Assessment of the Spoilage Microbiota during Refrigerated (4 °C) Vacuum-Packed Storage of Fresh Greek Anthotyros Whey Cheese without or with a Crude Enterocin A-B-P-Containing Extract
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of the Crude Enterocin A-B-P Biopreservative Extract
2.2. Commercial Anthotyros Whey Cheese Samples
2.3. Enterocin Addition and Cheese Storage and Sampling
2.4. Cheese Analyses
2.5. Isolation of Representative Colonies of the Dominant Whey Cheese Spoilage Microbiota
2.6. Biochemical Characterization of Whey Cheese Spoilage LAB Isolates
2.7. Molecular Identification of Representative Whey Cheese Spoilage LAB Isolates
2.8. Biochemical Identification of Whey Cheese Spoilage Gram-Negative Isolates
2.9. Statistical Analysis
3. Results and Discussion
3.1. Evolution of the Whey Cheese Spoilage Microbiota in Relation with the Cheese pH Changes during Storage
3.2. Biochemical Characterization and Distribution of the LAB Biota in Anthotyros Cheeses
3.3. Identification of Representative Whey Cheese Spoilage Biotypes of Leuconostoc and Carnobacterium at the Species Level by 16S rRNA Sequencing
3.4. Biochemical Identification of Gram-Negative Spoilage Bacterial Isolates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pintado, M.E.; Macedo, A.C.; Malcata, F.X. Review: Technology, chemistry and microbiology of whey cheeses. Food Sci. Technol. Int. 2001, 7, 105–116. [Google Scholar] [CrossRef]
- Alichanidis, E.; Polychroniadou, A. Characteristics of major traditional regional cheese varieties of East-Mediterranean countries: A review. Dairy Sci. Technol. 2008, 88, 495–510. [Google Scholar] [CrossRef]
- Samelis, J.; Kakouri, A.; Rogga, K.J.; Savvaidis, I.N.; Kontominas, M.G. Nisin treatments to control Listeria monocytogenes post-processing contamination on Anthotyros, a traditional Greek whey cheese, stored at 4 C in vacuum packages. Food Microbiol. 2003, 20, 661–669. [Google Scholar] [CrossRef]
- Hough, G.; Puglieso, M.L.; Sanchez, R.; Da Silva, O.M. Sensory and microbiological shelf-life of a commercial Ricotta cheese. J. Dairy Sci. 1999, 82, 454–459. [Google Scholar] [CrossRef]
- Pala, C.; Scarano, C.; Venusti, M.; Sardo, D.; Casti, D.; Cossu, F.; Lamon, S.; Spanu, V.; Ibba, M.; Marras, M. Shelf life evaluation of ricotta fresca sheep cheese in modified atmosphere packaging. Ital. J. Food Saf. 2016, 5, 5502. [Google Scholar] [CrossRef][Green Version]
- Pintado, M.E.; Malcata, F.X. Characterization of whey cheese packaged under vacuum. J. Food Prot. 2000, 63, 216–221. [Google Scholar] [CrossRef]
- Sattin, E.; Andreani, N.A.; Carraro, L.; Fasolato, L.; Balzan, S.; Novelli, E.; Squartini, A.; Telatin, A.; Simionati, B.; Cardazzo, B. Microbial dynamics during shelf-life of industrial Ricotta cheese and identification of a Bacillus strain as a cause of a pink discolouration. Food Microbiol. 2016, 57, 8–15. [Google Scholar] [CrossRef]
- Genigeorgis, C.; Carniciu, M.; Dutulescu, D.; Farver, T.B. Growth and survival of Listeria monocytogenes in market cheeses stored at 4 °C to 30 °C. J. Food Prot. 1991, 54, 662–668. [Google Scholar] [CrossRef] [PubMed]
