Evolution of Food Fermentation Processes and the Use of Multi-Omics in Deciphering the Roles of the Microbiota
Abstract
1. Introduction
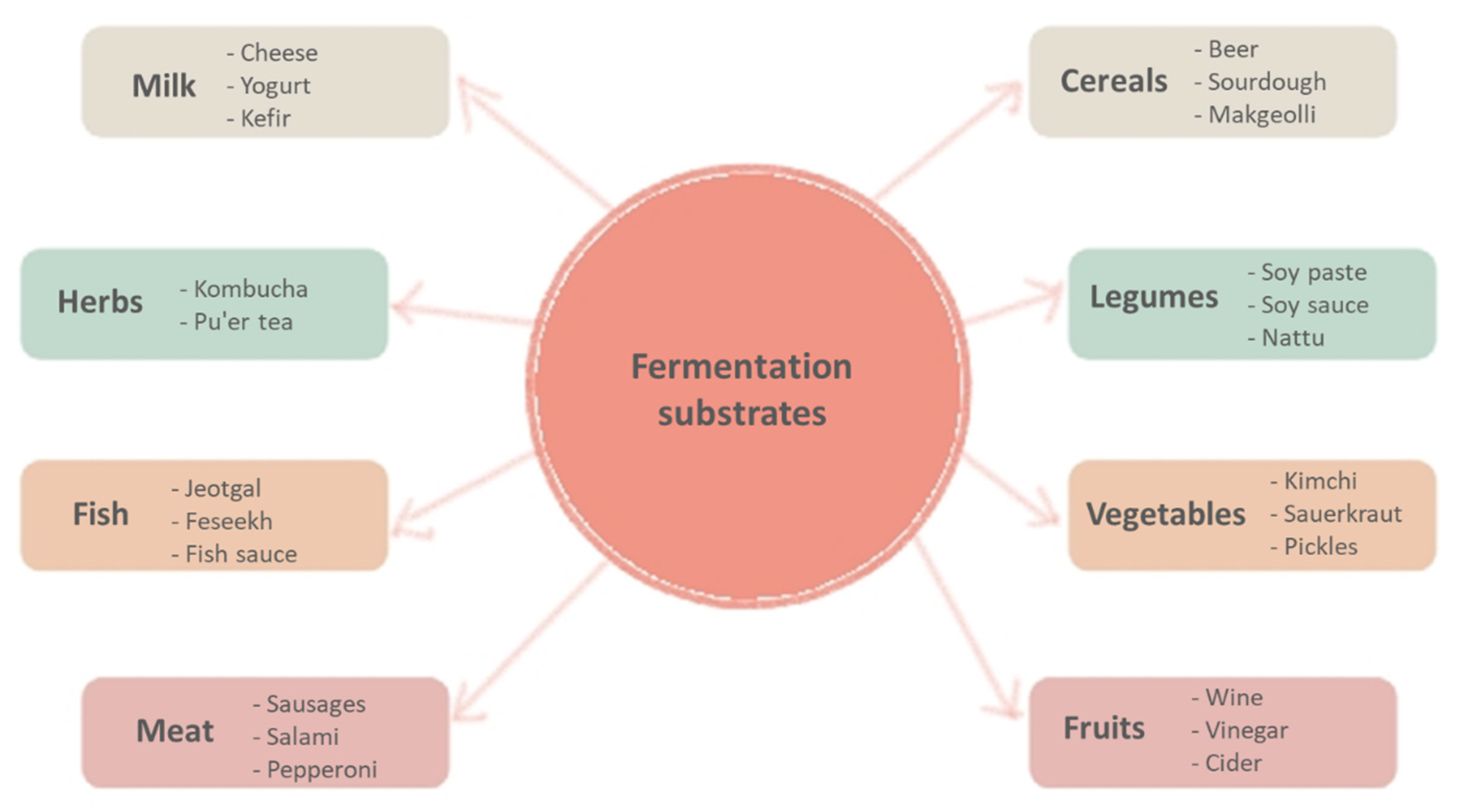
2. Classification of Major Types of Fermented Foods and Beverages
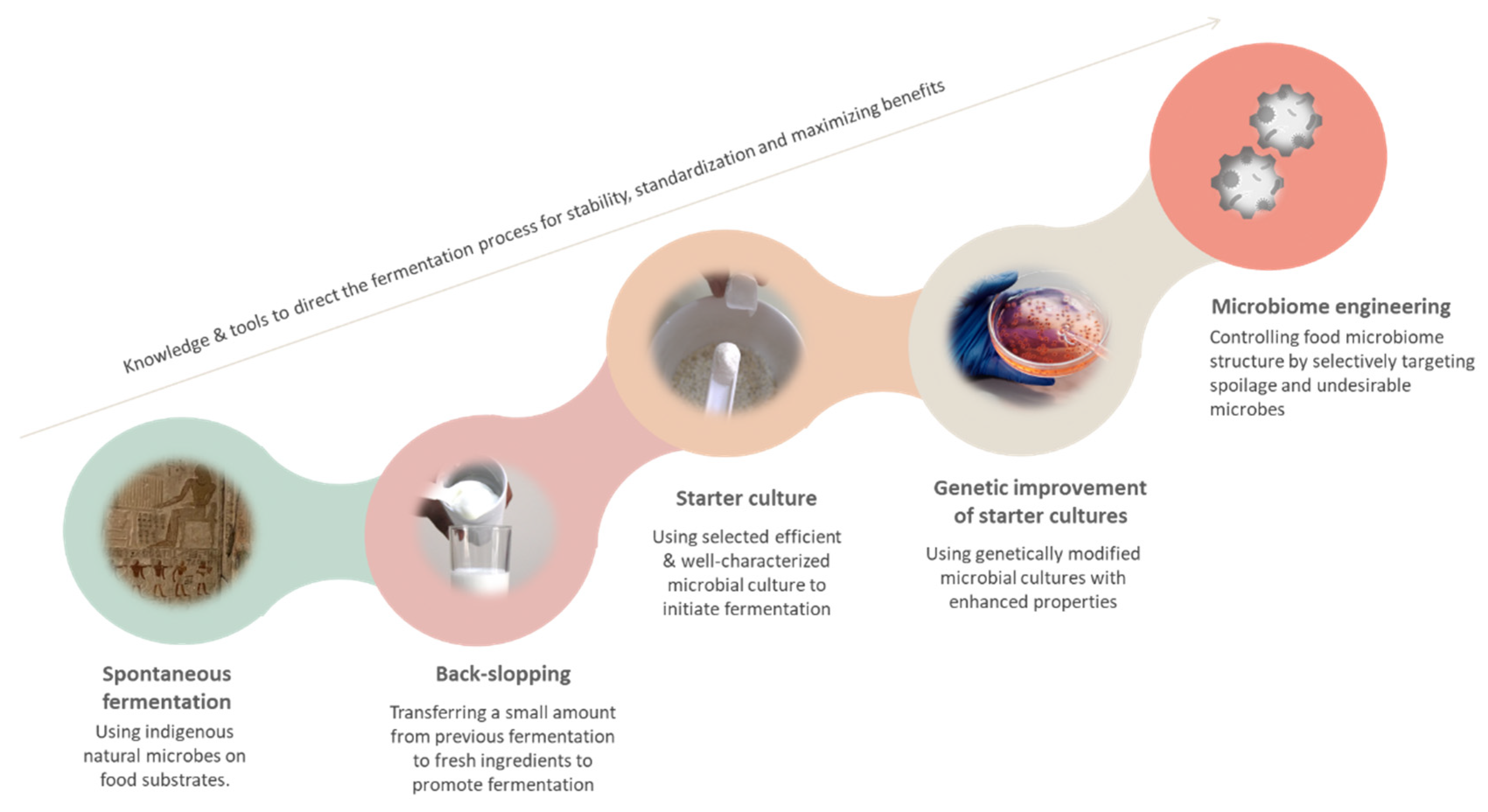
3. Evolution of the Process of Fermentation over the Years
3.1. Spontaneous Fermentation and Back-Slopping
3.2. Starter Cultures
3.3. Starter Cultures of Multiple Strains and Adaptation for Co-Existence
3.4. Genetic Improvement of Starter Cultures
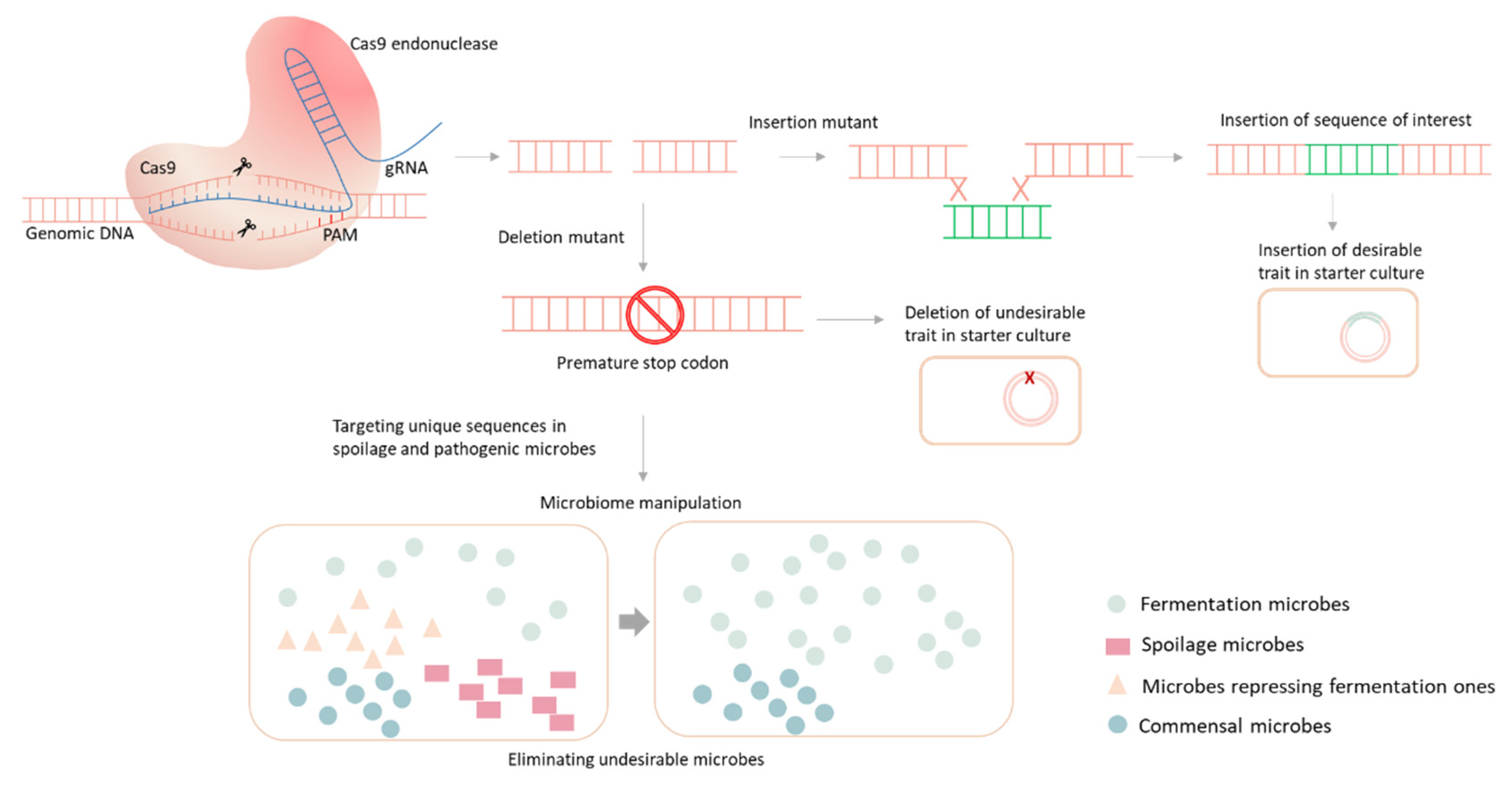
3.5. CRISPR/Cas9 Technology for Genetic Improvement of Starter Cultures
3.6. CRISPR-Mediated Microbiome Engineering and Fermentation
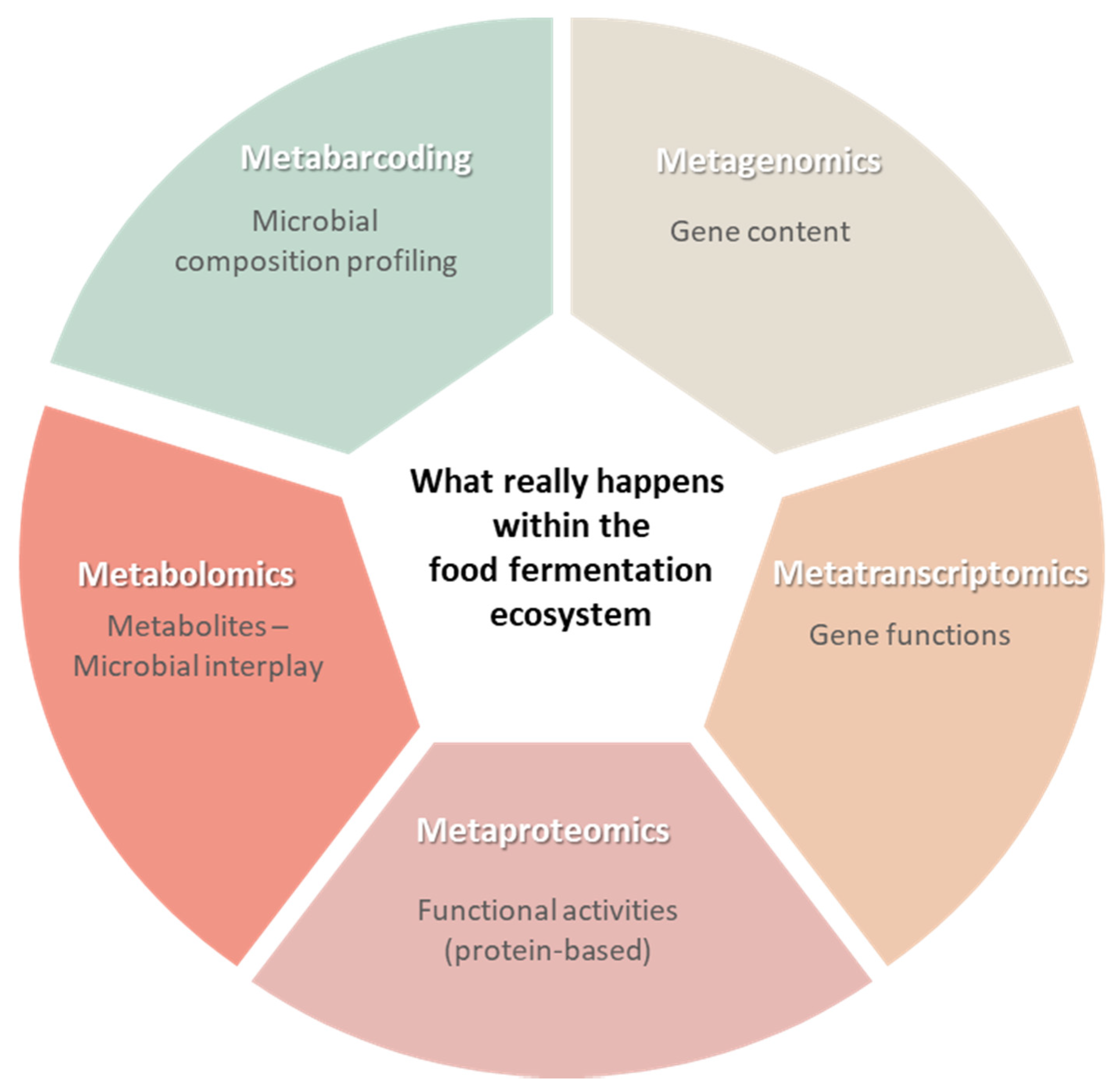
4. Multi-Omics and Microbiota Dynamics of Food Fermentation
4.1. Examples on the Use of Meta-Omics to Study Microbial Dynamics of Food Fermentation in Recent Studies
4.2. Functional Activity of the Food Fermentation Microbiome
5. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Marsh, A.J.; Hill, C.; Ross, R.P.; Cotter, P.D. Fermented beverages with health-promoting potential: Past and future perspectives. Trends. Food Sci. Technol. 2014, 38, 113–124. [Google Scholar] [CrossRef]
- Liu, L.; Wang, J.; Rosenberg, D.; Zhao, H.; Lengyel, G.; Nadel, D. Fermented beverage and food storage in 13,000 y-old stone mortars at Raqefet Cave, Israel: Investigating Natufian ritual feasting. J. Archaeol. Sci. 2018, 21, 783–793. [Google Scholar] [CrossRef]
- El-Gendy, S.M. Fermented foods of Egypt and the Middle East. J. Food Prot. 1983, 46, 358–367. [Google Scholar] [CrossRef]
- McGovern, P.E.; Zhang, J.; Tang, J.; Zhang, Z.; Hall, G.R.; Moreau, R.A.; Nuñez, A.; Butrym, E.D.; Richards, M.P.; Wang, C.S.; et al. Fermented beverages of pre-and proto-historic China. Proc. Natl. Acad. Sci. USA 2004, 101, 17593–17598. [Google Scholar] [CrossRef]
- Chojnacka, K. Fermentation products. Chem. Eng. Chem. Process Technol. 2010, 5, 1–12. [Google Scholar]
- Tamang, J.P.; Cotter, P.D.; Endo, A.; Han, N.S.; Kort, R.; Liu, S.Q.; Mayo, B.; Westerik, N.; Hutkins, R. Fermented foods in a global age: East meets West. Compr. Rev. Food Sci. Food Saf. 2020, 19, 184–217. [Google Scholar] [CrossRef]
- Waché, Y.; Do, T.L.; Do, T.B.H.; Do, T.Y.; Haure, M.; Ho, P.H.; Kumar Anal, A.; Le, V.V.M.; Li, W.J.; Licandro, H.; et al. Prospects for food fermentation in South-East Asia, topics from the tropical fermentation and biotechnology network at the end of the AsiFood Erasmus+ Project. Front. Microbiol. 2018, 9, 2278. [Google Scholar] [CrossRef]
- Galimberti, A.; Bruno, A.; Agostinetto, G.; Casiraghi, M.; Guzzetti, L.; Labra, M. Fermented food products in the era of globalization: Tradition meets biotechnology innovations. Curr. Opin. Biotechnol. 2021, 70, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Ojha, K.S.; Tiwari, B.K. Novel food fermentation technologies. In Novel Food Fermentation Technologies; Springer: Cham, Switzerland, 2016; pp. 1–5. [Google Scholar]
