CrossRef]
- Palmieri, G.; Tatè, R.; Gogliettino, M.; Balestrieri, M.; Rea, I.; Terracciano, M.; Proroga, Y.T.; Capuano, F.; Anastasio, A.; De Stefano, L. Small Synthetic Peptides Bioconjugated to Hybrid Gold Nanoparticles Destroy Potentially Deadly Bacteria at Submicromolar Concentrations. Bioconj. Chem. 2018, 29, 3877–3885. [Google Scholar] [CrossRef]
- Palmieri, G.; Balestrieri, M.; Capuano, F.; Proroga, Y.T.; Pomilio, F.; Centorame, P.; Riccio, A.; Marrone, R.; Anastasio, A. Bactericidal and antibiofilm activity of bactenecin-derivative peptides against the food-pathogen Listeria monocytogenes: New perspectives for food processing industry. Int. J. Food Microbiol. 2018, 279, 33–42. [Google Scholar] [CrossRef]
- Colagiorgi, A.; Festa, R.; Di Ciccio, P.A.; Gogliettino, M.; Balestrieri, M.; Palmieri, G.; Anastasio, A.; Ianieri, A. Rapid biofilm eradication of the antimicrobial peptide 1018-K6 against Staphylococcus aureus: A new potential tool to fight bacterial biofilms. Food Control. 2020, 107, 106815. [Google Scholar] [CrossRef]
- CLSI. M100-S25: Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; Informational Supplement; CLSI: Wayne, PA, USA, 2020. [Google Scholar]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, 2437. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to anti-microbial peptides. Drug Resist. Updat. 2016, 26, 43–57. [Google Scholar] [CrossRef]
- Yazgan, H.; Ozogul, Y.; Kuley, E. Antimicrobial influence of nanoemulsified lemon essential oil and pure lemon essential oil on food-borne pathogens and fish spoilage bacteria. Int. J. Food Microbiol. 2019, 306, 108266. [Google Scholar] [CrossRef]
- Lamas, A.; Miranda, J.M.; Regal, P.; Vazquez, B.; Franco, C.M.; Cepeda, A. A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiol. Res. 2018, 206, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Galiè, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 2018, 9, 898. [Google Scholar] [CrossRef] [PubMed]
- Lamas, A.; Miranda, J.; Vazquez, B.; Cepeda, A.; Franco, C. Biofilm formation, phenotypic production of cellulose and gene expression in Salmonella enterica decrease under anaerobic conditions. Int. J. Food Microbiol. 2016, 238, 63–67. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Komoda, E.; Ono, K.; Kumagai, S. Survival of biofilm-forming Salmonella on stainless steel bolt threads under dry conditions. J. Food Hyg. Soc. Jpn. 2011, 52, 299–303. [Google Scholar] [CrossRef]
- Marin, C.; Hernandiz, A.; Lainez, M. Biofilm development capacity of Salmonella strains isolated in poultry risk factors and their resistance against disinfectants. Poult. Sci. 2009, 88, 424–431. [Google Scholar] [CrossRef]
