Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Reagents
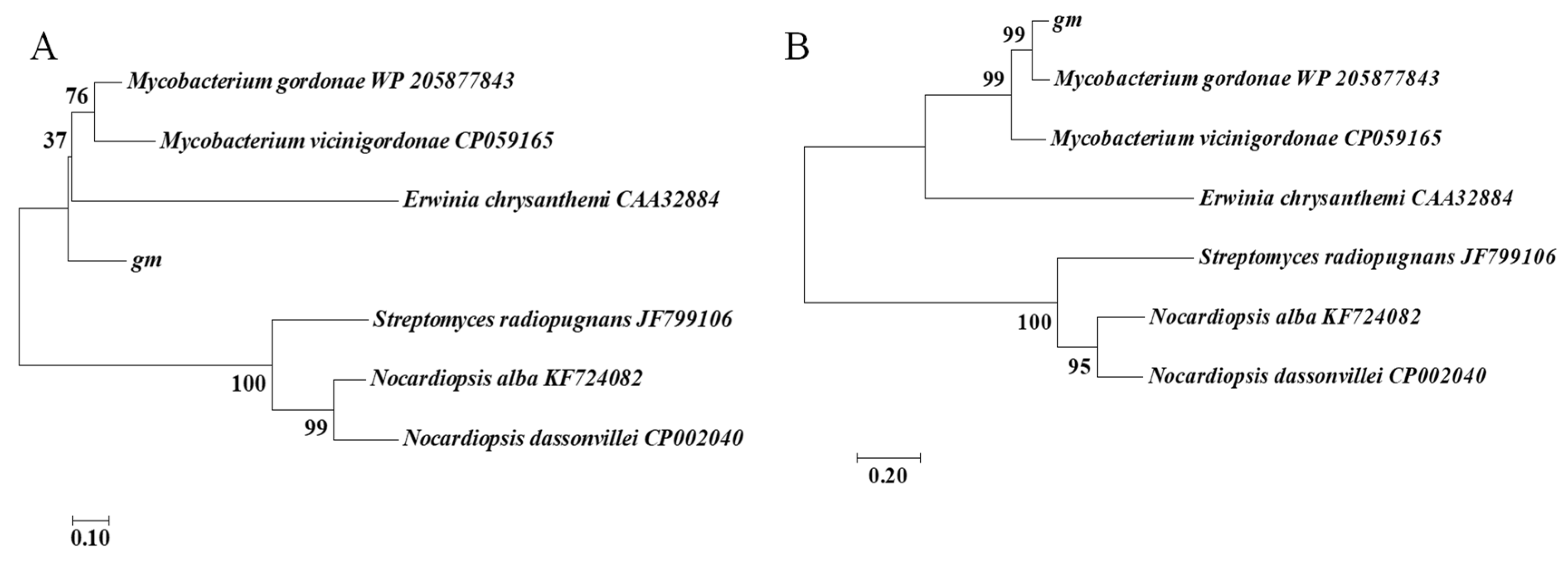
2.2. In Silico Sequence and Phylogenetic Analysis
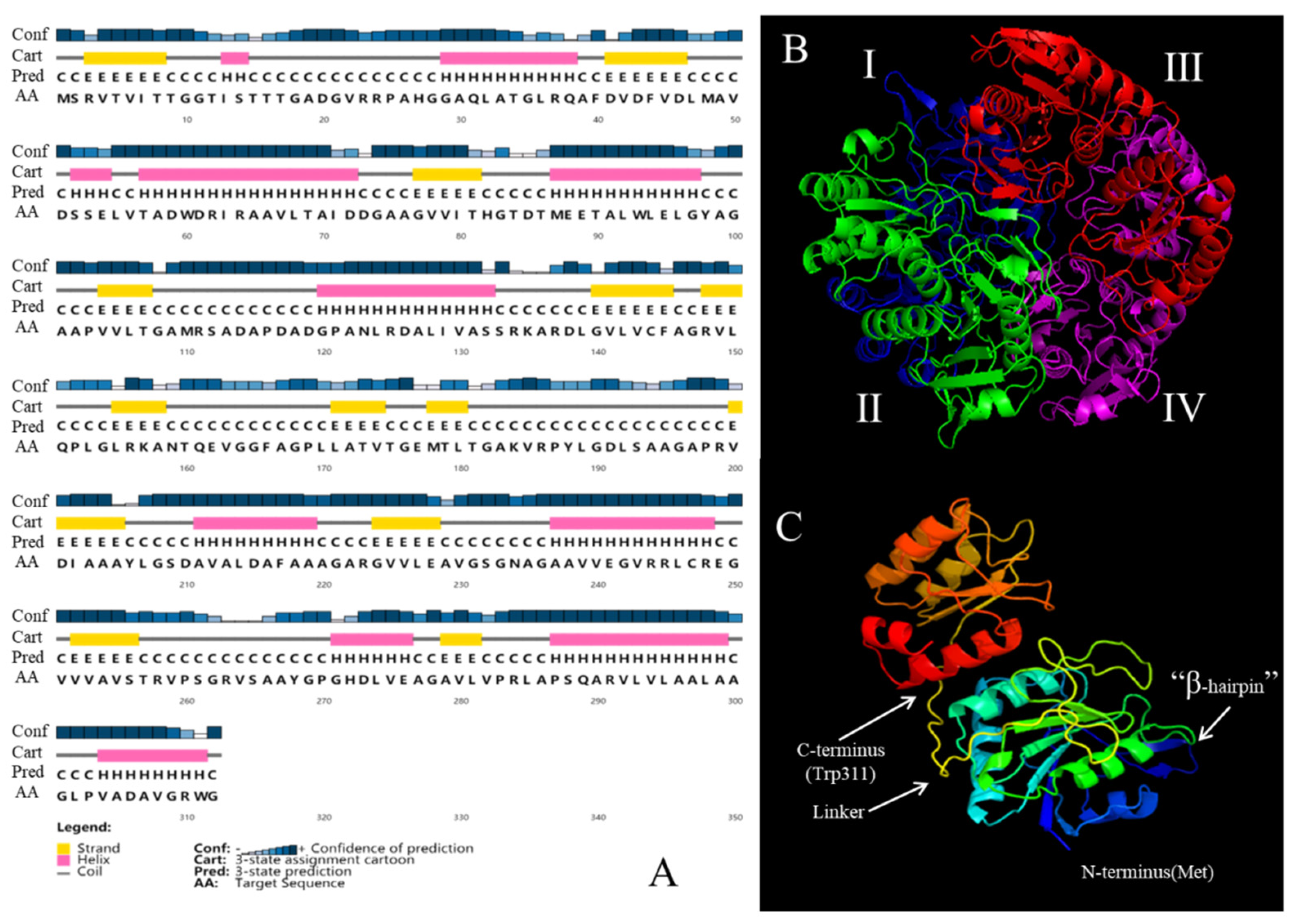
2.3. Structural Modeling of Mycobacterium Gordonae L-Asparaginase and Structure Analysis
2.4. Expression and Purification of Mycobacterium Gordonae L-Asparaginase in Escherichia coli
2.5. Protein Determination and Asparaginase Activity Assay
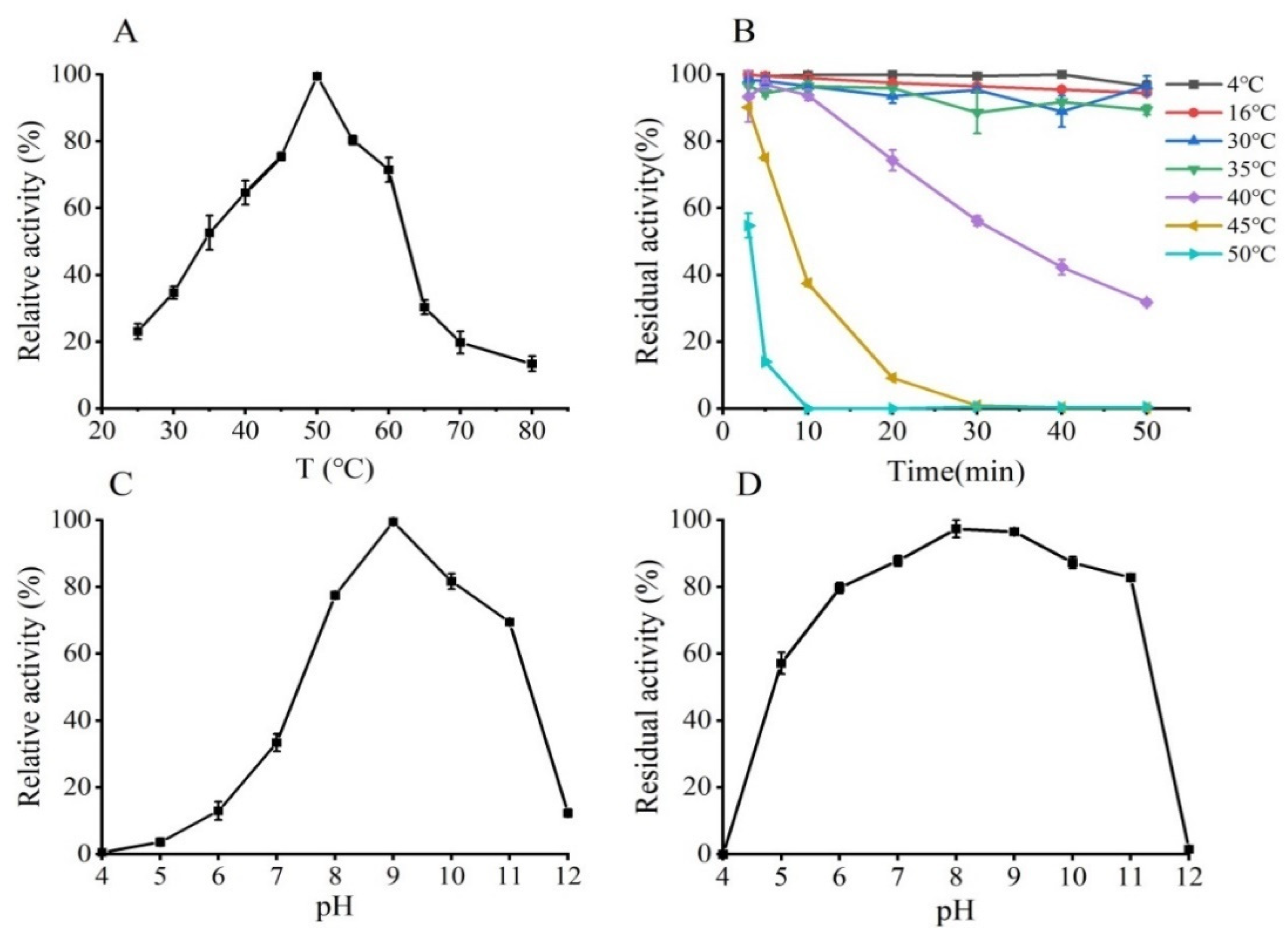
2.6. Effects of Temperatures and pHs on GmASNase Activity and Stability
2.7. Effect of Metal Ions and Inhibitors on GmASNase Activity
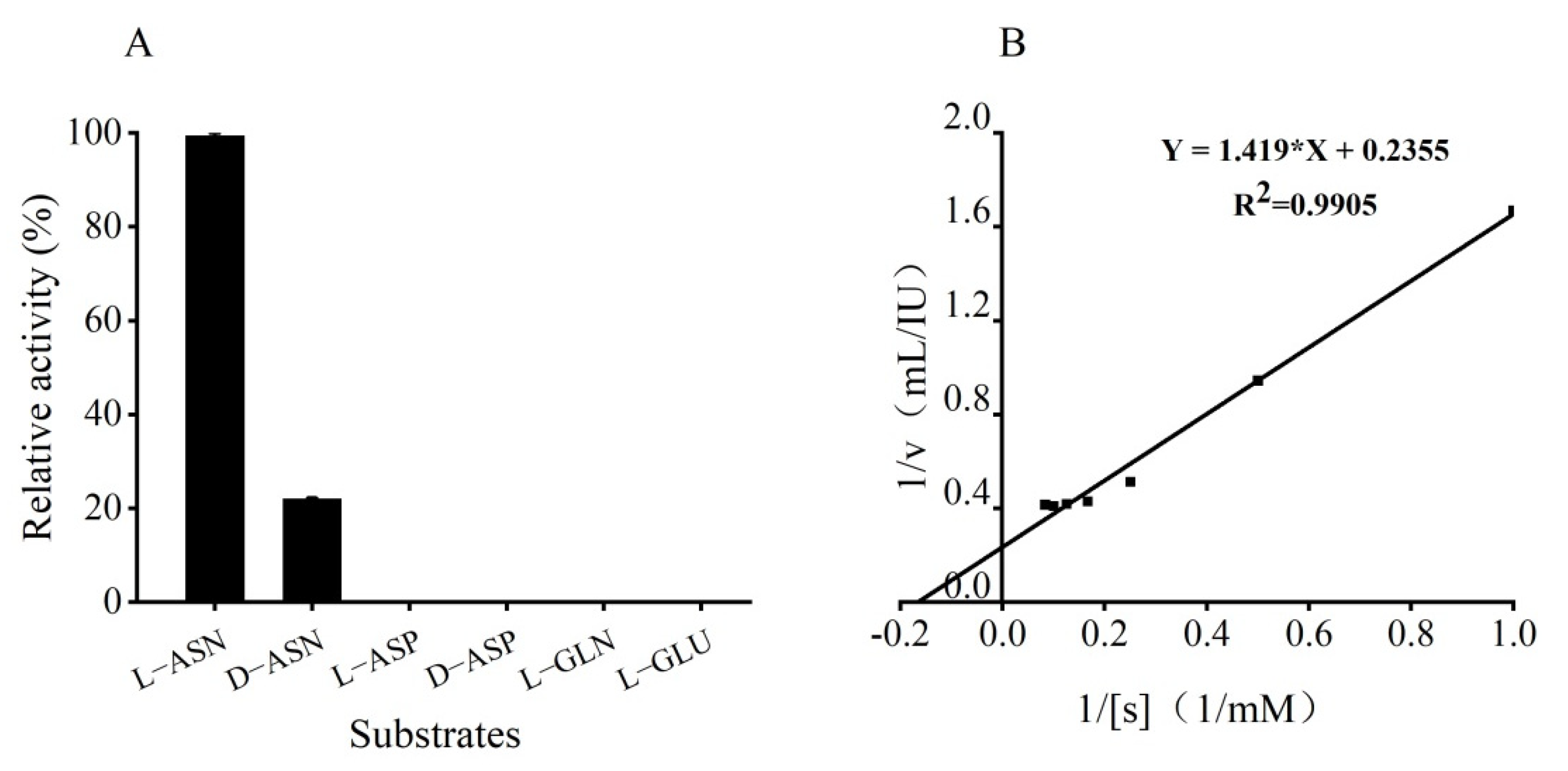
2.8. Substrate Specificity Assay and Determination of Kinetic Parameters of GmASNase
2.9. Application of GmASNase in Food
2.9.1. Preparation of Potato Chips
2.9.2. Separation and Purification of Acrylamide in the Sample
2.9.3. Detection of Acrylamide
2.10. Statistical Analysis
3. Results
3.1. In Silico Sequence and Phylogenetic Analysis
3.2. Structural Modeling of Mycobacterium Gordonae L-Asparaginase and Structure Analysis
3.3. Expression and Purification of Mycobacterium Gordonae L-Asparaginase in E. coli
3.4. Effects of pH and Temperature on the Activity and Stability of GmASNase
3.5. Effect of Metal Ions and Inhibitors on GmASNase Activity
3.6. Substrate Specificity Assay and Kinetic Parameters of GmASNase
3.7. Application of GmASNase in the Mitigation of Acrylamide in Fried Potato Chips
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shabana, A.M.; Shetaia, Y.M.; Abdelwahed, N.A.M.; Esawy, M.A.; Alfarouk, O.R. Optimization, purification and antitumor activity of Kodamaea ohmeri MRI ANOMY L-asparaginase isolated from banana peel. Curr. Pharm. Biotechnol. 2021, 22, 646–663. [Google Scholar] [CrossRef]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M.M. A comprehensive review on L-asparaginase and its applications. Appl. Biochem. Biotechnol. 2016, 178, 900–923. [Google Scholar] [CrossRef]