- Campbell-Platt, G. Fermented foods—A world perspective. Food Res. Int. 1994, 27, 253–257. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-Waterhouse, D.; Waterhouse, G.I.; Cui, C.; Ruan, Z. Fermentation-enabled wellness foods: A fresh perspective. Food Sci. Hum. Wellness 2019, 8, 203–243. [Google Scholar] [CrossRef]
- Navarrete-Bolaños, J.L. Improving traditional fermented beverages: How to evolve from spontaneous to directed fermentation. Eng. Life Sci. 2012, 12, 410–418. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef]
- De Roos, J.; De Vuyst, L. Acetic acid bacteria in fermented foods and beverages. Curr. Opin. Biotechnol. 2018, 49, 115–119. [Google Scholar] [CrossRef]
- Vīna, I.; Linde, R.; Patetko, A.; Semjonovs, P. Glucuronic acid from fermented beverages: Biochemical functions in humans and its role in health protection. Int. J. Recent Res. Appl. Stud. 2013, 14, 217–230. [Google Scholar]
- Kandler, O. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 1983, 49, 209–224. [Google Scholar] [CrossRef]
- Anal, A.K. Quality ingredients and safety concerns for traditional fermented foods and beverages from Asia: A review. Fermentation 2019, 5, 8. [Google Scholar] [CrossRef]
- Wang, J.; Fung, D.Y. Alkaline-fermented foods: A review with emphasis on pidan fermentation. Crit. Rev. Microbiol. 1996, 22, 101–138. [Google Scholar] [CrossRef] [PubMed]
- Borresen, E.; Henderson, A.; Kumar, A.; Weir, T.; Ryan, E. Fermented foods: Patented approaches and formulations for nutritional supplementation and health promotion. Recent Pat. Food Nutr. Agric. 2012, 4, 134–140. [Google Scholar] [CrossRef]
- Steinkraus, K.H. Classification of fermented foods: Worldwide review of household fermentation techniques. Food Cont. 1997, 8, 311–317. [Google Scholar] [CrossRef]
- Giraffa, G. Studying the dynamics of microbial populations during food fermentation. FEMS Microbiol. Rev. 2004, 28, 251–260. [Google Scholar] [CrossRef]
- Holzapfel, W.H. Appropriate starter culture technologies for small-scale fermentation in developing countries. Int. J. Food Microbiol. 2002, 75, 197–212. [Google Scholar] [CrossRef]
- Voidarou, C.; Antoniadou, Μ.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative Foods: Microbiology, biochemistry, potential human health benefits and public health issues. Foods 2021, 10, 69. [Google Scholar] [CrossRef]
- Whittington, H.D.; Dagher, S.F.; Bruno-Bárcena, J.M. Production and Conservation of Starter Cultures: From “Backslopping” to Controlled Fermentations. In How Fermented Foods Feed a Healthy Gut Microbiot; Springer: Cham, Switzerland, 2019; pp. 125–138. [Google Scholar]
- Hansen, E.B. Commercial bacterial starter cultures for fermented foods of the future. Int. J. Food Microbiol. 2002, 78, 119–131. [Google Scholar] [CrossRef]
- Gadaga, T.H.; Nyanga, L.K.; Mutukumira, A.N. The occurrence, growth and control of pathogens in African fermented foods. Afr. J. Food Agric. Nutr. Dev. 2004, 4, 5346. [Google Scholar] [CrossRef]
