Fingerprint Approaches Coupled with Chemometrics to Discriminate Geographic Origin of Imported Salmon in China’s Consumer Market
Abstract
:1. Introduction
2. Materials and Methods
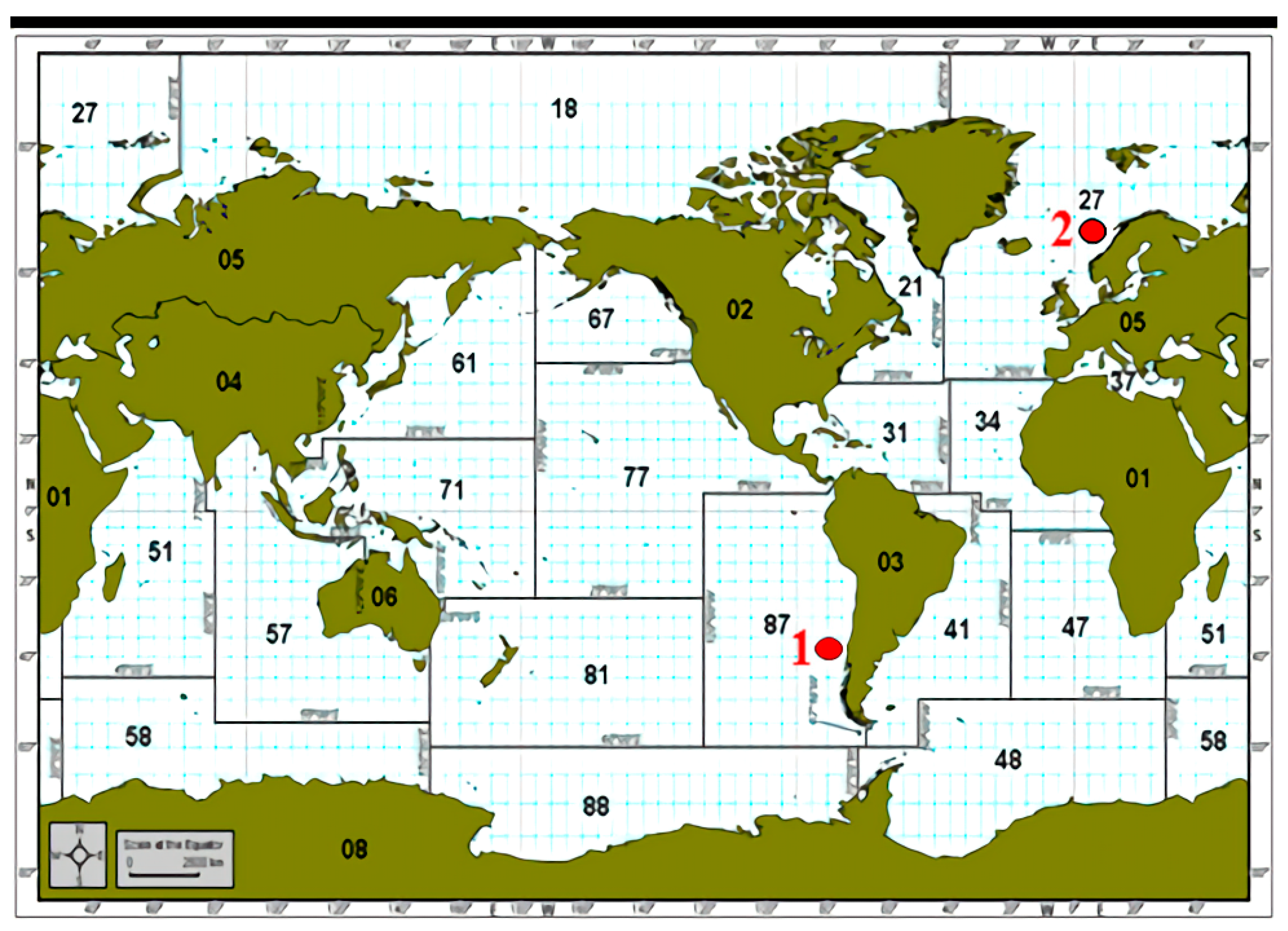
2.1. Salmon Samples, Reagents, Instruments and Equipments
2.2. Near-Infrared (NIR) Spectroscopy
