Abstract
Inhibition of Salmonella by Lactobacillus has been a popular research topic for decades; however, the inhibition potential of chicken-derived Salmonella by chicken-derived Lactobacillus has not yet been studied. In this study, 89 strains of Lactobacillus from chicken intestines were isolated by national standard method, Gram staining, physiological, and biochemical experiments and molecular sequencing; The inhibition characteristics of 89 strains of chicken derived Lactobacillus against 10 strains Salmonella (S. Enteritidis SE05, SC31, SC21, SC72 SC74, SC79, SC83, SC87; S. bongori SE47; S. Typhimurium, SC85) were detected by agar inhibition zone, The results showed that the inhibition zone of 24 strains of chicken derived Lactobacillus was more than 10 mm, which indicated that the isolated chicken derived Lactobacillus could effectively inhibit the growth of Salmonella; The drug resistance and bile salt tolerance of these 24 strains were analyzed, The results showed that the standard strains LG and L76 were not resistant, and the other 22 Lactobacillus strains showed different degrees of resistance. The strains LAB24, LAB26, LAB53, LAB69, and L76 showed good tolerance at the concentration of 3 g/L bile salt; Caco-2 cell experiment and flow cytometry were used to analyze the inhibitory effect of chicken derived Lactobacillus on the adhesion of Salmonella to Caco-2 cells, The results showed that 16 probiotics could effectively inhibit the adhesion of Salmonella to Caco-2 cells. Twelve probiotics were identified by molecular biology. The results showed that L76 was Enterococcus faecalis, and the other 11 strains were Lactobacillus.
1. Introduction
Salmonellosis is a zoonotic food borne disease which causes outbreaks and sporadic cases of gastroenteritis in human worldwide [1]. Chickens have been known as the major source of Salmonella contaminated food products such as chicken eggs and meat that cause human salmonellosis in many countries [2,3]. Although poultry are asymptomatic Salmonella carriers, and their production performance is unaffected, Salmonella can continuously colonize the cecum of broilers [4,5]. During slaughtering and processing, the infected broiler contaminates the meat products, thereby causing food poisoning to humans through the food chain. Infected laying hens can contaminate their eggs, thereby vertically transmitting the infection to the offspring. Salmonellosis may also be caused by ingesting egg products contaminated by Salmonella. Notably, in recent years, multiple resistant Salmonella strains have been detected. Moreover, the continued use of antibiotics as growth promoters and to control Salmonella colonization of broilers may result in more resistant strains. In many Chinese regions, meat products and poultry have been found positive for a variety of foodborne Salmonella strains [6,7].
Lactic acid bacteria (LAB) are widely used probiotic organisms, and their strains usually occur in human and animal intestinal microbiota. LAB promote the development of host-favorable microbiota reduce or prevent the colonization of harmful pathogens, enhance mucosal immunity, improve the digestibility of the gastrointestinal tract and reduce its pH, and enhance the maturity and integrity of the intestinal tissue [4]. Additionally, some LAB strains are tolerant to the effects of digestive products, such as gastric acid and bile salts [8], and they can adhere to the host intestinal epithelium [9], thereby gaining a competitive advantage and are important for bacterial maintenance [10]. In vitro models with intestinal cell lines are widely used. Laboratory models using human intestinal cell lines, such as Caco-2 [11], have been developed to study the adhesion of probiotic LAB and the competitive exclusion of pathogenic bacteria.
The aim of this study is to study the efficacy of chicken-derived probiotics on Salmonella (S. Enteritidis SE05, SC31, SC21, SC72 SC74, SC79, SC83, SC87; S. bongori SE47; S. Typhimurium, SC85) by preliminary screening. The isolated strains of chicken-derived probiotics with better antibacterial effect were selected, their surface characteristics, bile salt tolerance and drug resistance of lactic acid bacteria from chicken were studied; The possible reasons of lactic acid bacteria inhibiting Salmonella were further analyzed: Study on the adhesion of lactic acid bacteria and the effect of protecting cell membrane.
2. Materials and Methods
2.1. Strain Isolation, Culture Media, and Used Cell Lines
120 samples of chicken intestines from different slaughterhouses were collected, and Lactobacillus were isolated by national standard method [12]. Added 25 g mixed sample to 225 mL normal saline. Mixed at 37 °C for 30 min and serially diluted, coated on de Man, Rogosa, and Sharpe (MRS) agar solid medium, then the coated plates were incubated at 37 °C for 48 h. Typical samples with obvious sour taste were transferred to MRS liquid medium, incubated at 37 °C for 48 h, and then coated again. Then, 107 suspected Lactobacillus were selected for Gram staining, and 89 of them turned purple after adding Gram staining agent, microscopic observation showed spherical, short chain, rod-shaped, etc. [13], and then combined with the physiological and biochemical identification of Lactobacillus, finally the 89 strains of Lactobacillus were isolated. The result show in the Tables S1 and S2 and Figure S1. S. Enteritidis SE05, SC31, SC21, SC72 SC74, SC79, SC83, SC87; S. bongori SE47; S. Typhimurium, SC85 were isolated previously [14] and preserved in our laboratory, Caco-2 cells (Shanghai Bogu Biotechnology Co., Ltd., Shanghai, China) were purchased by our laboratory.
