Multifunctional Tyrosinase Inhibitor Peptides with Copper Chelating, UV-Absorption and Antioxidant Activities: Kinetic and Docking Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Chemicals
2.3. Determination of Tyrosinase Inhibitory Activities
2.4. Determination of Mode of Inhibition
2.5. Determination of Copper Chelating Activity
2.6. Structure–Activity Relationship (SAR) Analysis
2.7. Determination of Sun Protection Factor (SPF)
2.8. Determination of Antioxidant Activities
2.8.1. Ferric Reducing Antioxidant Power (FRAP) Assay
2.8.2. 2,2′-Azinobis(3-ethylbenzothiazoline-6-sulphonic acid) Diammonium Salt (ABTS) Free Radical Scavenging Assay
2.8.3. 2,2-Diphenyl-1-picrylhydrazyl (DPPH) Free Radical Scavenging Assay
2.9. Statistical Analysis
3. Results and Discussion
3.1. Tyrosinase Inhibitory Activities
3.2. Copper Chelating Activity
3.3. Structure–Activity Relationship (SAR) Analysis
3.4. Sun Protection Factor
3.5. Antioxidant Activities
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kobayashi, N.; Nakagawa, A.; Muramatsu, T.; Yamashina, Y.; Shirai, T.; Hashimoto, M.W.; Mori, T. Supranuclear melanin caps reduce ultraviolet induced DNA photoproducts in human epidermis. J. Investig. Dermatol. 1998, 110, 806–810. [Google Scholar] [CrossRef] [PubMed]
- Meredith, P.; Riesz, J. Radiative Relaxation Quantum Yields for Synthetic Eumelanin. Photochem. Photobiol. 2004, 79, 211–216. [Google Scholar] [CrossRef]
- Vashi, N.A.; Kundu, R.V. Facial hyperpigmentation: Causes and treatment. Br. J. Dermatol. 2013, 169, 41–56. [Google Scholar] [CrossRef] [PubMed]
- Stulberg, D.L.; Clark, N.; Tovey, D. Common hyperpigmentation disorders in adults: Part I. Diagnostic approach, cafe au lait macules, diffuse hyperpigmentation, sun exposure, and phototoxic reactions. Am. Fam. Physician 2003, 68, 1955–1960. [Google Scholar] [PubMed]
- Pandya, A.G.; Guevara, I.L. Disorders of hyperpigmentation. Dermatol. Clin. 2000, 18, 91–98. [Google Scholar] [CrossRef]
- Nakagawa, M.; Kawai, K.; Kawai, K. Contact allergy to kojic acid in skin care products. Contact Dermat. 1995, 32, 9–13. [Google Scholar] [CrossRef]
- Westerhof, W.; Kooyers, T.J. Hydroquinone and its analogues in dermatology—A potential health risk. J. Cosmet. Dermatol. 2005, 4, 55–59. [Google Scholar] [CrossRef]
- Yap, P.G.; Gan, C.Y. Chicken Egg White—Advancing from food to skin health therapy: Optimization of hydrolysis condition and identification of tyrosinase inhibitor peptides. Foods 2020, 9, 1312. [Google Scholar] [CrossRef]
- Kubglomsong, S.; Theerakulkait, C.; Reed, R.L.; Yang, L.; Maier, C.S.; Stevens, J.F. Isolation and identification of tyrosinase-inhibitory and copper-chelating peptides from hydrolyzed rice-bran-derived albumin. J. Agric. Food Chem. 2018, 66, 8346–8354. [Google Scholar] [CrossRef] [PubMed]
- PepFold3. Available online: https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/ (accessed on 1 October 2020).
- Lamiable, A.; Thévenet, P.; Rey, J.; Vavrusa, M.; Derreumaux, P.; Tufféry, P. PEP-FOLD3: Faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016, 44, W449–W454. [Google Scholar] [CrossRef]
- Protein Data Bank. Available online: https://www.rcsb.org/ (accessed on 1 October 2020).