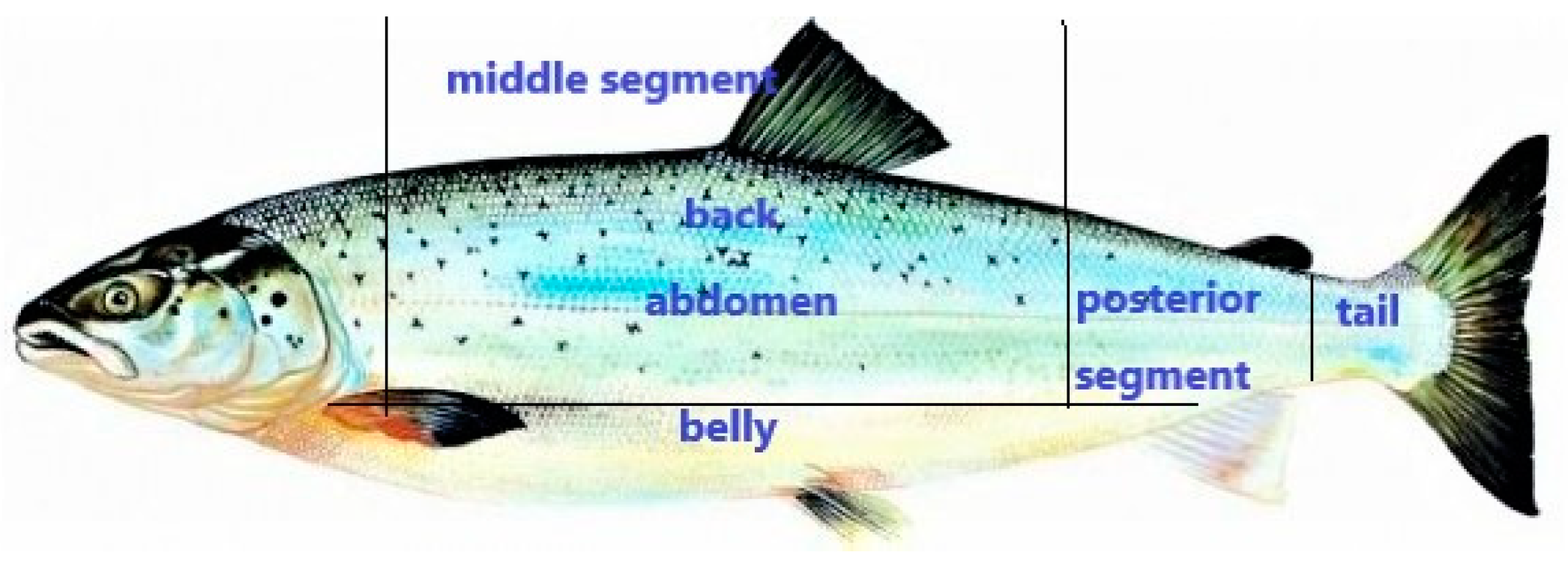
2.2.1. Sample Preparing and Detection
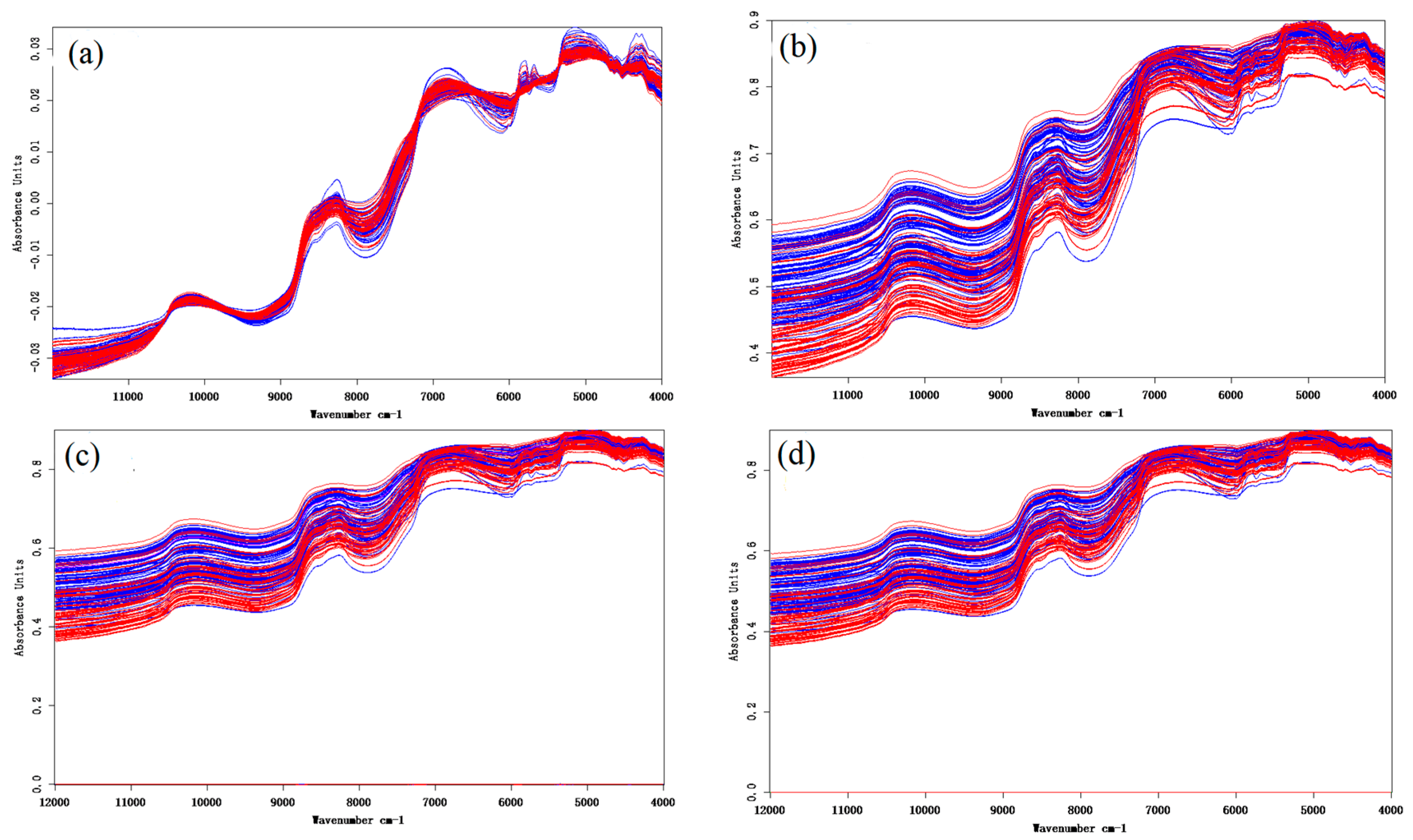
2.2.2. Spectral Data Preprocessing
2.3. Mineral Element Fingerprint (MEF)
2.3.1. Sample Preparing and Pretreatment
2.3.2. MEF Detection
2.4. Data Analysis Approaches