- Verma, N.; Kumar, K.; Kaur, G.; Anand, S. L-asparaginase: A promising chemotherapeutic agent. Crit. Rev. Biotechnol. 2007, 27, 45–62. [Google Scholar] [CrossRef] [PubMed]
- Lineback, D.R.; Coughlin, J.R.; Stadler, R.H. Acrylamide in foods: A review of the science and future considerations. Annu. Rev. Food Sci. Technol. 2012, 3, 15–35. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Qin, R.; Li, D.; Ji, K.; Wang, T.; Cui, Z.; Huang, Y. A novel bacterial type II L-asparaginase and evaluation of its enzymatic acrylamide reduction in French fries. Int. J. Biol. Macromol. 2016, 92, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Hendriksen, H.V.; Kornbrust, B.A.; Ostergaard, P.R.; Stringer, M.A. Evaluating the potential for enzymatic acrylamide mitigation in a range of food products using an asparaginase from Aspergillus oryzae. J. Agric. Food Chem. 2009, 57, 4168–4176. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, Y.; Zhang, C.; Bie, X.; Zhao, H.; Lu, F.; Lu, Z. Biochemical characterization of a novel L-asparaginase from Bacillus megaterium H-1 and its application in French fries. Food Res. Int. 2015, 77, 527–533. [Google Scholar] [CrossRef]
- Shi, R.; Liu, Y.; Mu, Q.; Jiang, Z.; Yang, S. Biochemical characterization of a novel L-asparaginase from Paenibacillus barengoltzii being suitable for acrylamide reduction in potato chips and mooncakes. Int. J. Biol. Macromol. 2017, 96, 93–99. [Google Scholar] [CrossRef]
- Kumar, N.S.M.; Manonmani, H.K. Purification, characterization and kinetic properties of extracellular L-asparaginase produced by Cladosporium sp. World J. Microbiol. Biotechnol. 2013, 29, 577–587. [Google Scholar] [CrossRef]