2.2. Inhibitory Effect of Lactobacillus Strains on Salmonella
The 89 Lactobacillus strains were cultured in MRS liquid medium at 37 °C for 48 h, centrifuged at 5000 rpm/min for 5 min, and the supernatant was taken for use. Next, we added 100 μL of Salmonella suspension (SE05, SE47, SC21, SC31, SC72, SC74, SC79, SC83, SC85, and SC87) to 100 mL of LB liquid medium, and cultured the mixture at 37 °C and 180 rpm/min for 24 h. Then, 100 mL bottles of solid LB culture medium melted completely and cooled to 40–50 °C, we added 100 μL of culture liquid that contained 10 Salmonella isolates, which were cultured overnight; then, the mixture was shaken and poured on the agar plates, which were marked in advance. After the plates cooled, we used a 5 mm punch to drill holes on them. The number of holes drilled depended on the plate size; the discarded agar pieces were picked out with sterilized toothpicks. We added the samples according to the marks on the backs of the plates, as well as 100 μL of LAB supernatant to each hole. Following this, we incubated the plates at 37 °C for 24 h [15]. Twenty-four Lactobacillus strains that had the best bacteriostatic effect were selected for subsequent experiments. Three parallel experiments were performed in each group
2.3. Antibiotics Resistance of Lactobacillus Strains
Antimicrobial susceptibility of the 24 Lactobacillus isolates was evaluated by the disk diffusion test, using Mueller–Hinton agar, according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2012) [16]. The isolates were screened for resistance to the following antibiotics: tetracycline (TET, 30 mg); amoxicillin (AMX, 30 mg); ceftriaxone (CRO, 30 μg); chloramphenicol (CHL, 30 μg); gentamicin (GEN, 10 μg); trimethoprim/sulfamethoxazole (SXT, 1.25/23.75 μg); kanamycin (KAN, 30 μg); erythromycin (ERY, 15 μg). Three parallel experiments were performed in each group. The results were interpreted according to the established CLSI guidelines (CLSI, 2012).
2.4. Lactobacillus Bile Salt Tolerance Test
The concentration of bile salt in most animals ranged from 0.3 g/L to 3.0 g/L. in this experiment, the concentration of bile salt was selected in the range of 1.0 g/L–3.0 g/L [17]. The activated bacterial suspension was centrifuged at 4000 rpm for 25 min, and the supernatant was discarded. The precipitated bacteria were washed three times with sterile MRS liquid medium. The bacteria were suspended in the medium, and the bacterial suspension concentration was adjusted to 3.0 × 108 CFU/mL. The reconstituted bacterial suspension was divided into different groups. Next, by adding 1.0, 2.0, and 3.0 g/L of pig and bovine bile salt, the bacterial suspensions were incubated at a constant temperature of 37 °C for 3 h and then removed the supernatant. Gradient dilution was performed immediately by PBS, 1 mL mixture was used to coat the MRS plate. After 48 h of culture at 37 °C, we determined the colony count. The survival rate was calculated as follows: survival rate (%) = number of “2 H” colonies/number of “0 h” colonies × 100% [18]. Three parallel experiments were performed in each group.
2.5. Adhesion of Different Bacteria to Caco-2 Cells
Firstly, 1 mL of each Lactobacillus or Salmonella suspension (2 × 108 CFU/mL) was added to the corresponding 6-well plate containing Caco-2 cells, and cultured at 37 ℃ and 5% CO2, Standard Lactobacillus strain LG (L. rhamnosus GG, FSMM22) as control. Each treatment was repeated three times. After 1 h culture, the culture medium was taken out, the supernatant was discarded, and Caco-2 cells were washed with PBS (pH 7.4) for three times to remove the non-adherent bacteria. The sterile 1% Triton X-100 PBS (1 mL, pH 7.4) solution was added into each well, and it was allowed to stand for 10 min. After the Caco-2 cells were completely removed, the supernatant were transferred into the sterile 1.5 mL centrifuge tube and mixed well. A total of 100 μL of the prepared mixture was diluted at a gradient of 10−1–10−6. Then Lactobacillus and Salmonella were counted on the medium plate [19,20]. The adhesion number of bacteria was calculated using Equation (1):
Adhesion index = number of bacteria/cells adhered
2.6. Inhibition of Salmonella Adhesion to Caco-2 Cells by Lactobacillus
Caco-2 cells were passaged for 3–4 generations in DMEM complete medium, and then lactic acid bacteria adhesion test was started. The density of Caco-2 cell suspension was adjusted to 2 × 105 cells/mL. A total of 16 strains of Lactobacillus were isolated and cultured for 24 h. After centrifugation, the supernatant was removed and the OD value of the suspension was adjusted to 0.6 with PBS. Three Salmonella strains were labeled with 1 mL FITC fluorescent solution and incubated in dark for 30 min. The supernatant was removed by centrifugation and resuspended with PBS. A 48 well plate, 200 μL/well was used. First, 48 wells were wetted with 100 μL DMEM solution, then 100 μL cell suspension was added, and 300 μL DMEM incomplete culture medium was added to 500 μL. In the competitive adhesion experiment group, 500 μL LAB + 500 μL fluorescent labeled Salmonella were added to each well at the same time; In the rejection adhesion experiment group, 500 μL LAB was added for incubation at 37 °C for 1 h, PBS was washed for 3 times, and then 500 μL fluorescent labeled Salmonella for 1 h was added for incubation at 37 °C for 1 h; In the replacement adhesion experiment group, 500 μL fluorescent labeled Salmonella was added for 1 h at 37 °C for 1 h, washed with PBS for 3 times, and then 500 μL LAB was added for 1 h at 37 °C for labeling. Finally, all treatments were washed with PBS three times, digested with 0.2 mL trypsin for 2 min, and then added with 0.4 mL DMEM complete culture medium to terminate the reaction. The liquid was collected, and the fluorescence values of each group were detected by the multi-function fluorescent enzyme reader (Infinite M200 Pro multi-function enzyme reader, TECAN, Männedorf, Switzerland). Three parallel experiments were performed in each group.