2.5. Other Approaches Employed and Software Implemented
3. Results and Discussion
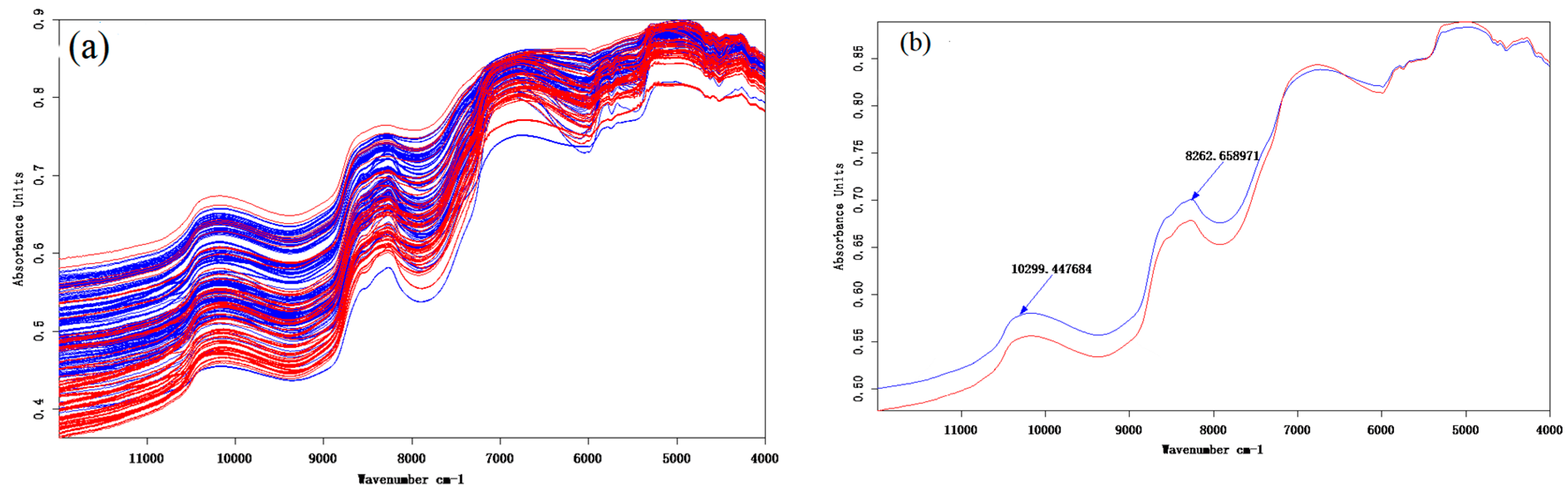
3.1. Near-Infrared (NIR) Spectrum Analysis
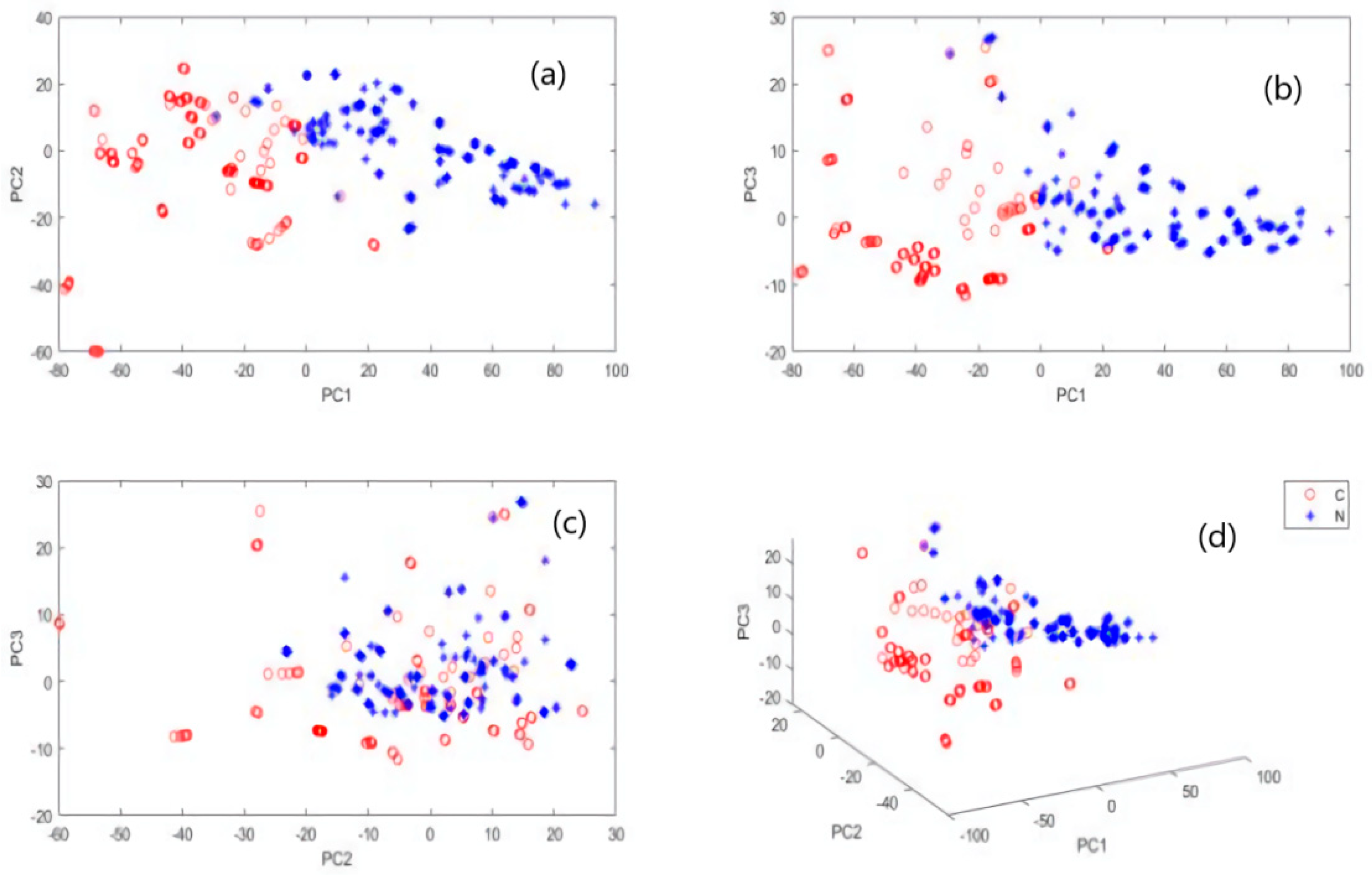
3.2. Principal Component Analysis (PCA)
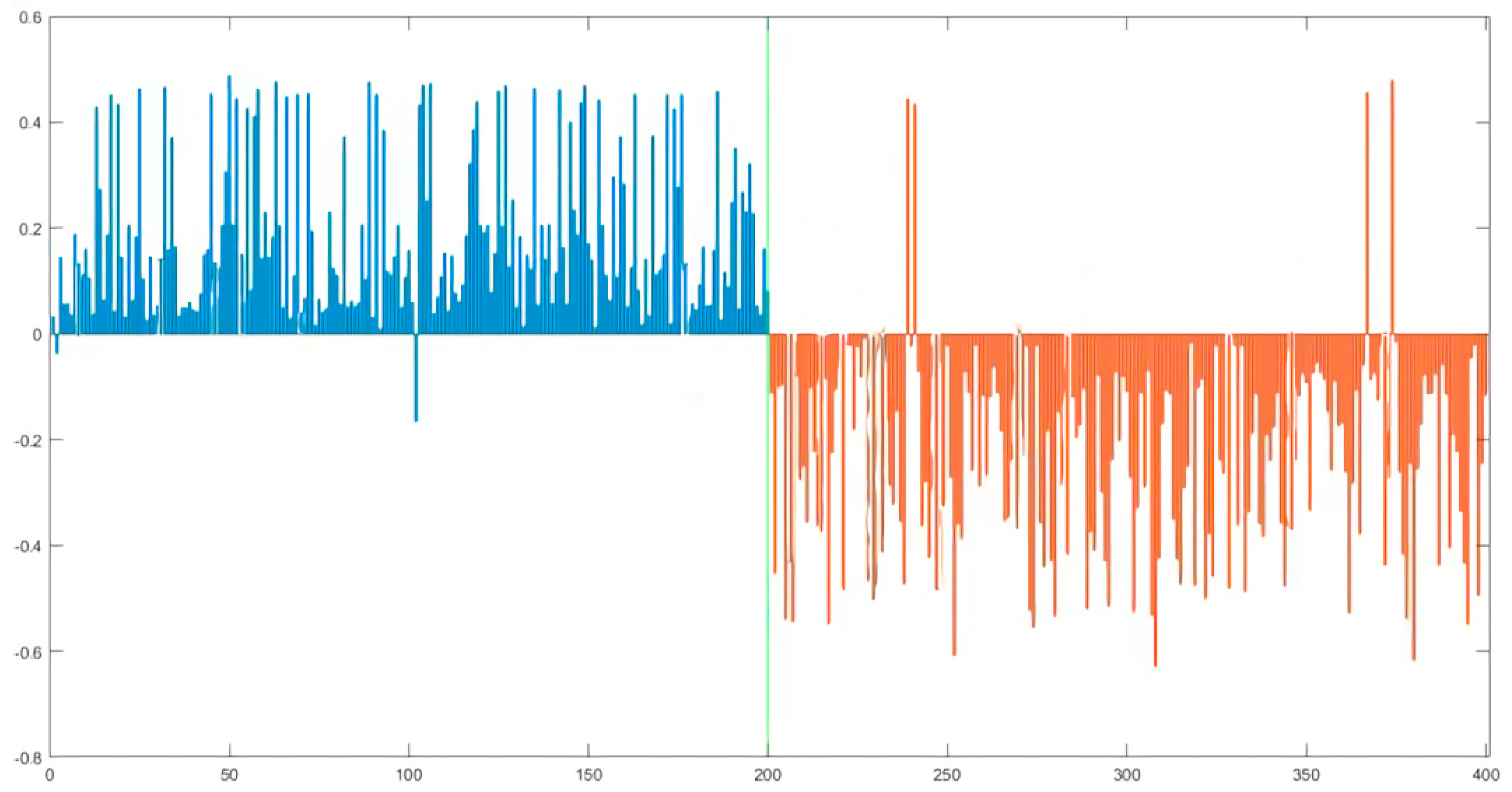
3.3. Partial Least Square (PLS) Discrimination Model