- Papageorgiou, D.K.; Bori, M.; Mantis, A. Growth of Listeria monocytogenes in the whey cheeses Myzithra, Anthotyros, and Manouri during storage at 5, 12, and 22 °C. J. Food Prot. 1996, 59, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.A.; Bevis, H.E.; Delves-Broughton, J. The use of bacteriocin nisin, as a preservative in ricotta-type cheeses to control the food-borne pathogen Listeria monocytogenes. Lett. Appl. Microbiol. 1997, 24, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Di Pierro, P.; Sorrentino, A.; Mariniello, L.; Giosafatto, C.V.L.; Porta, R. Chitosan/whey protein film as active coating to extend Ricotta cheese shelf-life. LWT-Food Sci. Technol. 2011, 44, 2324–2327. [Google Scholar] [CrossRef]
- Fernández, M.V.; Jagus, R.J.; Mugliaroli, S.L. Effect of combined natural antimicrobials on spoilage microorganisms and Listeria innocua in a whey cheese “Ricotta”. Food Bioprocess Technol. 2014, 7, 2528–2537. [Google Scholar] [CrossRef]
- Spanu, C.; Spanu, V.; Pala, C.; Virdis, S.; Scarano, C.; De Santis, E.P.L. Evaluation of a post-lethality treatment against Listeria monocytogenes on Ricotta salata cheese. Food Control. 2013, 30, 200–205. [Google Scholar] [CrossRef]
- Spanu, C.; Scarano, C.; Piras, F.; Spanu, V.; Pala, C.; Casti, D.; Lamon, S.; Cossu, F.; Ibba, M.; Nieddu, G. Testing commercial biopreservative against spoilage microorganisms in MAP packed Ricotta fresca cheese. Food Microbiol. 2017, 66, 72–76. [Google Scholar] [CrossRef]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological characteristics of Greek traditional cheeses. Small Rumin. Res. 2011, 101, 17–32. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Nychas, G.J.E.; Sofos, J.N. Types of traditional Greek foods and their safety. Food Cont. 2013, 29, 32–41. [Google Scholar] [CrossRef]
- Arvanitoyannis, I.S.; Kargaki, G.K.; Hadjichristodoulou, C. Effect of three MAP compositions on the physical and microbiological properties of a low fat Greek cheese known as “Anthotyros”. Anaerobe 2011, 17, 295–297. [Google Scholar] [CrossRef]
- Kalogridou-Vassiliadou, D.; Tzanetakis, N.; Litopoulou-Tzanetaki, E. Microbiological and physicochemical characteristics of “Anthotyro”, a Greek traditional whey cheese. Food Microbiol. 1994, 11, 15–19. [Google Scholar] [CrossRef]
- Lioliou, K.; Litopoulou-Tzanetaki, E.; Tzanetakis, N.; Robinson, R.K. Changes in the microflora of Manouri, a traditional Greek whey cheese, during storage. Int. J. Dairy Technol. 2001, 54, 100–106. [Google Scholar] [CrossRef]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. The microfloras of traditional Greek cheeses. Microbiol. Spectr. 2014, 2, CM-0009-2012. [Google Scholar] [CrossRef]
- Dermiki, M.; Ntzimani, A.; Badeka, A.; Savvaidis, I.N.; Kontominas, M.G. Shelf-life extension and quality attributes of the whey cheese “MyzithraKalathaki” using modified atmosphere packaging. LWT-Food Sci. Technol. 2008, 41, 284–294. [Google Scholar] [CrossRef]
- Papaioannou, G.; Chouliara, I.; Karatapanis, A.E.; Kontominas, M.G.; Savvaidis, I.N. Shelf-life of a Greek whey cheese under modified atmosphere packaging. Int. Dairy J. 2007, 17, 358–364. [Google Scholar] [CrossRef]
- Tsiraki, M.I.; Savvaidis, I.N. Effect of packaging and basil essential oil on the quality characteristics of whey cheese “Anthotyros”. Food Bioprocess Technol. 2013, 6, 124–132. [Google Scholar] [CrossRef]