- Panagou, E.Z.; Tassou, C.C.; Vamvakoula, P.; Saravanos, E.K.; Nychas, G.J.E. Survival of Bacillus cereus vegetative cells during Spanish-style fermentation of conservolea green olives. J. Food Prot. 2008, 71, 1393–1400. [Google Scholar] [CrossRef]
- Fuller, R. Probiotics in man and animals. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar] [PubMed]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, C.J.; Gerds, L.M. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Banjara, N.; Suhr, M.J.; Hallen-Adams, H.E. Diversity of yeast and mold species from a variety of cheese types. Curr. Microbiol. 2015, 70, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.T.; Hsieh, C.W.; Lo, Y.C.; Liou, B.K.; Lin, H.W.; Hou, C.Y.; Cheng, K.C. Isolation and identification of aroma-producing non-Saccharomyces yeast strains and the enological characteristic comparison in wine making. WT-Food Sci. Technol. 2021, 154, 112653. [Google Scholar] [CrossRef]
- Vinícius de Melo Pereira, G.; Soccol, V.T.; Brar, S.K.; Neto, E.; Soccol, C.R. Microbial ecology and starter culture technology in coffee processing. Crit. Rev. Food Sci. Nutr. 2017, 57, 2775–2788. [Google Scholar] [CrossRef]
- Dufresne, C.; Farnworth, E. Tea, Kombucha, and health: A review. Food Res. Int. 2000, 33, 409–421. [Google Scholar] [CrossRef]
- Villarreal-Soto, S.A.; Beaufort, S.; Bouajila, J.; Souchard, J.P.; Taillandier, P. Understanding kombucha tea fermentation: A review. J. Food Sci. 2018, 83, 580–588. [Google Scholar] [CrossRef]
- Jayabalan, R.; Malbaša, R.V.; Lončar, E.S.; Vitas, J.S.; Sathishkumar, M. A review on kombucha tea—microbiology, composition, fermentation, beneficial effects, toxicity, and tea fungus. Compr. Rev. Food Sci. Food Saf. 2014, 13, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, S.; Bhattacharya, S.; Chatzinotas, A.; Chakraborty, W.; Bhattacharya, D.; Gachhui, R. Kombucha tea fermentation: Microbial and biochemical dynamics. Int. J. Food Microbiol. 2016, 220, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Laavanya, D.; Shirkole, S.; Balasubramanian, P. Current challenges, applications and future perspectives of SCOBY cellulose of Kombucha fermentation. J. Clean. Prod. 2021, 295, 126454. [Google Scholar] [CrossRef]
- Iguchi, M.; Yamanaka, S.; Budhiono, A. Bacterial cellulose–A masterpiece of nature’s arts. J. Mater. Sci. 2000, 35, 261–270. [Google Scholar] [CrossRef]
- Podolich, O.; Kukharenko, O.; Haidak, A.; Zaets, I.; Zaika, L.; Storozhuk, O.; Palchikovska, L.; Orlovska, I.; Reva, O.; Borisova, T.; et al. Multimicrobial kombucha culture tolerates Mars-like conditions simulated on low Earth orbit. Astrobiology 2019, 19, 183–196. [Google Scholar] [CrossRef]
- Vinicius De Melo Pereira, G.; De Carvalho Neto, D.P.; Junqueira, A.C.D.O.; Karp, S.G.; Letti, L.A.; Magalhães Júnior, A.I.; Soccol, C.R. A review of selection criteria for starter culture development in the food fermentation industry. Food Rev. Int. 2020, 36, 135–167. [Google Scholar] [CrossRef]