3.4. Mineral Element Fingerprint (MEF) Analysis
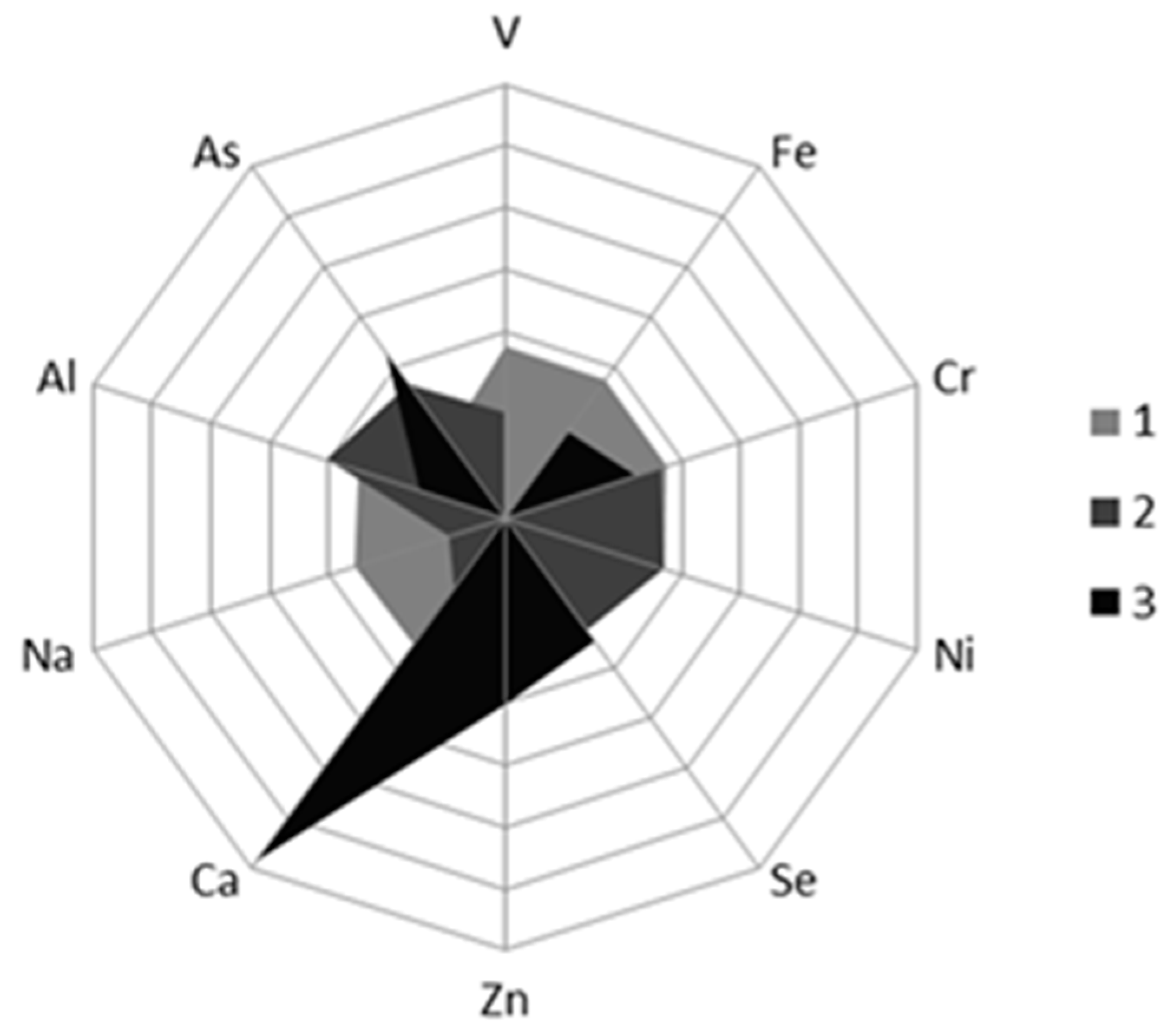
3.4.1. Differences in the Content of Mineral Element between Norwegian and Chilean Salmon
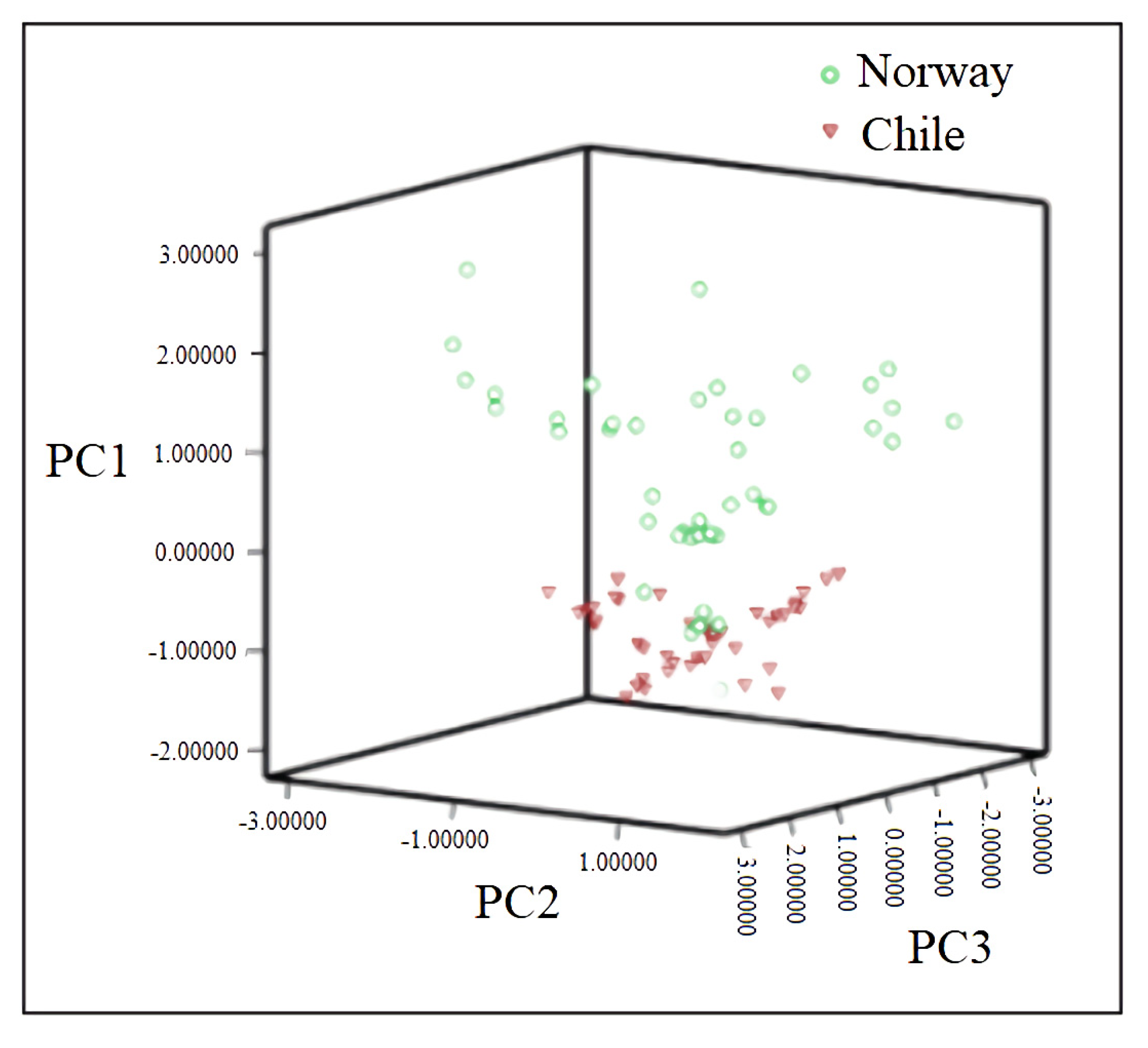
3.4.2. PCA of Mineral Elements in Norwegian and Chilean Salmons
3.4.3. Linear Discriminant Analysis (LDA) of Mineral Elements
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAO. Cultured Aquatic Species Information Programme Salmo salar (Linnaeus, 1758) [WWW Document]. Available online: http://www.fao.org/fishery/culturedspecies/Salmo_salar/en (accessed on 10 August 2019).