- Bagherzadeh, K.; Talari, F.S.; Sharifi, A.; Ganjali, M.R.; Saboury, A.A.; Amanlou, M. A new insight into mushroom tyrosinase inhibitors: Docking, pharmacophore-based virtual screening, and molecular modeling studies. J. Biomol. Struct. Dyn. 2015, 33, 487–501. [Google Scholar] [CrossRef] [PubMed]
- HADDOCK 2.2. Available online: https://wenmr.science.uu.nl/haddock2.4/ (accessed on 5 October 2020).
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Bonvin, A.M.J.J. The HADDOCK2. 2 web server: User-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- PRODIGY. Available online: https://wenmr.science.uu.nl/prodigy/ (accessed on 12 October 2020).
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef]
- Dutra, E.A.; Kedor-Hackmann, E.R.M.; Santoro, M.I.R.M. Determination of sun protection factor (SPF) of sunscreens by ultraviolet spectrophotometry. Rev. Bras. Ciênc. Farm. 2004, 40, 381–385. [Google Scholar] [CrossRef]
- Sayre, R.M.; Agin, P.P.; LeVee, G.J.; Marlowe, E. A comparison of in vivo and in vitro testing of sunscreening formulas. Photochem. Photobiol. 1979, 29, 559–566. [Google Scholar] [CrossRef]
- Benzie, I.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef]
- Cai, Y.; Luo, Q.; Sun, M.; Corke, H. Antioxidant activity and phenolic compounds of 112 traditional Chinese medicinal plants associated with anticancer. Life Sci. 2004, 74, 2157–2184. [Google Scholar] [CrossRef]
- Blois, M.S. Antioxidant determinations by the use of a stable free radical. Nature 1958, 181, 1199–1200. [Google Scholar] [CrossRef]
- Hsiao, N.W.; Tseng, T.S.; Lee, Y.C.; Chen, W.C.; Lin, H.H.; Chen, Y.R.; Tsai, K.C. Serendipitous discovery of short peptides from natural products as tyrosinase inhibitors. J. Chem. Inf. Model. 2014, 54, 3099–3111. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.S.; Tsai, K.C.; Chen, W.C.; Wang, Y.T.; Lee, Y.C.; Lu, C.K.; Hsu, H.J. Discovery of potent cysteine-containing dipeptide inhibitors against tyrosinase: A comprehensive investigation of 20 × 20 dipeptides in inhibiting dopachrome formation. J. Agric. Food Chem. 2015, 63, 6181–6188. [Google Scholar] [CrossRef]