2.7. Effect of Lactobacillus Adhesion to Caco-2 Cells on Their Physiological Metabolism
A total of 1 mL Caco-2 cell suspension (2 × 105 cells/mL) was transferred to a 6-well cell culture plate, and 1 mL DMEM complete culture medium was added and incubated at 37 °C and 5% CO2. After the second day of culture, the new DMEM solution was replaced. After the Caco-2 cells completely adhered to the 6-well plate, the used DMEM medium was discarded, and the Caco-2 cells were washed twice with PBS. In each well, 1 mL DMEM medium containing 10% fetal bovine serum was added to a 6-well cell culture plate. The components are as follows: The suspension of CT group (1 mL sterile PBS), LAB group (1 mL 2 × 108 CFU/mL Lactobacillus), SAL group (1 mL 2 × 108 CFU/mL Salmonella), and rejection group (1 mL of Lactobacillus was cultured for 1 h, then 1 mL of Salmonella was cultured for 1 h), each group was cultured for 2 h. After culture, the supernatant was collected, and the Caco-2 cells were washed three times with sterile PBS. The Caco-2 cells were dissolved with 1% Triton X-100 (1 mL per well), and the collected lysate stored at -20 °C. Then the alkaline phosphatase (AKP, BC2145, Solarbio, BeiJing, China) [21] and lactate dehydrogenase (LDH, BC0685, Solarbio, BeiJing, China) activities were tested according to the instructions of the sword test box. Three parallel experiments were performed in each group.
2.8. Adhesion of Lactobacillus and Salmonella Isolates to Caco-2 Cells and Apoptosis Test
Firstly, 5 mL Cultured Caco-2 cells ((1 − 5) × 106/mL per sample) were directly collected into 10 mL centrifuge tubes and centrifuged at 500–1000 r/min for 5 min. Then, the culture medium was discarded, and the cells were washed with the incubation buffer. Next, they were centrifuged again at 1000 r/min for 5 min. Then, they were labeled, re-suspended in a 100 μL labeling solution, incubated in the dark for 10–15 min at room temperature, centrifuged at 1000 r/min for 5 min, and washed with the incubation buffer. SA-FLOUS (Fluorescent solution) was added, and the mixture was incubated at 4 °C for 20 min, in the dark, with no shaking. Flow cytometry analysis (CytoFLEX LX, BECKMAN, Bria, Florida, USA) showed that the fluorochromes were excited with a 488 nm wavelength. Fluorescein isothiocyanate (FITC) fluorescence was detected with a passband filter at 515 nm wavelength, while PI was detected with a filter at a wavelength greater than 560 nm. Specific parameters of the instrument: Flow pressure: 0–100 psi, step-less adjustable. The liquid flow system can effectively filter 99.999% of the impurities >0.1 μm in the sheath. We used the automatic sample loading system with 1.5 mL sample tubes, which offers the advantage of automatic cleaning, backwashing, bubble removal, and temperature control. Separation speed: 100,000 cells/s, separation purity: >99% (using 70 μm nozzle, at the speed of 70,000 s, 60 psi pressure, the sample with 1% target cell content was separated under the separation mode). The recovery rate: >90% of the theoretically predicted cells. Effective acquisition speed: >100,000 cells/s. Effective sorting speed: >70,000 cells/s. Three parallel experiments were performed in each group.
2.9. Purification and Sequencing of PCR Products
Firstly, 12 strains of Lactobacillus with typical physiological and biochemical characteristics were screened out by acid-base, bile salt, drug resistance and cell adhesion tests, and then the molecular sequencing test was carried out. 27F(5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R(5′-GGTTACCTTGTTACGACTT-3′) primers (Primer production, Henan ShangYa Biotechnology Co., Ltd., Zhengzhou, China) were used for PCR amplification and sequencing identification All the PCR products were purified with a High Pure PCR purification kit (Roche, Mannheim, Germany) and sequenced by Nanjing GenScript Biotech Co., Ltd., China. The resultant DNA sequences were analysed using BLAST (http://www.ncbi.nih.gov/BLAST/. accessed date: 19 May 2020).
2.10. Statistical Analysis
The experiment was performed triplicate as a replicated test. Data were analyzed using one-way analysis of variance using the SPSS 16.0 software (SPSS Inc., Chicago, Illinois, USA) for Windows. The results are expressed as mean (M) ± standard deviation (SD). Mean separation was performed via Duncan’s multiple range tests (p < 0.05).
3. Results
3.1. Inhibition of Salmonella by Lactobacillus
Figure 1 shows the inhibition results obtained for 10 different Salmonella strains by using the bacteriostatic circle method. Different Lactobacillus strains had different bacteriostatic effects on different Salmonella strains. The diameter of inhibition circle is less than 5 mm, which indicates that the bacteriostatic effect of lactic acid bacteria is not obvious. The diameter of inhibition circle is more than 8 mm, which indicates that lactic acid bacteria have bacteriostatic effect. The diameter of inhibition circle is more than 10 mm, which indicates that the bacteriostatic effect of lactic acid bacteria is obvious. From the Figure 1, the average inhibition zone diameter of the strains LAB1, LAB4, LAB7, LAB16, LAB22, LAB24, LAB35, LAB44, LAB53, LAB54, LAB56, LAB60, LAB64, LAB69, LAB70, LAB73, LAB76, and LG were more than 8 mm and were selected. The inhibition zone of LAB69 (17.2 mm) and LAB76 (17.5 mm) is the largest, indicating that the antibacterial effect is the best.
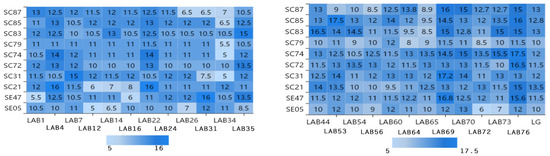
Figure 1.