- Kondo, J.K.; McKay, L.L. Transformation of Streptococcus lactis protoplasts by plasmid DNA. Appl. Environ. Microbiol. 1982, 43, 1213–1215. [Google Scholar] [CrossRef]
- Mercenier, A.; Pouwels, P.H.; Chassy, B.M. Genetic engineering of lactobacilli, leuconostocs and Streptococcus thermophilus. In Genetics and biotechnology of Lactic Acid Bacteria; Springer: Dordrecht, The Netherlands, 1994; pp. 252–293. [Google Scholar]
- Jarvis, A.W. Bacteriophages of lactic acid bacteria. J. Dairy Sci. 1989, 72, 3406–3428. [Google Scholar] [CrossRef]
- Walker, S.A.; Klaenhammer, T.R. An explosive antisense RNA strategy for inhibition of a lactococcal bacteriophage. Appl. Environ. Microbiol. 2000, 66, 310–319. [Google Scholar] [CrossRef]
- De Vos, W.M.; Hols, P.; van Kranenburg, R.; Luesink, E.; Kuipers, O.P.; van der Oost, J.; Kleerebezem, M.; Hugenholtz, J. Making more of milk sugar by engineering lactic acid bacteria. Int. Dairy J. 1998, 8, 227–233. [Google Scholar] [CrossRef][Green Version]
- Johansen, E. Future access and improvement of industrial lactic acid bacteria cultures. Microb. Cell Factories 2017, 16, 1–5. [Google Scholar] [CrossRef]
- Derkx, P.M.; Janzen, T.; Sørensen, K.I.; Christensen, J.E.; Stuer-Lauridsen, B.; Johansen, E. The art of strain improvement of industrial lactic acid bacteria without the use of recombinant DNA technology. Microb. Cell Factories 2014, 13, 1–13. [Google Scholar] [CrossRef]
- Margolles, A.; Sánchez, B. Selection of a Bifidobacterium animalis subsp. lactis strain with a decreased ability to produce acetic acid. Appl. Environ. Microbiol. 2012, 78, 3338–3342. [Google Scholar] [PubMed]
- Saarela, M.; Alakomi, H.L.; Matto, J.; Ahonen, A.M.; Puhakka, A.; Tynkkynen, S. Improving the storage stability of Bifidobacterium breve in low pH fruit juice. Int. J. Food Microbiol. 2011, 149, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Vilela, A. An Overview of CRISPR-Based Technologies in Wine Yeasts to Improve Wine Flavor and Safety. Fermentation 2021, 7, 5. [Google Scholar] [CrossRef]
- Coulon, J.; Husnik, J.I.; Inglis, D.L.; van der Merwe, G.K.; Lonvaud, A.; Erasmus, D.J.; van Vuuren, H.J. Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am. J. Enol. Vitic. 2006, 57, 113–124. [Google Scholar]
- The Nobel Prize in Chemistry. NobelPrize.org. Nobel Media A.B. 2020. Sat. 24 October 2020. Available online: https://www.nobelprize.org/prizes/chemistry/2020/summary/ (accessed on 24 October 2021).
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Cong, L.; Zhang, F. Genome engineering using CRISPR-Cas9 system. In Chromosomal Mutagenesis; Humana Press: New York, NY, USA, 2015; pp. 197–217. [Google Scholar]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef]
- Vigentini, I.; Gebbia, M.; Belotti, A.; Foschino, R.; Roth, F.P. CRISPR/Cas9 system as a valuable genome editing tool for wine yeasts with application to decrease urea production. Front. Microbiol. 2017, 8, 2194. [Google Scholar] [CrossRef]