- Lim, J.Y.; Ishiguro, K.; Kubo, I. Tyrosinase inhibitory p-Coumaric acid from Ginseng leaves. Phytother. Res. 1999, 13, 371–375. [Google Scholar] [CrossRef]
- Maddaluno, J.F.; Faull, K.F. Inhibition of mushroom tyrosinase by 3-amino-L-tyrosines: Molecular probing of the active site of the enzyme. Experientia 1988, 44, 885–887. [Google Scholar] [CrossRef]
- Karbassi, F.; Saboury, A.A.; Khan, M.T.H.; Choudhary, M.I.; Saifi, Z.S. Mushroom tyrosinase inhibition by two potent uncompetitive inhibitors. J. Enzym. Inhib. Med. Chem. 2004, 19, 349–353. [Google Scholar] [CrossRef]
- Park, Y.D.; Kim, S.Y.; Lyou, Y.J.; Lee, J.Y.; Yang, J.M. A new type of uncompetitive inhibition of tyrosinase induced by Cl–binding. Biochimie 2005, 87, 931–937. [Google Scholar] [CrossRef]
- Torres-Fuentes, C.; Alaiz, M.; Vioque, J. Affinity purification and characterisation of chelating peptides from chickpea protein hydrolysates. Food Chem. 2011, 129, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Castilla, J.; Hernández-Álvarez, A.J.; Jiménez-Martínez, C.; Jacinto-Hernández, C.; Alaiz, M.; Girón-Calle, J.; Dávila-Ortiz, G. Antioxidant and metal chelating activities of peptide fractions from phaseolin and bean protein hydrolysates. Food Chem. 2012, 135, 1789–1795. [Google Scholar] [CrossRef]
- Chang, T.S. An updated review of tyrosinase inhibitors. Int. J. Mol. Sci. 2009, 10, 2440–2475. [Google Scholar] [CrossRef] [PubMed]
- Yorita, H.; Otomo, K.; Hiramatsu, H.; Toyama, A.; Miura, T.; Takeuchi, H. Evidence for the cation- π interaction between Cu2+ and tryptophan. J. Am. Chem. Soc. 2008, 130, 15266–15267. [Google Scholar] [CrossRef]