Experimental results of inhibitory zone of 24 Lactobacillus isolates to 10 Salmonella isolates. Note: The vertical axis is 10 strains of Salmonella, the horizontal axis is 24 strains of Lactobacillus, and the intersection of the horizontal axis and the vertical axis is the diameter of inhibition zone of Lactobacillus inhibiting Salmonella. (The diameter of inhibition zone: mm).
3.2. Antibiotic Resistance of Lactobacillus Strains
The resistance of 24 strains of Lactobacillus to antibiotics TET, AMX, CRO, CHL, GEN, SXT, KAN, and ERY were shown in Table 1. The standard strains LG and LAB76 were not drug-resistant. The other 22 strains of LAB showed different degrees of drug resistance. LAB12, LAB14, LAB22, and LAB34 were resistant to 3 antibiotics, 5 strains of lactic acid bacteria were resistant to 2 kinds of antibiotics, and 14 strains were resistant to 1 kind of antibiotics. This may be related to the type and frequency of antibiotics used in poultry breeding. Lactobacillus resistant to three kinds of antibiotics were not used in subsequent experiments. Among the 24 strains of Lactobacillus, 25% (6/24) were resistant to CRO, 29.2% (7/24) to SXT, 20.8% (5/24) to CHL, and 25% (6/24) to TET. These indicators provide a data base for screening target lactic acid bacteria.

Table 1.
Drug resistance of 24 strains of lactic acid bacteria.
3.3. Lactobacillus Bile Salt Tolerance Test
Figure 2 shows the growth of 24 strains of LAB within 8 h under three different bile salt concentrations. It can be seen from Figure 2 that the growth of 24 Lactobacillus strains is not consistent with the increase of bile salt concentration. The OD600 value of LAB4, LAB7, LAB24, LAB26, LAB31, LAB35, LAB44, LAB56, LAB65, LAB69, LAB70, L76, and LG were more than 0.20, show that most of the lactic acid bacteria detected in this paper can maintain a high survival rate under the condition of 3 g/L bile salt, and the strains still show good tolerance to the bile salt. At present, many studies have proved that most of the lactic acid bacteria can survive well in the intestine (trypsin, pH 8.0, and 0.3% bile salt), and can return to the initial number of bacteria when passing through the small intestine [22,23].
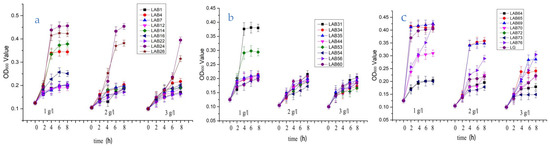
Figure 2.
The bile salt tolerance of 24 strains of Lactobacillus. Note: (a)-The growth of 8 strains of Lactobacillus (LAB1, LAB4, LAB7, LAB12, LAB14, LAB16, LAB22, LAB24) under the condition of 1 g/L, 2 g/L, 3 g/L bile salt concentration for 8 h; (b)-The growth of 9 strains of Lactobacillus (LAB26, LAB31, LAB34, LAB35, LAB44, LAB53, LAB54, LAB56,LAB60) under the condition of 1 g/L, 2 g/L, 3 g/L bile salt concentration for 8 h; (c)-The growth of 8 strains of Lactobacillus (LAB64, LAB65, LAB69, LAB70, LAB72, LAB73, LAB76, LG) under the condition of 1 g/L, 2 g/L, 3 g/L bile salt concentration for 8 h.
3.4. Bacterial Adhesion to Caco-2 Cells
Adhesion is an important basis for the colonization of Lactobacillus in the intestinal tract, and it is necessary to play the role of biological barrier, inhibit the growth and reproduction of pathogenic bacteria, and improve the structure of intestinal flora [24,25]. We screened the Lactobacillus with strong adhesion ability from 15 strains of chicken derived Lactobacillus. Figure 3 is the result about the vitro evaluation of the adhesion ability of 15 Lactobacillus isolates and 3 Salmonella strains using Caco-2 cells as a model. Adhesion index = number of bacteria/cells adhered, the higher the ratio of adhesion index, the stronger the adhesion ability. Fifteen Lactobacillus strains had significant differences in their ability to adhere to Caco-2 cells. L76 had the strongest adhesion ability (17.50 bacteria/cell), followed by LAB 69 (15.57 bacteria/cell). The adhesion ability of three Salmonella strains was 4.86, 6.03, and 7.57 bacteria/cell, respectively. We selected chicken-derived Lactobacillus with higher adhesion ability than Salmonella as the target strains of subsequent experiments.
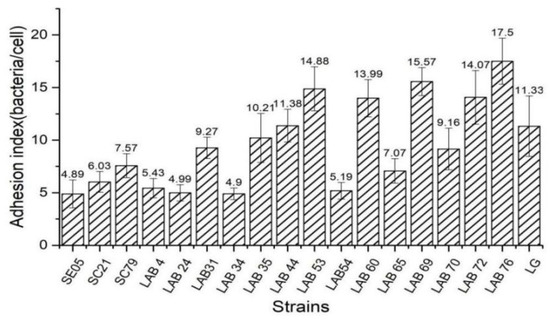
Figure 3.
Adhesion index of different bacteria to CaCO-2 cells.
3.5. Inhibition of Salmonella Adhesion to Caco-2 Cells by Lactobacillus
The treatment of Caco-2 cells by different bacterial combinations is shown in Table 2. Salmonella were incubated with fluorescence for 30 min, and Lactobacillus were not treated. The experimental group was incubated with Caco-2 cells according to different combinations. The stronger the fluorescence intensity was, the redder the color was, and the more Salmonella adhered to Caco-2 cells.

Table 2.
Treatment of Caco-2 cells with different combinations of bacteria.