- Temiz, H.; Aykut, U.; Hursit, A.K. Shelf life of Turkish whey cheese (Lor) under modified atmosphere packaging. Int. J. Dairy Technol. 2009, 62, 378–386. [Google Scholar] [CrossRef]
- Irkin, R. Shelf-life of unsalted and light ‘Lor’ whey cheese stored under various packaging conditions: Microbiological and sensory attributes. J. Food Process. Preserv. 2011, 35, 163–178. [Google Scholar] [CrossRef]
- Pintado, M.E.; Malcata, F.X. The effect of modified atmosphere packaging on the microbial ecology in Requeijao, a Portuguese whey cheese. J. Food Process. Preserv. 2000, 24, 107–124. [Google Scholar] [CrossRef]
- Gérard, A.; El-Hajjaji, S.; Niyonzima, E.; Daube, G.; Sindic, M. Prevalence and survival of Listeria monocytogenes in various types of cheese—A review. Int. J. Dairy Technol. 2018, 71, 825–843. [Google Scholar] [CrossRef]
- Silva, C.C.G.; Silva, S.P.M.; Ribeiro, S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018, 9, 594. [Google Scholar] [CrossRef]
- Spanu, C.; Piras, F.; Mocci, A.M.; Nieddu, G.; De Santis, E.P.L.; Scarano, C. Use of Carnobacterium spp protective culture in MAP packed Ricotta fresca cheese to control Pseudomonas spp. Food Microbiol. 2018, 74, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Falardeau, J.; Trmčić, A.; Wang, S. The occurrence, growth, and biocontrol of Listeria monocytogenes in fresh and surface-ripened soft and semisoft cheeses. Compr. Rev. Food Sci. Food Saf. 2021, 20, 4019–4048. [Google Scholar] [CrossRef]
- Dapkevicious, M.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current trends of enterococci in dairyproducts: A comprehensivereview of their multiple roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef] [PubMed]
- Aspri, M.; O’Connor, P.M.; Field, D.; Cotter, P.D.; Ross, P.; Hill, C.; Papademas, P. Application of bacteriocin-producing Enterococcus faecium isolated from donkey milk, in the bio-control of Listeria monocytogenes in fresh whey cheese. Int. Dairy J. 2017, 73, 1–9. [Google Scholar] [CrossRef]
- Giraffa, G. Functionality of enterococci in dairy products. Int. J. Food Microbiol. 2003, 88, 215–222. [Google Scholar] [CrossRef]
- Ribeiro, S.C.; Ross, R.P.; Stanton, C.; Silva, C.C.G. Characterization and application of antilisterialenterocins on model fresh cheese. J. Food Prot. 2017, 80, 1303–1316. [Google Scholar] [CrossRef]
- Giannou, E.; Lianou, A.; Kakouri, A.; Kallimanis, A.; Drainas, C.; Samelis, J. Identification and biopreservation potential of Enterococcus spp. isolated from fully ripened Graviera, a traditional hard Greek cheese. Ital. J. Food Sci. 2009, 21, 135–147. [Google Scholar]
- Vandera, E.; Parapouli, M.; Kakouri, A.; Koukkou, A.I.; Hatziloukas, E.; Samelis, J. Structural enterocin gene profiles and mode of antilisterial activity in synthetic liquid media and skim milk of autochthonous Enterococcus spp. isolates from artisan Greek Graviera and Galotyri cheeses. Food Microbiol. 2020, 86, 103335. [Google Scholar] [CrossRef] [PubMed]
- Tsanasidou, C.; Asimakoula, S.; Sameli, N.; Fanitsios, C.; Vandera, E.; Bosnea, L.; Koukou, A.I.; Samelis, J. Safety evaluation, biogenic amine formation, and enzymatic activity profiles of autochthonous enterocin-producing Greek cheese isolates of the Enterococcus faecium/durans group. Microorganisms 2021, 9, 777. [Google Scholar] [CrossRef]