- Ubeid, A.A.; Do, S.; Nye, C.; Hantash, B.M. Potent low toxicity inhibition of human melanogenesis by novel indole-containing octapeptides. Biochim. Biophys. Acta Gen. Subj. 2012, 1820, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Ismaya, W.T.; Rozeboom, H.J.; Weijn, A.; Mes, J.J.; Fusetti, F.; Wichers, H.J.; Dijkstra, B.W. Crystal structure of Agaricus bisporus mushroom tyrosinase: Identity of the tetramer subunits and interaction with tropolone. Biochemistry 2011, 50, 5477–5486. [Google Scholar] [CrossRef] [PubMed]
- Olivares, C.; García-Borrón, J.C.; Solano, F. Identification of active site residues involved in metal cofactor binding and stereospecific substrate recognition in mammalian tyrosinase. Implications to the catalytic cycle. Biochemistry 2002, 41, 679–686. [Google Scholar] [CrossRef]
- Ramsden, C.A.; Riley, P.A. Tyrosinase: The four oxidation states of the active site and their relevance to enzymatic activation, oxidation and inactivation. Bioorg. Med. Chem. 2014, 22, 2388–2395. [Google Scholar] [CrossRef]
- Hassani, S.; Haghbeen, K.; Fazli, M. Non-specific binding sites help to explain mixed inhibition in mushroom tyrosinase activities. Eur. J. Med. Chem. 2016, 122, 138–148. [Google Scholar] [CrossRef]
- Ochiai, A.; Tanaka, S.; Tanaka, T.; Taniguchi, M. Rice bran protein as a potent source of antimelanogenic peptides with tyrosinase inhibitory activity. J. Nat. Prod. 2016, 79, 2545–2551. [Google Scholar] [CrossRef]
- Si, Y.X.; Wang, Z.J.; Park, D.; Chung, H.Y.; Wang, S.F.; Yan, L.; Park, Y.D. Effect of hesperetin on tyrosinase: Inhibition kinetics integrated computational simulation study. Int. J. Biol. Macromol. 2012, 50, 257–262. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Guo, Z.; Tan, T.; Zhang, Y. Novel tyrosinase inhibitory peptide with free radical scavenging ability. J. Enzym. Inhib. Med. Chem. 2019, 34, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Aitken, A.; Learmonth, M.P. Protein determination by UV absorption. In The Protein Protocols Handbook; Humana Press: Totowa, NJ, USA, 2009; pp. 3–6. [Google Scholar]