On the contrary, the fluorescence intensity was weaker, and the color was blue, indicating that the number of Salmonella adhered to Caco-2 cells was less. Adding Lactobacillus and Salmonella labeled with fluorescence to Caco-2 cells can show the change of fluorescence intensity. The changes of cell adhesion of Lactobacillus and Salmonella in different treatments were studied by color change. In the competition experiment, Lactobacillus and Salmonella were added to Caco-2 cells at the same time. The results showed that Salmonella in SC31-LAB84, SC79-LAB34 and SC79-LAB72 groups adhered more to Caco-2 cells than other groups (Figure 4a). In the rejection experiment, Caco-2 cells were incubated with Lactobacillus for 1 h, and then incubated with Salmonella (Figure 4b). The results showed that Salmonella had more adhesion to Caco-2 cells in LAB54-SC31 group. In other treatments, Lactobacillus adhered more to Caco-2 cells. In the substitution experiment (Figure 4c), Caco-2 cells were incubated with Salmonella for 1 h, and then incubated with Lactobacillus. The results showed that seven groups of treatments (SC31-LAB35, SC31-LAB44, SC31-LAB53, SC31-LAB54, SC79-LAB24, SC79-LAB34, and SC79-LAB44) had stronger fluorescence intensity, and indicated more Salmonella adhesion to Caco-2 cells. While the adhesion rate in other groups was inhibited by Lactobacillus, based on the color change.
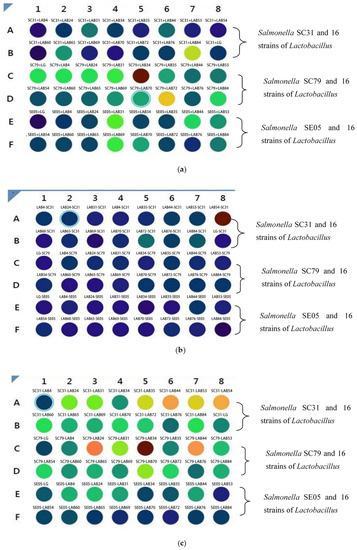
Figure 4.
(a) Competitive adhesion test of Caco-2 cells incubated with Lactobacillus and Salmonella. In Figure 4a, The target Salmonella are SC31, SC85, and SE05. In this experiment, Lactobacillus and Salmonella were added at the same time and incubated for 1 h, then the fluorescence intensity was measured. The stronger the fluorescence intensity was, the more red the color was. It showed that the number of Salmonella incubated on Caco-2 cells was more, while the fluorescence intensity was weak, the color was blue, indicating that the number of Salmonella incubated on Caco-2 cells was small. (b) Rejection and adhesion test of Caco-2 cells incubated by lactic acid bacteria first and then by Salmonella. In Figure 4b, the Lactobacillus isolated strains were added for incubation for 1 h, then the target Salmonella SC21, SC85 and SE05 were added for incubation for 1 h, and the results were measured by multi-functional enzyme labeling instrument. The stronger the fluorescence intensity was, the more red the color was. It showed that the number of Salmonella incubated on Caco-2 cells was more, while the fluorescence intensity was weak, the color was blue, indicating that the number of Salmonella incubated on Caco-2 cells was small. (c) Displacement adhesion test of Caco-2 cells incubated by Salmonella and then by lactic acid bacteria. In Figure 4c, the target Salmonella SC21, SC85 and SE05 were added to incubate for 1 h, and then the Lactobacillus isolate was added to incubate for 1 h, and then the results were measured by multi-functional enzyme labeling instrument. The stronger the fluorescence intensity was, the more red the color was. It showed that the number of Salmonella incubated on Caco-2 cells was more, while the fluorescence intensity was weak, the color was blue, indicating that the number of Salmonella incubated on Caco-2 cells was small.
3.6. Lactobacilli Adherence Effect on the Physiological Metabolism of Caco-2 Cells
Caco-2 cells incubated with PBS was the control group. LAB group were incubated with Lactobacillus, and the SAL group were incubated with Salmonella. The AKP activity of Caco-2 cell lysate and supernatant in the control group were 1378.21 U/gprot and 79.95 U/gprot, respectively (Table 3). LAB group and SAL group (SC31, SC79 and SE05) had different effects on the metabolism of Caco-2 cells. Compared with the control group, the AKP activity of Caco-2 cell supernatant in SAL group was significantly higher than that of Caco-2 cell lysate. SAL significantly inhibited the activity of AKP in Caco-2 cells, destroyed the integrity of cell membrane, and resulted in a large amount of AKP leakage into the cell culture supernatant. Compared with the control group, the AKP activity in the supernatant of Caco-2 cells treated with LAB decreased by 15–59.6% (p < 0.01), and the AKP activity in Caco-2 cells treated with LAB also decreased significantly, but it was still 3.45–4.92 times higher than that in SAL group (p < 0.01). The AKP activity of Caco-2 cells treated with LAB-SAL was slightly higher than that of SAL, while the AKP content in the supernatant of Caco-2 cells treated with LAB-SAL was significantly lower than that of control group. These results indicate that Lactobacillus can repel Salmonella and protect Caco-2 cell membrane.

Table 3.
Activity of alkaline phosphatase (AKP) in lysate and supernatant of Caco-2 cells incubated with 12 strains of Lactobacillus and 3 strains of Salmonella.