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef]
- Lamas, A.; Fernandez-No, I.C.; Miranda, J.M.; Vázquez, B.; Cepeda, A.; Franco, C.M. Biofilm Formation and Morphotypes of Salmonella enterica subsp. arizonae Differs from Those of Other Salmonella enterica Subspecies in Isolates from Poultry Houses. J. Food Prot. 2016, 79, 1127–1134. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhao, X.; Xia, X.; Zhu, C.; Qin, W.; Xu, Y.; Hang, B.; Sun, Y.; Chen, S.; Zhang, H.; et al. Antimicrobial Peptide JH-3 Effectively Kills Salmonella enterica Serovar Typhimurium Strain CVCC541 and Reduces Its Pathogenicity in Mice. Probiotics Antimicrob. Proteins 2019, 11, 1379–1390. [Google Scholar] [CrossRef] [PubMed]
- Dipaolo, J.A.; Takano, K.; Popescu, N.C. Quantitation of chemically induced neoplastic transformation of BALB-3T3 cloned cell lines. Cancer Res. 1972, 32, 2686–2695. [Google Scholar]
- Quarles, J.M.; Tennant, R.W. Effects of nitrosocarbaryl on BALB/3T3 cells. Cancer Res. 1975, 35, 2637–2645. [Google Scholar] [PubMed]
- Laverty, G.; Gorman, S.P.; Gilmore, B.F. The Potential of Antimicrobial Peptides as Biocides. Int. J. Mol. Sci. 2011, 12, 6566–6596. [Google Scholar] [CrossRef] [PubMed]
- Agrillo, B.; Balestrieri, M.; Gogliettino, M.; Palmieri, G.; Moretta, R.; Proroga, Y.T.; Rea, I.; Cornacchia, A.; Capuano, F.; Smaldone, G.; et al. Functionalized Polymeric Materials with Bio-Derived Antimicrobial Peptides for “Active” Packaging. Int. J. Mol. Sci. 2019, 20, 601. [Google Scholar] [CrossRef]
- Gogliettino, M.; Balestrieri, M.; Ambrosio, R.L.; Anastasio, A.; Smaldone, G.; Proroga, Y.T.R.; Moretta, R.; Rea, I.; De Stefano, L.; Agrillo, B.; et al. Extending the Shelf-Life of Meat and Dairy Products via PET-Modified Packaging Activated with the Antimicrobial Peptide MTP1. Front. Microbiol. 2020, 10, 2963. [Google Scholar] [CrossRef]
- Guo, Y.; Xun, M.; Han, J. A bovine myeloid antimicrobial peptide (BMAP-28) and its analogs kill pan-drug-resistant acinetobacter baumannii by interacting with outer membrane protein A (OmpA). Medicine 2018, 97, e12832. [Google Scholar] [CrossRef] [PubMed]
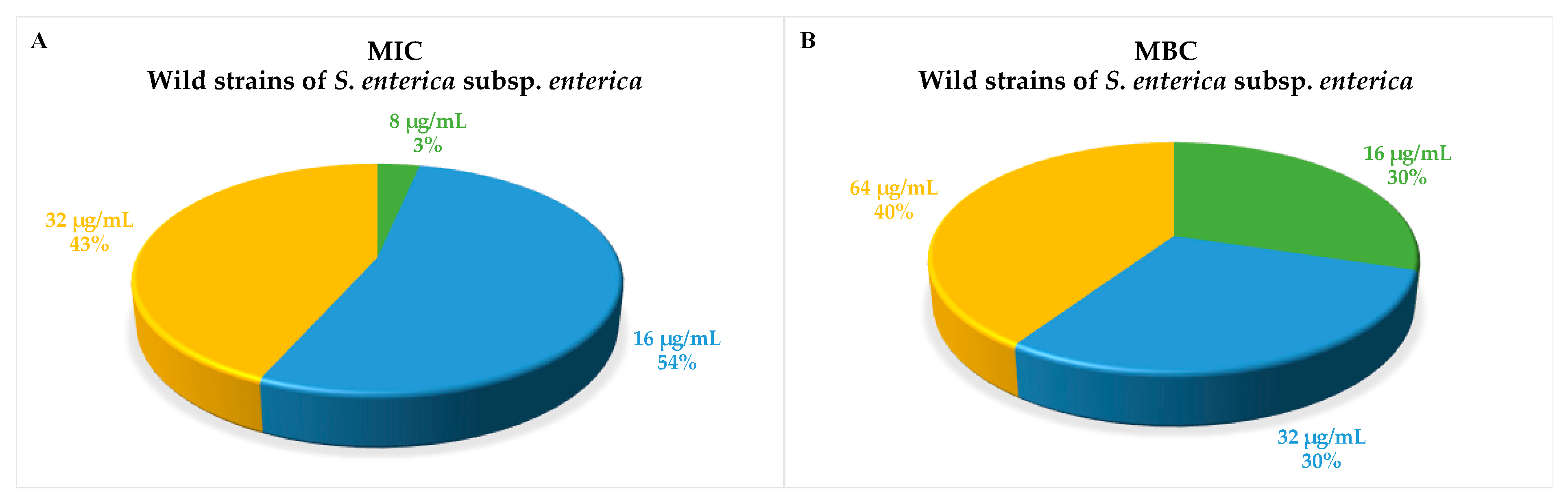
| MIC | MBC | Antimicrobial Resistance Profile | |
|---|---|---|---|
| Subspecies or serovar | µg/mL | µg/mL | Resistant to |
| S. Stanleyville | 8 | 32 | - |
| S. Agama | 16 | 32 | - |
| S. Anatum | 16 | 16 | - |
| S. Bredeney | 16 | 64 | - |
| S. Cerro | 16 | 16 | - |
| S. Dublin | 16 | 16 | - |