- Chiesa, L.M.; Pavlovic, R.; Nobile, M.; Cesare, F.D.; Malandra, R.; Pessina, D.; Panseri, S. Discrimination between Fresh and Frozen-Thawed Fish Involved in Food Safety and Fraud Protection. Foods 2020, 9, 1896. [Google Scholar] [CrossRef]
- Waagbø, R.; Hamre, K.; Bjerkas, E.; Berge, R.; Wathne, E.; Lie, O.; Torstensen, B. Cataract formation in Atlantic salmon, Salmo salar L., smolt relative to dietary pro- and antioxidants and lipid level. J. Fish. Dis. 2003, 26, 213–229. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, J.E.; Metcalfe, N.B.; Huntingford, F.A. Behavioural influence on life-history variation in juvenile Atlantic salmon, Salmo salar. Environ. Biol. Fish 1992, 33, 331–340. [Google Scholar] [CrossRef]
- Fidalgo, L.G. Microbial and physicochemical evolution during hyperbaric storage at room temperature of fresh Atlantic salmon (Salmo salar). Innov. Food. Sci. Emerg. Technol. 2018, 45, 264–272. [Google Scholar] [CrossRef]
- Cai, L. Effects of different freezing treatments on physicochemical responses and microbial characteristics of Japanese sea bass (Lateolabrax japonicas) fillets during refrigerated storage. LWT Food. Sci. Technol. 2014, 59, 122–129. [Google Scholar] [CrossRef]
- Haq, M. Modifications of Atlantic salmon by-product oil for obtaining different w-3 polyunsaturated fatty acids concentrates: An approach to comparative analysis. J. Food Drug Anal. 2017, 26, 545–556. [Google Scholar] [CrossRef] [Green Version]
- Jedermann, R.; Ruiz-Garcia, L.; Lang, W. Spatial temperature profiling by semi-passive RFID loggers for perishable food transportation. Comput. Electron. Agric. 2009, 65, 145–154. [Google Scholar] [CrossRef] [Green Version]
- Aung, M.M.; Chang, Y.S. Temperature management for the quality assurance of a perishable food supplychain. Food Control 2014, 40, 198–207. [Google Scholar] [CrossRef]
- FAO. Global Aquaculture Leaders’ Vision Summit 2020 annual meeting. In The State of World Fisheries and Aquaculture 2020; Sustainability in action; FAO: Rome, Italy, 2020. [Google Scholar] [CrossRef]
- Christiansen, H.; Fournier, N.; Hellemans, B.; Volckaert, F.A.M. Seafood substitution and mislabeling in Brussels’ restaurants and canteens. Food Control 2018, 85, 66–75. [Google Scholar] [CrossRef]
- Luque, G.M.; Donlan, C.J. The characterization of seafood mislabeling: A global meta-analysis. Biol. Conserv. 2019, 236, 556–570. [Google Scholar] [CrossRef]
- Deconinck, D.; Volckaert, F.A.M.; Hostens, K.; Panicz, R.; Eljasik, P.; Faria, M.; Monteiro, C.S.; Robbens, J.; Sofie, D. A high-quality genetic reference database for European commercial fishes reveals substitution fraud of processed Atlantic cod (Gadus morhua) and common sole (Solea solea) at different steps in the Belgian supply chain. Food Chem. Toxicol. 2020, 141, 111417. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Colmenero, M.; Juanes, F.; Dopico, E.; Martinez, J.L.; Garcia-Vazquez, E. Economy matters: A study of mislabeling in salmon products from two regions, Alaska and Canada (Northwest of America) and Asturias (Northwest of Spain). Fish. Res. 2017, 195, 180–185. [Google Scholar] [CrossRef]
- Jacquet, J.L.; Pauly, D. Trade secrets: Renaming and mislabeling of seafood. Mar. Policy 2008, 32, 309–318. [Google Scholar] [CrossRef]
- Armani, A.; Guardone, L.; La Castellana, R.; Gianfaldoni, D.; Guidi, A.; Castigliego, L. DNA barcoding reveals commercial and health issues in ethnic seafood sold on the Italian market. Food Control 2015, 55, 206–214. [Google Scholar] [CrossRef] [Green Version]
- Pardo, M.Á.; Jiménez, E.; Pérez-Villarreal, B. Misdescription incidents in seafood sector. Food Control 2016, 62, 277–283. [Google Scholar] [CrossRef]
- Quinto, C.A.; Tinoco, R.; Hellberg, R.S. DNA barcoding reveals mislabeling of game meat species on the U.S. commercial market. Food Control 2016, 59, 386–392. [Google Scholar] [CrossRef] [Green Version]
- Moretti, V.M.; Turchini, G.M.; Bellagamba, F.; Caprino, F. Traceability issues in fishery and aquaculture products. Vet. Res. Commun. 2003, 27, 497–505. [Google Scholar] [CrossRef]
- Grazina, L.; Rodrigues, P.J.; Igrejas, G.; Nunes, M.A.; Mafra, I.; Arlorio, M.; Amaral, J.S. Machine learning approaches applied to gc-fid fatty acid profiles to discriminate wild from farmed salmon. Foods 2020, 9, 1622. [Google Scholar] [CrossRef]
- Masoum, S.; Malabat, C.; Jalali-Heravi, M.; Guillou, C.; Rezzi, S.; Rutledge, D.N. Application of support vector machines to 1 h nmr data of fish oils: Methodology for the confirmation of wild and farmed salmon and their origins. Anal. Bioanal. Chem. 2007, 387, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Maldini, M.; Marzano, F.N.; Fortes, G.G.; Papa, R.; Gandolfi, G. Fish and seafood traceability based on aflp markers: Elaboration of a species database. Aquaculture 2006, 261, 487–494. [Google Scholar] [CrossRef]
- Thu, V.; Nguyen, T.V.; Amoroso, G.; Ventura, T.; Elizur, A. Deploying new generation sequencing for the study of flesh color depletion in atlantic salmon (Salmo salar). BMC Genom. 2021, 22, 545. [Google Scholar] [CrossRef]
- Khaksar, R.; Carlson, T.; Schaffner, D.W.; Ghorashi, M.; Best, D.; Jandhyala, S.; Traverso, J.; Amini, S. Unmasking seafood mislabeling in us markets: Dna barcoding as a unique technology for food authentication and quality control. Food Control 2015, 56, 71–76. [Google Scholar] [CrossRef]