- Samelis, J.; Kakouri, A. Hurdle factors minimizing growth of Listeria monocytogenes while counteracting in situ antilisterial effects of a novel nisin A-producing Lactococcus lactis subsp. cremoris costarter in thermized cheese milks. AIMS Microbiol. 2018, 4, 19–41. [Google Scholar] [CrossRef]
- Samelis, J.; Doulgeraki, A.I.; Bikouli, V.; Pappas, D.; Kakouri, A. Microbiological and metagenomic characterization of a retail delicatessen Galotyri-like fresh acid-curd cheese product. Fermentation 2021, 7, 67. [Google Scholar] [CrossRef]
- Tsafrakidou, P.; Sameli, N.; Bosnea, L.; Chorianopoulos, N.; Samelis, J. Assessment of the spoilage microbiota in minced free-range chicken meat during storage at 4 °C in retail modified atmosphere packages. Food Microbiol. 2021, 99, 103822. [Google Scholar] [CrossRef]
- Spanu, C.; Scarano, C.; Spanu, V.; Pala, C.; Casti, D.; Lamon, S.; Cossu, F.; Ibba, M.; Nieddu, G.; De Santis, E.P.L. Occurrence and behavior of Bacillus cereus in naturally contaminated ricotta salata cheese during refrigerated storage. Food Microbiol. 2016, 58, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Angelidis, A.S.; Chronis, E.N.; Papageorgiou, D.K.; Kazakis, I.I.; Arsenoglou, K.C.; Stathopoulos, G.A. Non-lactic acid, contaminating microbial flora in ready-to-eat foods: A potential food-quality index. Food Microbiol. 2006, 23, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Pappa, E.C.; Samelis, J.; Kondyli, E.; Pappas, A.C. Characterisation of Urda whey cheese: Evolution of main biochemical and microbiological parameters during ripening and vacuum packaged cold storage. Int. Dairy J. 2016, 58, 54–57. [Google Scholar] [CrossRef]
- Hemme, D.; Foucaud-Scheunemann, C. Leuconostoc, characteristics, use in dairy technology and prospects in functional foods. Int. Dairy J. 2004, 14, 467–494. [Google Scholar] [CrossRef]
- Holzapfel, W.H.; Bjorkroth, J.A.; Dicks, L.M.T. Genus I Leuconostoc van Tieghem 1878, 198AL. In Bergey’s Manual of Systematic Bacteriology, the Firmicutes, 2nd ed.; Whitman, W.B., Ed.; Springer: New York, NY, USA, 2009; Volume 3, pp. 624–635. [Google Scholar]
- Dicks, L.M.T.; Fantuzzi, L.; Gonzalez, F.C.; Du Toit, M.; Dellaglio, F. Leuconostoc argentinum sp. nov., isolated from Argentine raw milk. Int. J. Syst. Evol. Microbiol. 1993, 43, 347–351. [Google Scholar] [CrossRef]
- Vancanneyt, M.; Zamfir, M.; De Wachter, M.; Cleenwerck, I.; Hoste, B.; Rossi, F.; Dellaglio, F.; De Vuyst, L.; Swings, J. Reclassification of Leuconostoc argentinum as a later synonym of Leuconostoc lactis. Int. J. Syst. Evol. Microbiol. 2006, 56, 213–216. [Google Scholar] [CrossRef][Green Version]
- Gu, C.T.; Wang, F.; Li, C.Y.; Liu, F.; Huo, G.C. Leuconostoc mesenteroides subsp. suionicum subsp. nov. Int. J. Syst. Evol. Microbiol. 2012, 62, 1548–1551. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.H.; Kim, K.H.; Chun, B.H.; Ryu, B.H.; Han, N.S.; Jeon, C.O. A proposal of Leuconostoc mesenteroides subsp. jonggajibkimchii subsp. nov. and reclassification of Leuconostoc mesenteroides subsp. suionicum (Gu et al., 2012) as Leuconostoc suionicum sp. nov. based on complete genome sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2225–2230. [Google Scholar] [CrossRef]