- Jiao, L.; Chi, H.; Lu, Z.; Zhang, C.; Chia, S.R.; Show, P.L.; Tao, Y.; Lu, F. Characterization of a novel type I L-asparaginase from Acinetobacter soli and its ability to inhibit acrylamide formation in potato chips. J. Biosci. Bioeng. 2020, 129, 672–678. [Google Scholar] [CrossRef]
- Onishi, Y.; Prihanto, A.A.; Yano, S.; Takagi, K.; Umekawa, M.; Wakayama, M. Effective treatment for suppression of acrylamide formation in fried potato chips using L-asparaginase from Bacillus subtilis. 3 Biotech 2015, 5, 783–789. [Google Scholar] [CrossRef]
- Zuo, S.; Zhang, T.; Jiang, B.; Mu, W. Reduction of acrylamide level through blanching with treatment by an extremely thermostable L-asparaginase during French fries processing. Extremophiles 2015, 19, 841–851. [Google Scholar] [CrossRef]
- Chand, S.; Mahajan, R.V.; Prasad, J.P.; Sahoo, D.K.; Mihooliya, K.N.; Dhar, M.S.; Sharma, G. A comprehensive review on microbial L-asparaginase: Bioprocessing, characterization, and industrial applications. Biotechnol. Appl. Biochem. 2020, 67, 619–647. [Google Scholar] [CrossRef]
- Krishnapura, P.R.; Belur, P.D.; Subramanya, S. A critical review on properties and applications of microbiall-asparaginases. Crit. Rev. Microbiol. 2016, 42, 720–737. [Google Scholar] [CrossRef]
- Ali, U.; Naveed, M.; Ullah, A.; Ali, K.; Shah, S.A.; Fahad, S.; Mumtaz, A.S. L-asparaginase as a critical component to combat Acute Lymphoblastic Leukaemia (ALL): A novel approach to target ALL. Eur. J. Pharmacol. 2016, 771, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, W.; Wu, H.; Zhang, W.; Guang, C.; Mu, W. Microbial production, molecular modification, and practical application of L-asparaginase: A review. Int. J. Biol. Macromol. 