The effects of different treatments on LDH metabolism of Caco-2 cells were significantly different (Table 4). Compared with the control group, LDH activity was not detected in Caco-2 cells incubated with SAL, while LDH activity in the supernatant of Caco-2 cells incubated with SAL increased by 64.73% (p < 0.01). It may be that SAL treatment destroys the integrity of Caco-2 cell membrane and releases LDH into cell culture supernatant. The LDH activity in Caco-2 cells incubated with LAB increased significantly (p < 0.05), but decreased by 98.49% (p < 0.01). The LDH activity of Caco-2 cells incubated with LAB-SAL was significantly higher than that of Caco-2 cells incubated with SAL (p < 0.05); the LDH activity content in the supernatant of Caco-2 cells incubated with LAB-SAL increased by 35.26% (p < 0.01), but it was still lower than that of SAL. These results suggest that LAB can enhance the LDH metabolic activity of Caco-2 cells, which further confirms the protective effect of SAL on the integrity of Caco-2 cell membrane by preventing the destruction of Caco-2 cell membrane.

Table 4.
Lactate dehydrogenase (LDH) activity in lysate and supernatant of Caco-2 cells incubated with 12 strains of Lactobacillus and 3 strains of Salmonella.
3.7. Lactobacillus and Salmonella Isolate Adhesion to Caco-2 Cells and Apoptosis Test Results
Apoptosis was detected using flow cytometry (CytoFLEX LX, BECKMAN, USA). Propidium iodide (PI) is a type of nucleic acid dye. Under normal circumstances, PI cannot penetrate the entire cell membrane; this is not true for mid- and late apoptotic as well as dead cells, the cell membrane of which PI penetrates and dyes the nucleus red. Caco-2 cells treated with PBS were used as blank control group, multidrug resistant Salmonella SC79 was used as negative control group, and standard Lactobacillus strain LG was used as positive control group. Figure 5 shows the four quadrant method to observe the apoptosis of Caco-2 cells, that is, the scatter plot of bivariate flow cytometry. In the Figure 5, P1 gate showed the ratio of living cells, which shows the number of living cells detected by flow cytometry after Caco-2 cells were incubated with different bacteria. Compared with the blank control group, the number of Caco-2 cells in LAB69 group and LG group were significantly increased, while the number of Caco-2 cells in LAB69 group was higher than that in the positive control group; the number of Caco-2 cells in the negative control group was significantly decreased. P2 gate and P3 gate show the apoptotic and dead cells detected by flow cytometry after Caco-2 cells were incubated with different bacteria. Compared with the blank control group, the number of apoptotic cells and dead cells of Caco-2 cells in LAB69 group and LG group decreased, and the number of apoptotic cells and dead cells in LAB69 group was lower than that in the positive control group; while the number of apoptotic cells and dead cells in Caco-2 cells in SC79 group increased significantly. As can be seen from Figure 5, compared with the blank control group, the results of Caco-2 cells incubated with different bacteria were significantly different. The invasion and destruction of Caco-2 cells in Salmonella negative control group were more serious, Compared with positive control group LG and LAB69, the number of live cells increased and the number of dead cells decreased. These results indicate that LAB69 has a certain protective effect on Caco-2.
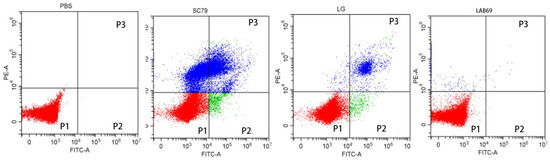
Figure 5.
Four quadrants results of Caco-2 cells treated with different bacteria in flow cytometry. Note: P1 gate, the red part of P1 gate is the ratio of the number of living cells. The higher the red ratio, the more living Caco-2 cells; P2 gate, the green part of P2 gate is the ratio of apoptotic cells. The larger the green area, the more apoptotic cells; P3 gate, The blue part of P3 gate is the ratio of death cells. The larger the blue area, the more death cells.
3.8. Molecular Identification of Lactobacillus Strains
After PCR amplification, electrophoresis results of Lactobacillus amplification products was show in the Figure S2. The sequencing results in Figure 6 and Table 5. In this experiment, 12 strains of chicken derived lactic acid bacteria with good inhibitory effect on Salmonella, weak drug resistance, strong bile salt resistance, and strong adhesion to Caco-2 cells were selected for molecular sequencing analysis, according to the experimental results in Table 5, LAB53, LAB60, and LAB64 are the same strain; that is, Lactobacillus salinus strain. Except Lactobacillus reuteri strain LAB4 and Enterococcus faecium strain IAH L76, the rest were Lactobacillus plantarum strains.
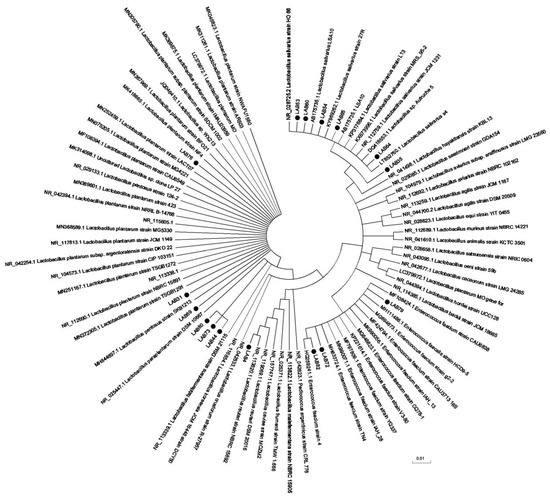
Figure 6.
Neighbor-joining phylogenetic tree based on 16 S rRNA gene sequences showing relationship between Lactobacillus isolates.

Table 5.
16S rDNA sequence analysis results of BLAST.