- Hammes, W.P.; Hertel, C.; Genus, I. Lcarnobacterium Collins, Farrow, Philips, Feresu and Jones 1987, 314VP. In Bergey’s Manual of Systematic Bacteriology, the Firmicutes, 2nd ed.; Whitman, W.B., Parte, A.C., Eds.; Springer: New York, NY, USA, 2009; Volume 3, pp. 549–557. [Google Scholar]
- Laursen, B.G.; Bay, L.; Cleenwerck, I.; Vancanneyt, M.; Swings, J.; Dalgaard, P.; Leisner, J.J. Carnobacterium divergens and Carnobacteriummaltaromaticum as spoilers or protective cultures in meat and seafood: Phenotypic and genotypic characterization. Syst. Appl. Microbiol. 2005, 28, 151–164. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Nisic, A.; Hyden, P.; Cabal, A.; Sucher, J.; Stöger, A.; Allerberger, F.; Martinović, A. Genetic Diversity of Leuconostoc mesenteroides isolates from traditional Montenegrin brine cheese. Microorganisms 2021, 9, 1612. [Google Scholar] [CrossRef] [PubMed]
- Bozoudi, D.; Pavlidou, S.; Kotzamanidis, C.; Georgakopoulos, P.; Torriani, S.; Kondyli, E.; Claps, S.; Belibasaki, S.; Litopoulou-Tzanetaki, E. Graviera Naxou and Graviera Kritis Greek PDO cheeses: Discrimination based on microbiological and physicochemical criteria and volatile organic compounds profile. Small Rum. Res. 2016, 136, 161–172. [Google Scholar] [CrossRef]
- Moschetti, G.; Blaiotta, G.; Villani, F.; Coppola, S. Specific detection of Leuconostoc mesenteroides subsp. mesenteroides with DNA primers identified by randomly amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 2000, 66, 422–424. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, A.; Storti, L.V.; Zotta, T.; Felis, G.E.; Parente, E. Analysis of rpoB polymorphism and PCR-based approaches for the identification of Leuconostoc mesenteroides at the species and subspecies level. Int. J. Food Microbiol. 2020, 318, 108474. [Google Scholar] [CrossRef] [PubMed]
- Pogačić, T.; Chuat, V.; Madec, M.-N.; Samaržija, D.; Lortal, S.; Valence, F. Phenotypic traits of genetically closely related Leuconostoc spp. Int. Dairy J. 2014, 39, 96–101. [Google Scholar] [CrossRef]
- Nogarol, C.; Acutis, P.L.; Bianchi, D.M.; Maurella, C.; Peletto, S.; Gallina, S.; Adriano, D.; Zuccon, F.; Borrello, S.; Caramelli, M. Molecular characterization of Pseudomonas fluorescens isolates involved in the Italian “blue mozzarella” event. J. Food Prot. 2013, 76, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Carminati, D.; Bonvini, B.; Rossetti, L.; Zago, M.; Tidona, F.; Giraffa, G. Investigation on the presence of blue pigment-producing Pseudomonas strains along a production line of fresh mozzarella cheese. Food Cont. 2019, 100, 321–328. [Google Scholar] [CrossRef]
- Bassi, D.; Gazzola, S.; Sattin, E.; Dal Bello, F.; Simionati, B.; Cocconcelli, P.S. Lactic acid bacteria adjunct cultures exert a mitigation effect against spoilage microbiota in fresh cheese. Microorganisms 2020, 8, 1199. [Google Scholar] [CrossRef]
- Madureira, A.R.; Pintado, M.E.; Gomes, A.M.P.; Malcata, F.X. Incorporation of probiotic bacteria in whey cheese: Decreasing the risk of microbial contamination. J. Food Prot. 2011, 74, 1194–1199. [Google Scholar] [CrossRef]
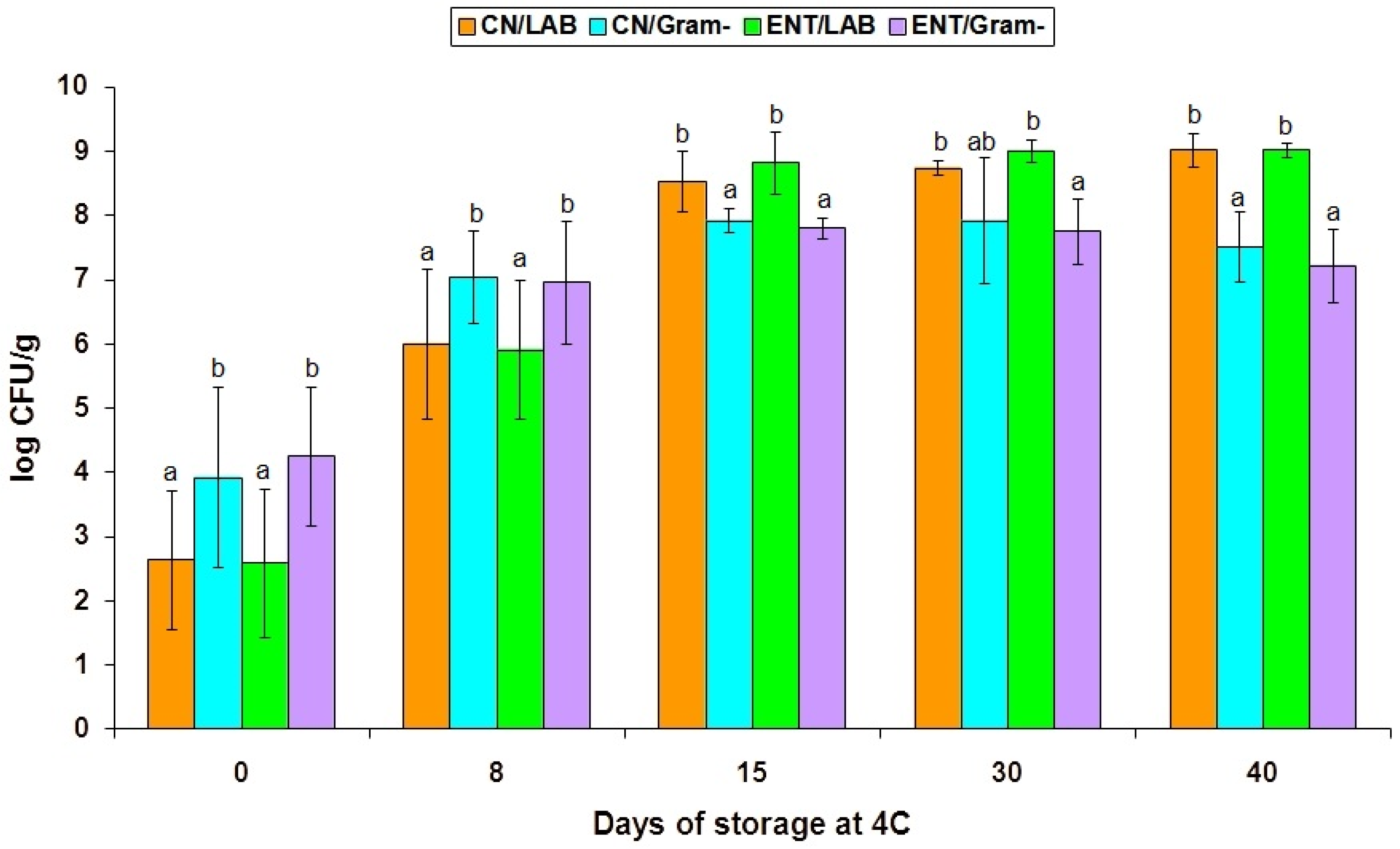
| Bacterial Group | Cheese Treatment | Days of Storage | ||||
|---|---|---|---|---|---|---|
| 0 | 8 | 15 | 30 | 40 | ||