- Taggart, J.B.; Mclaren, I.S.; Hay, D.W.; Webb, J.H.; Youngson, A.F. Spawning success in Atlantic salmon (Salmo salar L.): A long-term DNA profiling-based study conducted in a natural stream. Mol. Ecol. 2010, 10, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Deconinck, D.; Hostens, K.; Taverniers, I.; Volckaert, F.A.; Robbens, J.; Derycke, S. Identification and semi-quantification of Atlantic salmon in processed and mixed seafood products using Droplet Digital PCR (ddPCR). Food Chem. Toxicol. 2021, 154, 112329. [Google Scholar] [CrossRef] [PubMed]
- Kg, A.; Dmab, C.; Jsa, B.; Ns, C. Determining the provenance and authenticity of seafood: A review of current methodologies. Trends Food Sci. Tech. 2019, 91, 294–304. [Google Scholar] [CrossRef]
- Carter, J.F.; Tinggi, U.; Yang, X.; Fry, B. Stable isotope and trace metal compositions of Australian prawns as a guide to authenticity and wholesomeness. Food Chem. 2015, 170, 241–248. [Google Scholar] [CrossRef]
- Ricardo, F.; Génio, L.; Costa Leal, M.; Albuquerque, R.; Queiroga, H.; Rosa, R.; Calado, R. Trace element fingerprinting of cockle (Cerastoderma edule) shells can reveal harvesting location in adjacent areas. Sci. Rep. 2015, 5, 11932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorte, C.J.B.; Etter, R.J.; Spackman, R.; Boyle, E.E.; Hannigan, R.E. Elemental fingerprinting of mussel shells to predict population sources and redistribution potential in the Gulf of Maine. PLoS ONE 2013, 8, e80868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Boyd, C.E.; Odom, J.; Dong, S. Identification of ictalurid catfish fillets to rearing location using elemental profiling. J. World Aquacult. Soc. 2013, 44, 405–414. [Google Scholar] [CrossRef]
- Anderson, K.A.; Hobbie, K.A.; Smith, B.W. Chemical profiling with modeling differentiates wild and farm-raised salmon. Agric. Food Chem. 2010, 58, 11768–11774. [Google Scholar] [CrossRef] [PubMed]
- Dunphy, B.J.; Millet, M.A.; Jeffs, A.G. Elemental signatures in the shells of early juvenile green-lipped mussels (Perna canaliculus) and their potential use for larval tracking. Aquaculture 2011, 311, 187–192. [Google Scholar] [CrossRef]
- Cubadda, F.; Raggi, A.; Coni, E. Element fingerprinting of marine organisms by dynamic reaction cell inductively coupled plasma mass spectrometry. Anal. Bioanal. Chem. 2006, 384, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Costas-Rodríguez, M.; Lavilla, I.; Bendicho, C. Classification of cultivated mussels from Galicia (Northwest Spain) with European Protected Designation of Origin using trace element fingerprint and chemometric analysis. Anal. Chim. Acta 2010, 664, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Boyd, C.E.; Odom, J. Identification of Pacific white shrimp (Litopenaeus vannamei) to rearing location using elemental profiling. Food Control 2014, 45, 70–75. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N.; Crawford, J.; Gadd, P. Combined use of stable isotope analysis and elemental profiling to determine provenance of black tiger prawns (Penaeus monodon). Food Control 2019, 95, 242–248. [Google Scholar] [CrossRef]
- Gopi, K.; Mazumder, D.; Sammut, J.; Saintilan, N.; Crawford, J.; Gadd, P. Isotopic and elemental profiling to trace the geographic origins of farmed and wild-caught Asian seabass (Lates calcarifer). Aquaculture 2019, 502, 56–62. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, J.; Han, D.; Zhao, X.; Chen, X.; Liu, Y. Geographical origin traceability and species identification of three scallops (Patinopecten yessoensis, Chlamys farreri, and Argopecten irradians) using stable isotope analysis. Food Chem. 2019, 299, 125107. [Google Scholar] [CrossRef]
- Christopher, S.J.; Ellisor, D.L.; Davis, W.C. Investigating the feasibility of ICP-MS/MS for differentiating NIST salmon reference materials through determination of Sr and S isotope ratios. Talanta 2021, 231, 122363. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Wang, Z.; Xia, L.; Robbat, A. Application of feature selection algorithms to select elements that distinguish regional differences in chinese grown winter jujube fruit (Zizyphus jujuba mill. cv. Huanghua dongzao). Food Anal. Method 2020, 14, 1–10. [Google Scholar] [CrossRef]
- Espinasse, B.; Hunt, B.P.; Finney, B.P.; Fryer, J.K.; Bugaev, A.V.; Pakhomov, E.A. Using stable isotopes to infer stock-specific high-seas distribution of maturing sockeye salmon in the North Pacific. Ecol. Evol. 2020, 10, 13555–13570. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Cai, C.B.; Cui, H.F.; Ye, Z.H.; Yu, X.P. Rapid discrimination of pork in halal and non-halal chinese ham sausages by fourier transform infrared (FTIR) spectroscopy and chemometrics. Meat Sci. 2012, 92, 506–510. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least-squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Xu, L.; Cai, C.B.; Deng, D.H. Multivariate quality control solved by one-class partial least squares regression: Identification of adulterated peanut oils by mid-infrared spectroscopy. Chemometrics 2011, 25, 568–574. [Google Scholar] [CrossRef]