4. Discussion
In this experiment, 89 strains of lactic acid bacteria were isolated from Chicken Intestines of different slaughterhouses according to the national standard method and Gram staining. Using Salmonella as indicator bacteria and agar diffusion inhibition zone method, 24 strains of chicken-derived lactic acid bacteria with an average inhibition diameter of more than 10 mm were preliminarily screened. Due to the cost, all 24 strains of chicken-derived lactic acid bacteria were not identified by molecular sequencing. The drug resistance of these 24 strains of lactic acid bacteria was analyzed. Except for LAB 76 and LG, the drug resistance rate of 24 strains of lactic acid bacteria was 33%, gentamicin 21%, chloramphenicol 21%, ceftriaxone sodium 25% and tetracycline 25%. LAB12, LAB22, and LAB34 were resistant to three kinds of antibiotics, so the selected lactic acid bacteria were selected from the remaining 21 strains. At the same time, the bile salt tolerance of 24 lactic acid bacteria isolates was detected, the OD600 value of LAB4, LAB7, LAB24, LAB26, LAB31, LAB35, LAB44, LAB56, LAB65, LAB69, LAB70, LAB76, and LG were more than 0.20, and the strains still show good tolerance to the bile salt. The cell adhesion ability of 15 LAB strains differed depending on the species and strain. Murphy (2009) [26] also confirmed great adhesion differences among different strains and even within the same strain. Thus, adhesion ability is suggested to have an inevitable relationship with LAB characteristics. Adhesion can be non-specific (related to surface properties) or specific (related to adhesives) [27]. The existing adhesives include surface proteins, pili, peptidoglycans, and lipopolysaccharides. Studies have shown that the S-layer protein on the surface of Lactobacillus strains may be the active adhesion site [28]. Based on the aforementioned considerations, the adhesion of Lactobacilli to Caco-2 cells may result from the orchestrated action of various adhesives. The mechanism of Lactobacillus antagonizing the adhesion of Salmonella to Caco-2 cells was explored by three ways of exclusion, competition and substitution; the protective effect of Lactobacillus on intestinal cells was preliminarily analyzed by flow cytometry to detect the permeability of cell membrane; and the biological function of Lactobacillus SLP was preliminarily explored These results suggest that LAB group bacteria protected the AKP metabolism activity and Caco-2 cell membrane integrity by reducing the damage caused by the SAL group to the Caco-2 cell membrane.
Our results showed that the bacteriostatic zones of LAB69 and L76 were the widest (17.2 mm and 17.5 mm, respectively). Additionally, most of the 24 Lactobacillus strains inhibited the adhesion of Salmonella to the Caco-2 cell surface bacteria effectively. Moreover, after attaching to the cell surface, Lactobacillus effectively controlled the reattachment of Salmonella bacteria to cells. Further, 16 Lactobacillus strains could replace Salmonella strains effectively and adhere to the surface of Caco-2 cells. In general, incubation with Lactobacillus had protective effects on Caco-2 cell membranes, and the detection results were close to that of the control group.
5. Conclusions
In this experiment, we isolated chicken-derived Lactobacillus, which can effectively inhibit the colonization and adhesion of Salmonella, from chicken intestines. The LAB53, LAB60, LAB69, LAB72, and LAB76 Lactobacillus isolates had slightly higher tolerance and adhesion capacity, as well as probiotic potential than the standard LG strain; however, the probiotic effect of Lactobacillus isolates on the host after adhering to the intestinal tract should be studied further. We speculate that the incorporation of chicken-derived LAB during the later stage of chicken feeding might curtail Salmonella outbreaks in the breeding and production processes of chicken eggs, ultimately improving food safety, Additionally, the LAB69 and LAB76 isolates had good adhesion ability and may be rich in surface proteins. Their adhesion properties and mechanism need to be studied in detail in the future.
Supplementary Materials
The following are available online at https://www.mdpi.com/2304-8158/10/3/569/s1, Figure S1: Gram staining results of strains isolated from Lactobacillus., Figure S2: Electrophoresis results of Lactobacillus amplification products., Table S1: Colony characteristics and Bacterial forms of different strains., Table S2: Test results of physiological and biochemical characteristics of Lactobacillus.
Author Contributions
Conceptualization and study design: D.H. and X.B.; Performing experiments: D.H., Z.L. and X.H.; Review and final edit: F.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R&D Program of China, Grant No. 2018YFC1602500, NSFC (Grant No. 31972174), the Agricultural Independent Innovation Fund of Jiangsu Province (Grant number CX (18)3053), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no competing financial interest.