| Total mesophilic dairy bacteria | CN | 4.45 a (1.20) | 6.98 b (0.92) | 7.79 bc (0.71) | 8.48 c (0.28) | 8.68 c (0.52) |
| CEntE | 4.89 a (1.38) | 6.29 b (1.21) | 8.37 c (0.60) | 8.81 c (0.40) | 8.66 c (0.30) | |
| Total psychrotrophic bacteria | CN | 3.92 a (1.40) | 7.27 b (0.49) | 8.64 cd (0.40) | 8.87 d (0.15) | 9.03 d (0.28) |
| CEntE | 4.25 a (1.08) | 7.20 b (0.61) | 8.93 c (0.38) | 9.05 c (0.14) | 9.04 c (0.12) | |
| Total lactic acid bacteria (LAB) | CN | 2.64 a (1.08) | 5.57 b (1.23) | 7.21 c (0.85) | 8.49 cd * (0.34) | 8.86 d (0.24) |
| CEntE | 2.58 a (1.15) | 5.49 b (1.35) | 7.42 c (0.86) | 8.94 d * (0.19) | 8.89 d (0.12) | |
| Pseudomonas-like and related Gram-negative bacteria | CN | 3.35 a (1.29) | 6.37 b (0.76) | 7.84 c (0.42) | 7.41 cb (1.48) | 6.68 cb * (1.37) |
| CEntE | 3.40 a (1.25) | 6.28 b (0.75) | 7.79 c (0.33) | 7.21 cb (0.61) | 5.51 ab * (2.05) | |
| Whey cheese pH | CN | 6.80 d (0.18) | 6.84 d (0.28) | 6.21 c (0.13) | 5.51 b * (0.23) | 5.14 a * (0.21) |
| CEntE | 6.83 c (0.14) | 6.82 c (0.23) | 5.98 b (0.54) | 4.87 a * (0.28) | 4.63 a * (0.21) | |
| LAB Genus/Phenotypic Group | Basic Differentiating Characteristics | Whey Cheese Batch/Treatment | Total Isolates | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | (% frequency) | |||||||||||||
| MA | CO2 | NH3 | 15 °C | 37 °C | 45 °C | 6.5% | SL | CN | Ent | CN | Ent | CN | Ent | CN | Ent | ||
| Leuconostoc-likebacteria Unable to form slime | CB | + | − | + | + | − | +/++ | − | 10 | 8 | 11 | 10 | 7 | 11 | 10 | 12 | 79 (65.8) |
| Leuconostoc-likebacteria Slime producers | CB | + | − | + | + | − | ++ | + | 4 | 7 | 2 | 2 | 2 | 17 (14.2) | |||
| Carnobacterium | SR | (+)/− | +/(+) | + | V | − | − | − | 2 | 3 | 4 | 4 | 13 (10.9) | ||||
| Thermophilic Streptococcus spp. | LC | − | − | − | + | + | − | − | 5 | 2 | 7 (5.9) | ||||||
| Enterococcus spp. | C | − | + | + | + | + | ++ | − | 1 | 1 (0.8) | |||||||
| Mesophilic, homofermentative arginine-negative LAB cocci | C | − | − | + | + | − | − | − | 2 | 2 (1.6) | |||||||
| Mesophilic, homofermentative arginine-negative lactobacilli | R | − | − | + | + | − | + | − | 1 | 1 (0.8) | |||||||
| Total isolates from each batch | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 15 | 120 | ||||||||
| Leuconostoc Biotypes | SL | Acid Production from (Key Sugar Fermentation Reactions) | Whey Cheese Batch | Total | Representative Strains’ Codes | 16S rRNA Identification | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LARA | GAL | LAC | RAF | TRE | DXYL | A | B | C | D | WM prefix | ||||
| L1 | − | − | + | + | − | + | + | 4 | 6 | 10 | 19 | 39 | 106, 109B, 123, 136, 137, 153 | Leuconostoc mesenteroides |
| L2 | − | + | + | + | + | + | + | 10 | 9 | 5 | 1 | 25 | 105, 110A, 122A | Leuconostoc mesenteroides |
| L3 | − | + | + | + | − | + | + | 4 | 6 | 1 | 0 | 11 | 121 | Leuconostoc mesenteroides |
| L4 | + | 15/17 | + | + | 15/17 | + | + | 11 | 4 | 2 | 0 | 17 | 107, 108 | Leuconostoc mesenteroides |
| L5 | − | − | + | + | + | − | − | 0 | 0 | 2 | 2 | 4 | 118 | Leuconostoc lactis |
| Total isolates | 29 | 25 | 20 | 22 | 96 | |||||||||
| Biochemical Test | Carnobacterium Biotypes | |||
|---|---|---|---|---|
| C1 | C2 | C3 | C4 | |
| CO2 from glucose | (+) | − | − | − |
| Arginine hydrolysis | + | −/(+) | −/(+) | + |
| Esculin hydrolysis | + | + | + | + |