| S. Eboko | 16 | 16 | - |
| S. Enteritidis | 16 | 64 | Amp |
| S. Hadar | 16 | 32 | Tet, sulf |
| S. Infantis | 16 | 16 | - |
| S. Jerusalem | 16 | 16 | Sulf |
| S. Mbandaka | 16 | 64 | - |
| S. Mikawasima | 16 | 16 | Amp |
| S. Montevideo | 16 | 64 | Sulf |
| S. Newport | 16 | 16 | Sulf |
| S. Richmond | 16 | 16 | - |
| S. Seftenberg | 16 | 16 | Strep |
| S. Typhimurium monophasic | 16 | 16 | Tet, Strep, sulf, Amp |
| S. Typhimurium | 16 | 64 | |
| S. Typhimurium 1 | 16 | 64 | Tet, strep, amp |
| S. Typhimurium 2 | 16 | 64 | Amp |
| S. Typhimurium 3 | 16 | 64 | Amp, Strep |
| S. Typhimurium 4 | 16 | 64 | Strep |
| S. Virchow | 16 | 32 | Na |
| S. Isangi | 32 | 64 | Sulf |
| S. Meleagridis | 32 | 32 | Strep, sulf, Amp |
| S. Barro | 32 | 32 | - |
| S. Dabou | 32 | 32 | - |
| S. Drac | 32 | 32 | - |
| S. Enterica 4:b | 32 | 32 | Strep |
| S. Enteritidis CECT 4300 | 32 | 64 | - |
| S. Ndolo | 32 | 32 | - |
| S. Poona | 32 | 32 | - |
| S. Thompson | 32 | 64 | Amp |
| S. Typhimurium CECT 4594 | 32 | 64 | - |
| S. Typhimurium 5 | 32 | 64 | Strep |
| S. Typhimurium 6 | 32 | 64 | - |
| S. Typhimurium 7 | 32 | 64 | - |
| S. Typhimurium 8 | 32 | 64 | - |
| S. arizonae 48:z4,z23 | 32 | >128 | - |
| S. arizonae 48:z4,z23,z32 | 32 | 128 | - |
| S. salamae 4, 12: b- | 32 | 64 | Sulf |
| S. salamae 4,5,12:b | 32 | 64 | Sulf |
| S. salamae 6,8: g, m, t | 64 | 64 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Festa, R.; Ambrosio, R.L.; Lamas, A.; Gratino, L.; Palmieri, G.; Franco, C.M.; Cepeda, A.; Anastasio, A. A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food-Pathogen Salmonella enterica. Foods 2021, 10, 1372. https://doi.org/10.3390/foods10061372
Festa R, Ambrosio RL, Lamas A, Gratino L, Palmieri G, Franco CM, Cepeda A, Anastasio A. A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food-Pathogen Salmonella enterica. Foods. 2021; 10(6):1372. https://doi.org/10.3390/foods10061372
Chicago/Turabian StyleFesta, Rossella, Rosa Luisa Ambrosio, Alexandre Lamas, Lorena Gratino, Gianna Palmieri, Carlos Manuel Franco, Alberto Cepeda, and Aniello Anastasio. 2021. "A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food-Pathogen Salmonella enterica" Foods 10, no. 6: 1372. https://doi.org/10.3390/foods10061372
APA StyleFesta, R., Ambrosio, R. L., Lamas, A., Gratino, L., Palmieri, G., Franco, C. M., Cepeda, A., & Anastasio, A. (2021). A Study on the Antimicrobial and Antibiofilm Peptide 1018-K6 as Potential Alternative to Antibiotics against Food-Pathogen Salmonella enterica. Foods, 10(6), 1372. https://doi.org/10.3390/foods10061372