2021, 186, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Pedreschi, F.; Kaack, K.; Granby, K. The effect of asparaginase on acrylamide formation in French fries. Food Chem. 2008, 109, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Vala, A.K.; Sachaniya, B.; Dudhagara, D.; Panseriya, H.Z.; Gosai, H.; Rawal, R.; Dave, B.P. Characterization of L-asparaginase from marine-derived Aspergillus niger AKV-MKBU, its antiproliferative activity and bench scale production using industrial waste. Int. J. Biol. Macromol. 2018, 108, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Shivakumar, S. Bioprospecting of the agaricomycete Ganoderma australe GPC191 as novel source for L-asparaginase production. Sci. Rep. 2021, 11, 6192. [Google Scholar] [CrossRef]
- Saeed, H.; Ali, H.; Soudan, H.; Embaby, A.; El-Sharkawy, A.; Farag, A.; Hussein, A.; Ataya, F. Molecular cloning, structural modeling and production of recombinant Aspergillus terreus L-asparaginase in Escherichia coli. Int. J. Biol. Macromol. 2018, 106, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Knape, M.J.; Ahuja, L.G.; Bertinetti, D.; Burghardt, N.C.G.; Zimmermann, B.; Taylor, S.S.; Herberg, F.W. Divalent metal ions Mg2+ and Ca2+ have distinct effects on protein kinase a activity and regulation. ACS Chem. Biol. 2015, 10, 2303–2315. [Google Scholar] [CrossRef]
- Meena, B.; Anburajan, L.; Vinithkumar, N.V.; Shridhar, D.; Raghavan, R.V.; Dharani, G. Molecular expression of L-asparaginase gene from Nocardiopsis alba NIOT-VKMA08 in Escherichia coli: A prospective recombinant enzyme for leukaemia chemotherapy. Gene 2016, 590, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.; Choubey, P.; Agashe, M. Studies on optimization of growth parameters for L-asparaginase production by Streptomyces ginsengisoli. Sci. World J. 2014, 2014, 895167. [Google Scholar] [CrossRef][Green Version]