| Acid produced from: | ||||
| Amygdalin | + | + | + | + |
| Arabinose | − | − | − | − |
| Galactose | +/+d | (+) | (+) | +d |
| Gluconate | ((+)) | − | − | ((+)) |
| Glycerol | + | + | + | + |
| Inulin | − | − | − | − |
| Lactose | + | + | + | + |
| Mannitol | +/(+) | ((+)) | ((+)) | (+) |
| Melezitose | − | − | − | − |
| Melibiose | − | − | + | + |
| Methy-D-glucoside | +/+d | − | − | − |
| Ribose | + | + | + | + |
| Tagatose | − | − | − | − |
| Trehalose | + | − | − | + |
| Turanose | − | − | − | + |
| Xylose | − | − | − | + |
| Total isolates | 8 | 2 | 1 | 2 |
| Batch A | 0 | 0 | 0 | 0 |
| Batch B | 5 | 0 | 0 | 0 |
| Batch C | 3 | 2 | 1 | 2 |
| Batch D | 0 | 0 | 0 | 0 |
| Species | Whey Cheese Batch Isolates (%) | Total Isolates (%) | Enumeration/Isolation Agar Medium/ Incubation Temperature | |||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | MPCA/37 °C | MRS/30 °C | TSAYE/12 °C | ||
| Leuconostoc mesenteroides | 29 (96.7) | 25 (83.3) | 18 (60.0) | 20 (66.7) | 92 (76.7) | 28 | 39 | 25 |
| Leuconostoc lactis | 2 (6.7) | 2 (6.7) | 4 (3.3) | 1 | 3 | |||
| Carnobacterium maltaromaticum | 5 (16.7) | 8 (26.6) | 13 (10.9) | 4 | 9 | |||
| Streptococcus thermophilus | 7 (23.3) | 7 (5.9) | 7 | |||||
| Enterococcus faecium | 1 (3.3) | 1 (0.8) | 1 | |||||
| Lactococcus lactis | 2 (6.7) | 2 (1.6) | 2 | |||||
| Mesophilic Lactobacillus sp. | 1 (3.3) | 1 (0.8) | 1 | |||||
| Total isolates | 30 | 30 | 30 | 30 | 120 | 40 | 40 | 40 |
| Test | Reactions/Enzymes | Hafnia alvei I | Hafnia alvei II | Serratia liquefaciens I | Serratia liquefaciens II | Rahnella Aquatilis | Pantoea sp. | Klebsiella oxytoca | Enterobacter/ E. cloacae |
|---|---|---|---|---|---|---|---|---|---|
| Number of isolates 1 | 11 (30) | 6 (11) | 4 (4) | 11 (13) | 8 (8) | 1 (1) | 2 (3) | 4 (4) | |
| ONPG | Β-galactosidase | + | + | + | + | + | + | + | + |
| ADH | Arginine dihydrolase | − | − | − | − | − | − | − | + |
| LDC | Lysine decarboxylase | + | + | + | + | − | − | + | − |
| ODC | Ornithine decarboxylase | + | + | + | + | − | − | − | + |
| CIT | Citrate utilization | + | − | + | + | − | − | + | + |
| H2S | H2S production | − | − | − | − | − | − | − | − |
| URE | Urease | − | − | − | − | − | − | −/+ | − |
| TDA | Tryptophane deaminase | − | − | − | − | − | − | − | − |
| IND | Indole production | − | − | − | − | − | − | + | − |
| VP | Acetoin production | −/+ | −/+ | + | + | + | + | + | + |
| GEL | Gelatinase | − | − | − | + | − | − | − | − |
| GLU | Glusose (F/O) | + | + | + | + | + | + | + | + |
| MAN | Mannitol (F/O) | + | + | + | + | + | + | + | + |
| INO | Inositol (F/O) | − | − | + | + | − | − | + | - |
| SOR | Sorbitol(F/O) | − | − | + | + | +/− | − | + | −/+ |
| RHA | Rhamnose (F/O) | + | + | − | − | + | − | + | + |
| SAC | Saccharose (F/O) | − | − | + | + | + | + | + | + |
| MEL | Melibiose (F/O) | − | − | − | + | + | + | + | + |
| AMY | Amygdalin (F/O) | − | − | + | + | + | + | + | + |
| ARA | Arabinose (F/O) | + | + | + | + | + | − | + | + |
| OX | Oxidase reaction | − | − | − | − | − | − | − | − |
| API code | 5304112 5305112 | 5104112 5105112 | 5305723 | 5307763 | 1005573 1005173 | 1005161 | 5245773 5255773 | 3305173 3305573 | |
| Identification accuracy | Excellent | Excellent | Very good | Very good | Low | Acceptable | Good | Excellent/Good |