- Muysson, J.; Miller, L.; Allie, R.; Inglis, D.L. The use of CRISPR-Cas9 genome editing to determine the importance of glycerol uptake in wine yeast during Icewine fermentation. Fermentation 2019, 5, 93. [Google Scholar] [CrossRef]
- Wu, D.; Xie, W.; Li, X.; Cai, G.; Lu, J.; Xie, G. Metabolic engineering of Saccharomyces cerevisiae using the CRISPR/Cas9 system to minimize ethyl carbamate accumulation during Chinese rice wine fermentation. Appl. Microbiol. Biotechnol. 2020, 104, 4435–4444. [Google Scholar] [CrossRef]
- Jang, Y.J.; Seo, S.O.; Kim, S.A.; Li, L.; Kim, T.J.; Kim, S.C.; Jin, Y.S.; Han, N.S. Elimination of the cryptic plasmid in Leuconostoc citreum by CRISPR/Cas9 system. J. Biotechnol. 2017, 251, 151–155. [Google Scholar] [CrossRef]
- Katayama, T.; Tanaka, Y.; Okabe, T.; Nakamura, H.; Fujii, W.; Kitamoto, K.; Maruyama, J.I. Development of a genome editing technique using the CRISPR/Cas9 system in the industrial filamentous fungus Aspergillus Oryzae. Biotechnol. Lett. 2016, 38, 637–642. [Google Scholar] [CrossRef] [PubMed]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
- Urnov, F.D.; Ronald, P.C.; Carroll, D. A call for science-based review of the European court’s decision on gene-edited crops. Nat. Biotechnol. 2018, 36, 800–802. [Google Scholar] [CrossRef]
- Ramachandran, G.; Bikard, D. Editing the microbiome the CRISPR way. Philos. Trans. R. Soc. 2019, 374, 20180103. [Google Scholar] [CrossRef]
- Barrangou, R.; Notebaart, R.A. CRISPR-directed microbiome manipulation across the food supply chain. Trends Microbiol. 2019, 27, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Gomaa, A.A.; Klumpe, H.E.; Luo, M.L.; Selle, K.; Barrangou, R.; Beisel, C.L. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio 2014, 5, e00928-13. [Google Scholar] [CrossRef] [PubMed]
- Citorik, R.J.; Mimee, M.; Lu, T.K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014, 32, 1141–1145. [Google Scholar] [CrossRef]
- Bikard, D.; Euler, C.W.; Jiang, W.; Nussenzweig, P.M.; Goldberg, G.W.; Duportet, X.; Fischetti, V.A.; Marraffini, L.A. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 2014, 32, 1146–1150. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E.; Dutton, R.J. Fermented foods as experimentally tractable microbial ecosystems. Cell 2015, 161, 49–55. [Google Scholar] [CrossRef]
- Millet, V.; Lonvaud-Funel, A. The viable but non-culturable state of wine micro-organisms during storage. Lett. Appl. Microbiol. 2000, 30, 136–141. [Google Scholar] [CrossRef]
- Zhao, M.; Su, X.Q.; Nian, B.; Chen, L.J.; Zhang, D.L.; Duan, S.M.; Wang, L.Y.; Shi, X.Y.; Jiang, B.; Jiang, W.W.; et al. Integrated meta-omics approaches to understand the microbiome of spontaneous fermentation of traditional Chinese pu-erh tea. Msystems 2019, 4, e00680–e00719. [Google Scholar] [CrossRef]