- Xu, L.; Yan, S.M.; Ye, Z.H.; Yu, X.P. Combining Electronic Tongue Array and Chemometrics for Discriminating the Specific Geographical Origins of Green Tea. J. Anal. Methods Chem. 2013, 2013, 350801. [Google Scholar] [CrossRef]
- Fu, X.S.; Xu, L.; Yu, X.P.; Cui, H.F. Robust and automated internal quality grading of a Chinese green tea (Longjing) by Near-Infrared spectroscopy and chemometrics. J. Spectrosc. 2013, 2013, 139347. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Yan, S.M.; Cai, C.B.; Yu, X.P. One-class partial least squares (OCPLS) classifier. Chemom. Intell. Lab. Syst. 2013, 126, 1–5. [Google Scholar] [CrossRef]
- Oliveri, P. Class-modelling in food analytical chemistry: Development, sampling, optimisation and validation issues—A tutorial. Anal. Chim. Acta 2017, 982, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Goodarzi, M.; Shi, W.; Cai, C.B.; Jiang, J.H. A MATLAB toolbox for class modeling using one-class partial least squares (OCPLS) classifiers. Chemom. Intell. Lab. Syst. 2014, 139, 58–63. [Google Scholar] [CrossRef]
- Wold, S. Pattern recognition by means of disjoint principal components models. Pattern Recognit. 1976, 8, 127–139. [Google Scholar] [CrossRef]
- Xu, Q.S.; Liang, Y.Z. Monte Carlo cross validation. Chemom. Intell. Lab. Syst. 2001, 56, 1–11. [Google Scholar] [CrossRef]
- Han, C.; Dong, S.; Li, L.; Wei, F.; Zhou, Y.; Gao, Q. The effect of the seasons on geographical traceability of salmonid based on multi-element analysis. Food Control 2020, 109, 106893. [Google Scholar] [CrossRef]
- Giraudo, A.; Grassi, S.; Savorani, F.; Gavoci, G.; Casiraghi, E.; Geobaldo, F. Determination of the geographical origin of green coffee beans using NIR spectroscopy and multivariate data analysis. Food Control 2018, 99, 137–145. [Google Scholar] [CrossRef] [Green Version]
| Parameter | ICP-MS | ICP-OES |
|---|---|---|
| Setting Value | Setting Value | |
| Radio-frequency power | 1300 W | 1300 W |
| Scan times | 100 times | 100 times |
| scan patterns | Peak height | Peak height |
| Dwell time | 10 ms | 10 ms |
| Acquisition time | 20 s | 20 s |
| Sample uptake rate | 1 mL/min | 1.5 mL/min |
| Plasma gas flow | 13 L/min | 15 L/min |
| Coolant gas flow | 15 L/min | 12 L/min |
| Auxiliary gas flow | 1.2 L/min | 0.2 L/min |
| Atomizer gas flow | 0.87 L/min | 0.55 L/min |
| Pretreatment Methods | RMSE | R2 |
|---|---|---|
| Original Spectra | 0.198 | 0.879 |
| VN | 0.173 | 0.968 |
| SG9 | 0.167 | 0.974 |
| FD + SG9 | 0.159 | 0.983 |
| SD + SG9 | 0.163 | 0.976 |
| Element | Index | Norway | Chile | Significant Difference |
|---|---|---|---|---|
| Pb (ppb) | content | 0.61 ± 0.14 | 0.53 ± 0.28 | No |
| variable coefficient (%) | 22.4 | 53.5 | ||
| Fe (ppb) | content | 115 ± 35.9 | 102 ± 24.7 | Yes |
| variable coefficient (%) | 31.3 | 24.1 | ||
| Mn (ppb) | content | 1.29 ± 2.16 | 0.71 ± 1.16 | No |
| variable coefficient (%) | 168 | 164 | ||
| Cu (ppb) | content | 6.10 ± 0.99 | 6.40 ± 1.19 | No |
| variable coefficient (%) | 16.2 | 18.4 | ||
| Zn (ppb) | content | 63.9 ± 11.7 | 57.9 ± 7.65 | Yes |
| variable coefficient (%) | 18.4 | 13.2 | ||
| Al (ppb) | content | 44.6 ± 20.1 | 30.9 ± 17.0 | Yes |
| variable coefficient (%) | 45.2 | 55.1 | ||
| Sr (ppb) | content | 0.21 ± 0.79 | 0.03 ± 0.02 | No |
| variable coefficient (%) | 369 | 92.3 | ||
| Ni (ppb) | content | 2.39 ± 1.28 | 1.28 ± 0.83 | Yes |
| variable coefficient (%) | 53.6 | 64.8 | ||
| As (ppb) | content | 3.07 ± 1.03 | 3.99 ± 0.70 | Yes |
| variable coefficient (%) | 33.6 | 17.5 | ||
| Cr (ppb) | content | 16.3 ± 3.23 | 12.6 ± 2.90 | Yes |
| variable coefficient (%) | 19.9 | 23.1 | ||
| V (ppb) | content | 0.35 ± 0.24 | 0.09 ± 0.14 | Yes |
| variable coefficient (%) | 70.2 | 167 | ||
| Se (ppb) | content | 4.18 ± 0.63 | 6.82 ± 0.49 | Yes |
| variable coefficient (%) | 15.4 | 7.13 | ||
| K (ppm) | content | 65.7 ± 7.78 | 65.9 ± 6.37 | No |
| variable coefficient (%) | 11.8 | 9.66 | ||
| Ca (ppm) | content | 2.28 ± 2.20 | 1.14 ± 0.52 | Yes |
| variable coefficient (%) | 96.8 | 45.6 | ||
| Na (ppm) | content | 13.8 ± 4.05 | 9.26 ± 3.11 | Yes |
| variable coefficient (%) | 29.3 | 33.6 | ||
| Mg (ppm) | content | 4.79 ± 0.77 | 5.00 ± 0.37 | No |
| variable coefficient (%) | 16.1 | 7.41 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Hong, X.; Liao, J.; Ji, Q.; Li, C.; Zhang, M.; Ye, Z.; Yu, X. Fingerprint Approaches Coupled with Chemometrics to Discriminate Geographic Origin of Imported Salmon in China’s Consumer Market. Foods 2021, 10, 2986. https://doi.org/10.3390/foods10122986
Fu X, Hong X, Liao J, Ji Q, Li C, Zhang M, Ye Z, Yu X. Fingerprint Approaches Coupled with Chemometrics to Discriminate Geographic Origin of Imported Salmon in China’s Consumer Market. Foods. 2021; 10(12):2986. https://doi.org/10.3390/foods10122986
Chicago/Turabian StyleFu, Xianshu, Xuezhen Hong, Jinyan Liao, Qingge Ji, Chaofeng Li, Mingzhou Zhang, Zihong Ye, and Xiaoping Yu. 2021. "Fingerprint Approaches Coupled with Chemometrics to Discriminate Geographic Origin of Imported Salmon in China’s Consumer Market" Foods 10, no. 12: 2986. https://doi.org/10.3390/foods10122986
APA StyleFu, X., Hong, X., Liao, J., Ji, Q., Li, C., Zhang, M., Ye, Z., & Yu, X. (2021). Fingerprint Approaches Coupled with Chemometrics to Discriminate Geographic Origin of Imported Salmon in China’s Consumer Market. Foods, 10(12), 2986. https://doi.org/10.3390/foods10122986