| Test | Reactions/Enzymes | Aeromonas sp./ A. salmonicida | Pseudomonas sp. I | Pseudomonas sp. II | Flavibacterium oryzihabitans |
|---|---|---|---|---|---|
| Number of isolates 1 | 4 (9) | 4 (9) | 2 (2) | 1 (2) | |
| ONPG | β-galactosidase | − | − | − | − |
| ADH | Arginine dihydrolase | − | + | + | − |
| LDC | Lysine decarboxylase | − | − | − | − |
| ODC | Ornithine decarboxylase | − | − | − | − |
| CIT | Citrate utilization | − | −/(+) | − | + |
| H2S | H2S production | − | − | − | − |
| URE | Urease | − | − | − | − |
| TDA | Tryptophane deaminase | − | − | − | − |
| IND | Indole production | − | − | − | − |
| VP | Acetoin production | − | + | − | − |
| GEL | Gelatinase | + | − | − | − |
| GLU | Glusose (F/O) | −/(+) | −/+ | + | − |
| MAN | Mannitol (F/O) | −/(+) | − | − | − |
| INO | Inositol (F/O) | − | − | − | − |
| SOR | Sorbitol(F/O) | − | − | − | − |
| RHA | Rhamnose (F/O) | − | − | − | − |
| SAC | Saccharose (F/O) | − | − | − | − |
| MEL | Melibiose (F/O) | − | − | + | − |
| AMY | Amygdalin (F/O) | − | − | − | − |
| ARA | Arabinose (F/O) | − | − | + | − |
| OX | Oxidase reaction | + | + | + | − |
| API code | 0002004 0006104 | 2001004 2205004 | 2004046 | 0200000 | |
| Identification accuracy | Low/Very good | Acceptable/Very good | Very good | Acceptable |
| Genus/Species | Biotype | Whey Cheese Batch | Total Isolates | |||
|---|---|---|---|---|---|---|
| A | B | C | D | |||
| Hafnia alvei | I | 30 | 30 | |||
| Hafnia alvei | II | 11 | 11 | |||
| Serratia liquefaciens | I | 4 | 4 | |||
| Serratia liquefaciens | II | 3 | 10 | 13 | ||
| Rahnellaaquatilis | 5 | 3 | 8 | |||
| Pantoea sp. | 1 | |||||
| Klebsiella oxytoca | 1 | 2 | 3 | |||
| Enterobacter sp./E. cloacae | 2 | 2 | 4 | |||
| Aeromonas sp. | 9 | 9 | ||||
| Pseudomonas sp. | I | 5 | 4 | 9 | ||
| Pseudomonas sp. | II | 2 | 2 | |||
| Flavibacterium sp. | 2 | 2 | ||||
| Total isolates/batch | 15 | 17 | 38 | 26 | 96 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sameli, N.; Sioziou, E.; Bosnea, L.; Kakouri, A.; Samelis, J. Assessment of the Spoilage Microbiota during Refrigerated (4 °C) Vacuum-Packed Storage of Fresh Greek Anthotyros Whey Cheese without or with a Crude Enterocin A-B-P-Containing Extract. Foods 2021, 10, 2946. https://doi.org/10.3390/foods10122946
Sameli N, Sioziou E, Bosnea L, Kakouri A, Samelis J. Assessment of the Spoilage Microbiota during Refrigerated (4 °C) Vacuum-Packed Storage of Fresh Greek Anthotyros Whey Cheese without or with a Crude Enterocin A-B-P-Containing Extract. Foods. 2021; 10(12):2946. https://doi.org/10.3390/foods10122946
Chicago/Turabian StyleSameli, Nikoletta, Eleni Sioziou, Loulouda Bosnea, Athanasia Kakouri, and John Samelis. 2021. "Assessment of the Spoilage Microbiota during Refrigerated (4 °C) Vacuum-Packed Storage of Fresh Greek Anthotyros Whey Cheese without or with a Crude Enterocin A-B-P-Containing Extract" Foods 10, no. 12: 2946. https://doi.org/10.3390/foods10122946
APA StyleSameli, N., Sioziou, E., Bosnea, L., Kakouri, A., & Samelis, J. (2021). Assessment of the Spoilage Microbiota during Refrigerated (4 °C) Vacuum-Packed Storage of Fresh Greek Anthotyros Whey Cheese without or with a Crude Enterocin A-B-P-Containing Extract. Foods, 10(12), 2946. https://doi.org/10.3390/foods10122946