- Ferrocino, I.; Cocolin, L. Current perspectives in food-based studies exploiting multi-omics approaches. Curr. Opin. Food Sci. 2017, 13, 10–15. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012, 45, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Alessandria, V.; Ferrocino, I.; De Filippis, F.; Fontana, M.; Rantsiou, K.; Ercolini, D.; Cocolin, L. Microbiota of an Italian Grana-like cheese during manufacture and ripening, unraveled by 16S rRNA-based approaches. Appl. Environ. Microbiol. 2016, 82, 3988–3995. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Yang, X.; Noyes, N.R.; Doster, E.; Martin, J.N.; Linke, L.M.; Magnuson, R.J.; Yang, H.; Geornaras, I.; Woerner, D.R.; Jones, K.L.; et al. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 2016, 82, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Zepeda, A.; Sanchez-Flores, A.; Baruch, M.Q. Metagenomic analysis of a Mexican ripened cheese reveals a unique complex microbiota. Food Microbiol. 2016, 57, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Nalbantoglu, U.; Cakar, A.; Dogan, H.; Abaci, N.; Ustek, D.; Sayood, K.; Can, H. Metagenomic analysis of the microbial community in kefir grains. Food Microbiol. 2014, 41, 42–51. [Google Scholar] [CrossRef]
- Tamang, J.P.; Watanabe, K.; Holzapfel, W.H. Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed]
- Park, K.Y.; Cho, E.J.; Rhee, S.H.; Jung, K.O.; Yi, S.J.; Jhun, B.H. Kimchi and an active component, β-sitosterol, reduce oncogenic H-Rasv12-induced DNA synthesis. J. Med. Food 2003, 6, 151–156. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Kim, J.M.; Park, M.S.; Bae, J.W.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 2011, 77, 2264–2274. [Google Scholar] [CrossRef]
- Lee, M.; Song, J.H.; Jung, M.Y.; Lee, S.H.; Chang, J.Y. Large-scale targeted metagenomics analysis of bacterial ecological changes in 88 kimchi samples during fermentation. Food Microbiol. 2017, 66, 173–183. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Jeon, C.O. Kimchi microflora: History, current status, and perspectives for industrial kimchi production. Appl. Microbiol. Biotechnol. 2014, 98, 2385–2393. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Paramithiotis, S.; Shin, H.S. Kimchi and other widely consumed traditional fermented foods of Korea: A review. Front. Microbiol. 2016, 7, 1493. [Google Scholar] [CrossRef]
- Jeong, S.E.; Chun, B.H.; Kim, K.H.; Park, D.; Roh, S.W.; Lee, S.H.; Jeon, C.O. Genomic and metatranscriptomic analyses of Weissella koreensis reveal its metabolic and fermentative features during kimchi fermentation. Food Microbiol. 2018, 76, 1–10. [Google Scholar] [CrossRef]
- Mannaa, M.; Seo, Y.S.; Park, I. Effect of seafood (gizzard shad) supplementation on the chemical composition and microbial dynamics of radish kimchi during fermentation. Sci. Rep. 2019, 9, 1–12. [Google Scholar]
- Nam, H.G.; Jang, M.-S.; Seo, K.-C.; Nam, K.-H.; Park, H.-Y. Changes in the taste compounds of Kimchi with seafood added during its fermentation. Korean J. Food Preserv. 2013, 20, 404–418. [Google Scholar] [CrossRef]