- Stevanato, R.; Bertelle, M.; Fabris, S. Photoprotective characteristics of natural antioxidant polyphenols. Regul. Toxicol. Pharmacol. 2014, 69, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Esfandi, R.; Walters, M.E.; Tsopmo, A. Antioxidant properties and potential mechanisms of hydrolyzed proteins and peptides from cereals. Heliyon 2019, 5, e01538. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef] [PubMed]
- Khositanon, P.; Panya, N.; Roytrakul, S.; Krobthong, S.; Chanroj, S.; Choksawangkarn, W. Effects of fermentation periods on antioxidant and angiotensin I-converting enzyme inhibitory activities of peptides from fish sauce by-products. LWT 2021, 135, 110122. [Google Scholar] [CrossRef]
- Zou, T.B.; He, T.P.; Li, H.B.; Tang, H.W.; Xia, E.Q. The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef]
- Cai, L.; Wu, X.; Zhang, Y.; Li, X.; Ma, S.; Li, J. Purification and characterization of three antioxidant peptides from protein hydrolysate of grass carp (Ctenopharyngodon idella) skin. J. Funct. Foods 2015, 16, 234–242. [Google Scholar] [CrossRef]
- Park, J.; Jung, H.; Jang, B.; Song, H.K.; Han, I.O.; Oh, E.S. D-tyrosine adds an anti-melanogenic effect to cosmetic peptides. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.D.; Meinardi, M.M. The 500 Dalton rule for the skin penetration of chemical compounds and drugs. Exp. Dermatol. 2000, 9, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, Y.B.; Naik, A.; Guy, R.H.; Kalia, Y.N. Emerging strategies for the transdermal delivery of peptide and protein drugs. Expert Opin. Drug Deliv. 2005, 2, 533–548. [Google Scholar] [CrossRef]
- Lim, S.H.; Sun, Y.; Madanagopal, T.T.; Rosa, V.; Kang, L. Enhanced skin permeation of anti-wrinkle peptides via molecular modification. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]





| Peptide Sequence | Tyrosinase Activity Inhibition # (%) | Mode of Inhibition | Copper Chelating Activity # (%) | SPF # | Antioxidant Activity # | ||||