References
- Nagappa, K.; Tamuly, S.; Saxena, M.; Singh, S. Isolation of salmonella typhimurium from poultry eggs and meat of tarai region of uttaranchal. Indian J. Biotechnol. 2007, 6, 407–409. [Google Scholar]
- Jalali, M.; Abedi, D.; Pourbakhsh, S.A.; Ghoukasin, K. Prevalence of Salmonella spp. in raw and cooked foods in Isfahan-Iran. J. Food Saf. 2008, 28, 442–452. [Google Scholar] [CrossRef]
- Akiba, M.; Kusumoto, M.; Iwata, T. Rapid identification of Salmonella enterica serovars, typhimurium, choleraesuis, infantis, hadar, enteritidis, dublin and gallinarum, by multiplex PCR. J. Microbiol. Methods 2011, 85, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Tellez, G.; Pixley, C.; Wolfenden, R.E.; Layton, S.L.; Hargis, B.M. Probiotics/direct fed microbials for Salmonella control in poultry. Food Res. Int. 2012, 45, 628–633. [Google Scholar] [CrossRef]
- Chambers, J.R.; Gong, J. The intestinal microbiota and its modulation for Salmonella control in chickens. Food Res. Int. 2011, 44, 3159. [Google Scholar] [CrossRef]
- Revolledo, L.; Ferreira, A.J.P.; Mead, G.C. Prospects in salmonella control: Competitive exclusion, probiotics, and enhancement of avian intestinal immunity. J. Appl. Poult. Res. 2006, 15, 341–351. [Google Scholar] [CrossRef]
- Sadler, W.W.; Brownell, J.R.; Fanelli, M.J. Influence of age and inoculum level on shed pattern of Salmonella typhimurium in chickens. Avian Dis. 1969, 13, 793–803. [Google Scholar] [CrossRef]
- Gast, R.K.; Holt, P.S. Persistence of Salmonella enteritidis from one day of age until maturity in experimentally infected layer chickens. Poult. Sci. 1998, 77, 1759–1762. [Google Scholar] [CrossRef]
- Yang, B.; Qu, D.; Zhang, X.; Shen, J.; Cui, S.; Shi, Y.; Xia, M.; Shenga, M.; Zhi, S.; Mengad, J. Prevalence and characterization of Salmonella serovars in retail meats of marketplace in Shaanxi, China. Int. J. Food Microbiol. 2010, 141, 63–72. [Google Scholar] [CrossRef]
- Yin, M.; Yang, B.; Wu, Y.; Wang, L.; Wu, H.; Zhang, T.; Tuohetaribayi, G. Prevalence and characterization of Salmonella enterica serovar in retail meats in market place in Uighur, Xinjiang, China. Food Control 2016, 64, 165–172. [Google Scholar] [CrossRef]
- Chou, S.L.; Weimer, B. Isolation and characterization of acid- and bile-tolerant isolates from strains of lactobacillus aci-dophilus. J. Dairy Sci. 1999, 82, 23–31. [Google Scholar] [CrossRef]
- Xu, Z. Update of national inspection standard for lactic acid bacteria. Mod. Food 2018, 4, 47–49. [Google Scholar]
- Yin, Q.; Zheng, Q. Isolation and identification of the dominant lactobacillus in gut and faeces of pigs using carbohydrate fermentation and 16S rDNA analysis (microbial physiology and biotechnology). J. Biosci. Bioeng. 2005, 99, 68–71. [Google Scholar] [CrossRef]
- Hai, D.; Yin, X.; Lu, Z.; Lv, F.; Zhao, H.; Bie, X. Occurrence, drug resistance, and virulence genes of Salmonella isolated from chicken and eggs. Food Control 2020, 113, 107109. [Google Scholar] [CrossRef]
- Aween, M.M.; Hassan, Z.; Muhialdin, B.J.; Noor, H.M.; Eljamel, Y.A. Evaluation on antibacterial activity of lactobacillus acidophilus strains isolated from honey. Am. J. Appl. Sci. 2012, 9, 807–817. [Google Scholar] [CrossRef]
- Fothergill, A.W. Antifungal susceptibility testing: Clinical Laboratory and Standards Institute (CLSI) methods. In Interactions of Yeasts, Moulds, and Antifungal Agents; Humana Press: Totowa, NJ, USA, 2012. [Google Scholar]
- Lei, R.; Wu, H.; Yongxing, H. Screening of lactic acid bacteria with high antibacterial activity from Campylobacter. Chin. Anim. Husb. Vet. 2013, 40, 129–134. [Google Scholar]
- Stuart, M.R.; Chou, L.S.; Weimer, B.C. Influence of carbohydrate starvation and arginine on culturability and amino acid utilization of lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 1999, 65, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Jankowska, A.; Laubitz, D.; Antushevich, H.; Zabielski, R.; Grzesiuk, E. Competition of Lactobacillus paracasei with Salmonella enterica for Adhesion to Caco-2 Cells. J. Biomed. Biotechnol. 2008, 2008, 357964. [Google Scholar] [CrossRef] [PubMed]
- Adlerberth, I.; Ahrne, S.I.V.; Johansson, M.L.; Molin, G.; Hanson, L.A.; Wold, A.E. A mannose-specific adherence mechanism in Lactobacillus plantarum con-ferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 1996, 62, 2244–2251. [Google Scholar] [CrossRef]
- Chu, Y.H.; Yu, X.X.; Jin, X.; Wang, Y.T.; Zhao, D.J.; Zhang, P.; Sun, G.-M.; Zhang, Y.-H. Purification and characterization of alkaline phosphatase from lactic acid bacteria. RSC Adv. 2018, 9, 354–360. [Google Scholar] [CrossRef]
- Jensen, H.; Grimmer, S.; Naterstad, K.; Axelsson, L. In vitro testing of commercial and potential probiotic lactic acid bacteria. Int. J. Food Microbiol. 2012, 153, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.F.; Boris, S.; Barbés, C. Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J. Appl. Microbiol. 2003, 94, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Tuo, Y.; Yu, H.; Ai, L.; Wu, Z.; Guo, B.; Chen, W. Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 2013, 96, 4252–4257. [Google Scholar] [CrossRef]
- Sidira, M.; Kourkoutas, Y.; Kanellaki, M.; Charalampopoulos, D. In vitro study on the cell adhesion ability of immobilized lactobacilli on natural supports. Food Res. Int. 2015, 76, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L. Assessment of potential probiotic strains: Evaluation of their establishment, persistence, and localisation in the mu-rine gastrointestinal tract. Microb. Ecol. Health Dis. 2009, 11, 149–157. [Google Scholar]
- García-Cayuela, T.; Korany, A.M.; Bustos, I.; Gómez de Cadiñanos, L.P. Adhesion abilities of dairy Lactobacillus plantarum, strains showing an aggre-gation phenotype. Food Res. Int. 2014, 57, 44–50. [Google Scholar] [CrossRef]
- Taverniti, V.; Stuknyte, M.; Minuzzo, M.; Taverniti, V.; Stuknyte, M.; Minuzzo, M.; Arioli, S.; De Noni, I.; Scabiosi, C.; Martinez Cordova, Z.; et al. MIMLh5 on innate immunity. Appl. Environ. Microbiol. 2013, 79, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).