- Woo, M.; Choi, J.R.; Kim, M.; Jang, M.S.; Cho, E.J.; Song, Y.O. Physicochemical characteristics of seafood-added kimchi during fermentation and its sensory properties. J. Korean Soc. Food Sci. Nutr. 2012, 41, 1771–1777. [Google Scholar] [CrossRef]
- Jung, J.Y.; Lee, S.H.; Jin, H.M.; Hahn, Y.; Madsen, E.L.; Jeon, C.O. Metatranscriptomic analysis of lactic acid bacterial gene expression during kimchi fermentation. Int. J. Food Microbiol. 2013, 163, 171–179. [Google Scholar] [CrossRef]
- Wisselink, H.; Weusthuis, R.; Eggink, G.; Hugenholtz, J.; Grobben, G. Mannitol production by lactic acid bacteria: A review. Int. Dairy J. 2002, 12, 151–161. [Google Scholar] [CrossRef]
- Kim, J.; Bang, J.; Beuchat, L.R.; Kim, H.; Ryu, J.H. Controlled fermentation of kimchi using naturally occurring antimicrobial agents. Food Microbiol. 2012, 32, 20–31. [Google Scholar] [CrossRef]
- Jeong, S.H.; Lee, H.J.; Jung, J.Y.; Lee, S.H.; Seo, H.Y.; Park, W.S.; Jeon, C.O. Effects of red pepper powder on microbial communities and metabolites during kimchi fermentation. Int. J. Food Microbiol. 2013, 160, 252–259. [Google Scholar] [CrossRef]
- Lim, S.B.; Shin, S.Y.; Moon, J.S.; Otgonbayar, G.E.; Joo, W.; Lee, S.J.; Jeon, C.O.; Han, N.S. Garlic is a source of major lactic acid bacteria for early-stage fermentation of cabbage-kimchi. Food Sci. Biotechnol. 2015, 24, 1437–1441. [Google Scholar] [CrossRef]
- Chung, H.J.; Sohn, K.H. The changes of component in traditional Korean soy sauce during ripening period (I). Korean J. Soc. Food Sci. 1994, 10, 29–34. [Google Scholar]
- Cho, K.M.; Seo, W.T. Bacterial diversity in a Korean traditional soybean fermented foods (doenjang and ganjang) by 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 2007, 16, 320–324. [Google Scholar]
- Mannaa, M.; Seo, Y.S.; Park, I. Addition of Coriander during Fermentation of Korean Soy Sauce (Gangjang) Causes Significant Shift in Microbial Composition and Reduction in Biogenic Amine Levels. Foods 2020, 9, 1346. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Lee, Y.B.; Lee, C.H.; Park, I. Effects of the Addition of Herbs on the Properties of Doenjang. Foods 2021, 10, 1307. [Google Scholar] [CrossRef]
- Mannaa, M.; Cho, S.S.; Seo, Y.S.; Park, I. Microbial Composition of Fermented Korean Soy Paste (Doenjang) Prepared by Adding Different Herbs during Fermentation. Fermentation 2021, 7, 93. [Google Scholar] [CrossRef]
- Arıkan, M.; Mitchell, A.L.; Finn, R.D.; Gürel, F. Microbial composition of Kombucha determined using amplicon sequencing and shotgun metagenomics. J. Food Sci. 2020, 85, 455–464. [Google Scholar] [CrossRef]
- Góes-Neto, A.; Kukharenko, O.; Orlovska, I.; Podolich, O.; Imchen, M.; Kumavath, R.; Kato, R.B.; De Carvalho, D.S.; Tiwari, S.; Brenig, B.; et al. Shotgun metagenomic analysis of kombucha mutualistic community exposed to Mars-like environment outside the International Space Station. Environ. Microbiol. 2021, 23, 3727–3742. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, C.; Lei, Z. Meta-omics insights in the microbial community profiling and functional characterization of fermented foods. Trend Food Sci. Technol. 2017, 65, 23–31. [Google Scholar] [CrossRef]