|---|---|---|---|---|---|---|---|---|---|
| FRAP (mMFeSO4) | ABTS (%) | DPPH (%) | |||||||
| (a) | MIP1 | ADHPF | 35.86 ± 2.26 d | Competitive | 94.33 ± 0.04 e | 1.80 ± 0.02 b | 0.035 ± 0.004 a | 2.90 ± 0.28 b | n.d. |
| MIP2 | ILELPFASGDLLML | 17.26 ± 2.10 a | Competitive | 11.41 ± 0.13 a | 3.12 ± 0.13 c | n.d. | 1.44 ± 0.27 ab | n.d. | |
| MIP3 | FDKLPGFGD | 22.78 ± 1.12 b | Mixed | 94.43 ± 0.46 e | 1.36 ± 0.02 a | 0.034 ± 0.002 a | 2.11 ± 0.41 ab | n.d. | |
| (b) | DIP1 | GYSLGNWVCAAK | 80.04 ± 2.79 e | Competitive | 36.27 ± 1.17 d | 11.9 ± 0.24 e | 5.09 ± 0.13 b | 11.34 ± 0.90 c | 29.14 ± 1.36 a |
| DIP2 | HIATNAVLFFGR | 17.70 ± 2.66 a | Competitive | 19.13 ± 2.22 b | 6.45 ± 0.40 d | 0.018 ± 0.001 a | n.d. | n.d. | |
| DIP3 | FMMFESQNKDLLFK | 28.95 ± 1.47 c | Uncompetitive | 26.23 ± 1.03 c | 1.98 ± 0.05 b | n.d. | 0.99 ± 0.56 ab | n.d. | |
| Gallic acid | 16.6 ± 0.36 c | 83.14 ± 3.02 d | 81.67 ± 0.03 b | ||||||
| EDTA | 96.06 ± 0.03e | ||||||||
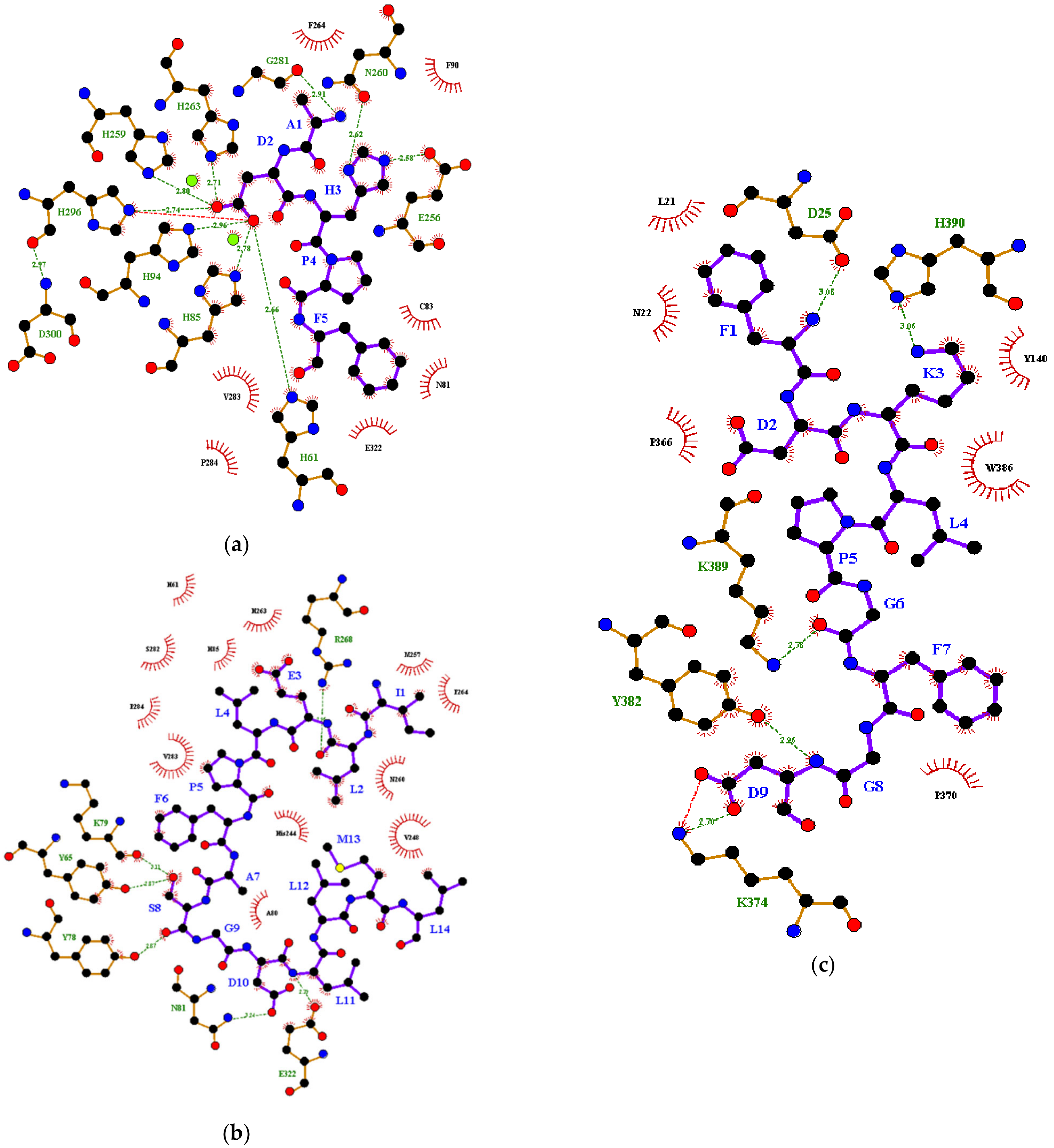
| Free Binding Energy (kcal/mol) | Hydrophobic Interaction | Salt Bridge | Covalent Bond | Hydrogen Bond | ||
|---|---|---|---|---|---|---|
| (a) | MIP1 | −9.0 | CuA | H296 | n.d. | H61 |
| CuB | H85 | |||||
| N81 | H94 | |||||
| C83 | E256 | |||||
| F90 | H259 | |||||
| F264 | N260 | |||||
| V283 | H263 | |||||
| P284 | G281 | |||||
| E322 | H296 | |||||
| MIP2 | −9.5 | H61 | n.d. | n.d. | Y65 | |
| Y65 | Y78 | |||||
| Y78 | K79 | |||||
| K79 | N81 | |||||
| A80 | R268 | |||||
| N81 | E322 | |||||
| H85 | ||||||
| V248 | ||||||
| M257 | ||||||
| N260 | ||||||
| H263 | ||||||
| F264 | ||||||
| R268 | ||||||
| S282 | ||||||
| V283 | ||||||
| P284 | ||||||
| E322 | ||||||
| MIP3 | −7.0 | L21 | K374 | n.d. | D25 | |
| N22 | K374 | |||||
| D25 | Y382 | |||||
| Y140 | K389 | |||||
| P366 | H390 | |||||
| P370 | ||||||
| K374 | ||||||
| Y382 | ||||||
| W386 | ||||||
| K389 | ||||||