- Saxena, A.; Upadhyay, R.; Kango, N. Isolation and identification of actinomycetes for production of novel extracellular glutaminase free L-asparaginase. Indian J. Exp. Biol. 2015, 53, 786–793. [Google Scholar] [PubMed]
- El-Naggar, E.A.; Deraz, S.F.; Soliman, H.M.; El-Deeb, N.M.; El-Ewasy, S.M. Purification, characterization, cytotoxicity and anticancer activities of L-asparaginase, anti-colon cancer protein, from the newly isolated alkaliphilic Streptomyces fradiae NEAE-82. Sci. Rep. 2016, 6, 32926. [Google Scholar] [CrossRef]
- Amena, S.; Vishalakshi, N.; Prabhakar, M.; Dayanand, A.; Lingappa, K. Production, purification and characterization of L-asparaginase from Streptomyces gulbargensis. Braz. J. Microbiol. 2010, 41, 173–178. [Google Scholar] [CrossRef]
- Delano, W.L. The pymol molecular graphics system. Proteins Struct. Funct. Bioinform. 2002, 30, 442–454. [Google Scholar]
- Lu, X.; Chen, J.; Jiao, L.; Zhong, L.; Lu, Z.; Zhang, C.; Lu, F. Improvement of the activity of L-asparaginase I improvement of the catalytic activity of L-asparaginase I from Bacillus megaterium H-1 by in vitro directed evolution. J. Biosci. Bioeng. 2019, 128, 683–689. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Kumar, S.; Dasu, V.V.; Pakshirajan, K. Localization and production of novel L-asparaginase from Pectobacterium carotovorum MTCC 1428. Process Biochem. 2010, 45, 223–229. [Google Scholar] [CrossRef]
- Yim, S.; Kim, M. Purification and characterization of thermostable L-asparaginase from Bacillus amyloliquefaciens MKSE in Korean soybean paste. LWT-Food Sci. Technol. 2019, 109, 415–421. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, H.; Zhang, W.; Xu, W.; Mu, W. Efficient control of acrylamide in French fries by an extraordinarily active and thermo-stable L-asparaginase: A lab-scale study. Food Chem. 2021, 360, 130046. [Google Scholar] [CrossRef]