- Song, Z.; Du, H.; Zhang, Y.; Xu, Y. Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Front. Microbiol. 2021, 8, 1294. [Google Scholar] [CrossRef] [PubMed]
- De Filippis, F.; Genovese, A.; Ferranti, P.; Gilbert, J.A.; Ercolini, D. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Sci. Rep. 2016, 6, 1–11. [Google Scholar]
- Afshari, R.; Pillidge, C.J.; Dias, D.A.; Osborn, A.M.; Gill, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef]
- Yang, L.; Fan, W.; Xu, Y. Metaproteomics insights into traditional fermented foods and beverages. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2506–2529. [Google Scholar] [CrossRef]
- Hettich, R.L.; Sharma, R.; Chourey, K.; Giannone, R.J. Microbial metaproteomics: Identifying the repertoire of proteins that microorganisms use to compete and cooperate in complex environmental communities. Curr. Opin. Microbiol. 2012, 15, 373–380. [Google Scholar] [CrossRef]
- Haange, S.B.; Jehmlich, N. Proteomic interrogation of the gut microbiota: Potential clinical impact. Expert Rev. Proteom. 2016, 13, 535–537. [Google Scholar] [CrossRef] [PubMed]
- Schiebenhoefer, H.; Van Den Bossche, T.; Fuchs, S.; Renard, B.Y.; Muth, T.; Martens, L. Challenges and promise at the interface of metaproteomics and genomics: An overview of recent progress in metaproteogenomic data analysis. Expert Rev. Proteom. 2019, 16, 375–390. [Google Scholar] [CrossRef] [PubMed]

| Type | Biosynthetic Pathway | Responsible Microbes | Fermented Food |
|---|---|---|---|
| Lactic | Sugars are converted into lactic acid | Lactic acid bacteria | Yoghurt and kimchi |
| Acetic | Several substrates are converted into acetic acid | Acetobacter | Vinegar and water kefir |
| Alcoholic | Sugars are converted to alcohols and CO2 | Yeast | Wine and beer |
| Alkali | Proteins are converted into amino acids, peptides, and ammonia | Bacillus and Staphylococcus spp. | Japanese nattu |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mannaa, M.; Han, G.; Seo, Y.-S.; Park, I. Evolution of Food Fermentation Processes and the Use of Multi-Omics in Deciphering the Roles of the Microbiota. Foods 2021, 10, 2861. https://doi.org/10.3390/foods10112861
Mannaa M, Han G, Seo Y-S, Park I. Evolution of Food Fermentation Processes and the Use of Multi-Omics in Deciphering the Roles of the Microbiota. Foods. 2021; 10(11):2861. https://doi.org/10.3390/foods10112861
Chicago/Turabian StyleMannaa, Mohamed, Gil Han, Young-Su Seo, and Inmyoung Park. 2021. "Evolution of Food Fermentation Processes and the Use of Multi-Omics in Deciphering the Roles of the Microbiota" Foods 10, no. 11: 2861. https://doi.org/10.3390/foods10112861
APA StyleMannaa, M., Han, G., Seo, Y.-S., & Park, I. (2021). Evolution of Food Fermentation Processes and the Use of Multi-Omics in Deciphering the Roles of the Microbiota. Foods, 10(11), 2861. https://doi.org/10.3390/foods10112861