| H390 | ||||||
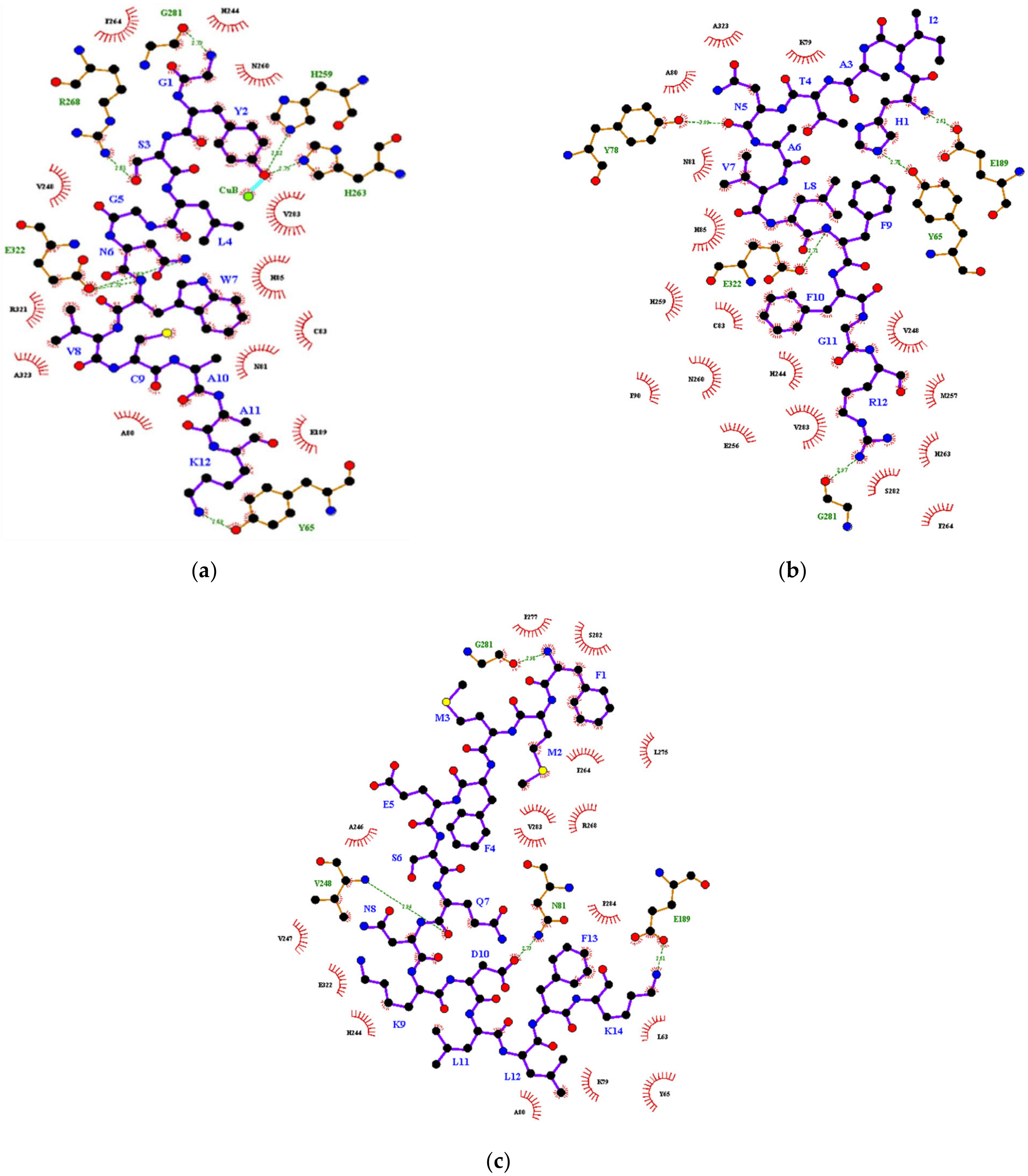
| (b) | DIP1 | −9.4 | A80 | n.d. | Cu(B) | Y65 |
| N81 | H259 | |||||
| C83 | H263 | |||||
| H85 | R268 | |||||
| E189 | G281 | |||||
| H244 | E322 | |||||
| V248 | ||||||
| N260 | ||||||
| F264 | ||||||
| V283 | ||||||
| R321 | ||||||
| A323 | ||||||
| DIP2 | −8.3 | K79 | n.d. | n.d. | Y65 | |
| A80 | Y78 | |||||
| N81 | E189 | |||||
| C83 | G281 | |||||
| H85 | E322 | |||||
| F90 | ||||||
| H244 | ||||||
| V248 | ||||||
| E256 | ||||||
| M257 | ||||||
| H259 | ||||||
| N260 | ||||||
| H263 | ||||||
| F264 | ||||||
| S282 | ||||||
| V283 | ||||||
| A323 | ||||||
| DIP3 | −9.3 | L63 | n.d. | n.d. | N81 | |
| Y65 | E189 | |||||
| K79 | V248 | |||||
| A80 | G281 | |||||
| N81 | ||||||
| E189 | ||||||
| H244 | ||||||
| A246 | ||||||
| V247 | ||||||
| V248 | ||||||
| F264 | ||||||
| R268 | ||||||
| L275 | ||||||
| P277 | ||||||
| G281 | ||||||
| S282 | ||||||
| V283 | ||||||
| P284 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yap, P.-G.; Gan, C.-Y. Multifunctional Tyrosinase Inhibitor Peptides with Copper Chelating, UV-Absorption and Antioxidant Activities: Kinetic and Docking Studies. Foods 2021, 10, 675. https://doi.org/10.3390/foods10030675
Yap P-G, Gan C-Y. Multifunctional Tyrosinase Inhibitor Peptides with Copper Chelating, UV-Absorption and Antioxidant Activities: Kinetic and Docking Studies. Foods. 2021; 10(3):675. https://doi.org/10.3390/foods10030675
Chicago/Turabian StyleYap, Pei-Gee, and Chee-Yuen Gan. 2021. "Multifunctional Tyrosinase Inhibitor Peptides with Copper Chelating, UV-Absorption and Antioxidant Activities: Kinetic and Docking Studies" Foods 10, no. 3: 675. https://doi.org/10.3390/foods10030675
APA StyleYap, P.-G., & Gan, C.-Y. (2021). Multifunctional Tyrosinase Inhibitor Peptides with Copper Chelating, UV-Absorption and Antioxidant Activities: Kinetic and Docking Studies. Foods, 10(3), 675. https://doi.org/10.3390/foods10030675