- Xian, L.; Wang, F.; Luo, X.; Feng, Y.L.; Feng, J.X. Purification and characterization of a highly efficient calcium-independent alpha-amylase from Talaromyces pinophilus 1-95. PLoS ONE 2015, 10, e0121531. [Google Scholar]
- Suo, H.; Xu, L.; Xue, Y.; Qiu, X.; Huang, H.; Hu, Y. Ionic liquids-modified cellulose coated magnetic nanoparticles for enzyme immobilization: Improvement of catalytic performance. Carbohydr. Polym. 2020, 234, 115914. [Google Scholar] [CrossRef]
- Sharma, D.; Uniyal, S.; Tewari, L.; Sharma, D. Biocatalytic transesterification of algal oil employing a heterogenous methanol tolerant lipase enzyme aggregate from Bacillus mycoides strain CV18. Process Biochem. 2021, 111, 43–52. [Google Scholar] [CrossRef]
- Patil, S.; Coutsouvelis, J.; Spencer, A. Asparaginase in the management of adult acute lymphoblastic leukaemia: Is it used appropriately? Cancer Treat. Rev. 2011, 37, 202–207. [Google Scholar] [CrossRef]
- Sarion, C.; Codina, G.G.; Dabija, A. Acrylamide in bakery products: A review on health risks, legal regulations and strategies to reduce its formation. Int. J. Environ. Res. Public Health 2021, 18, 4332. [Google Scholar] [CrossRef]
- Ran, T.; Jiao, L.; Wang, W.; Chen, J.; Chi, H.; Lu, Z.; Zhang, C.; Xu, D.; Lu, F. Structures of L-asparaginase from Bacillus licheniformis reveal an essential residue for its substrate stereoselectivity. J. Agric. Food Chem. 2021, 69, 223–231. [Google Scholar] [CrossRef]
- Desai, S.S.; Hungund, B.S. Submerged fermentation, purification, and characterization of L-asparaginase from Streptomyces sp. isolated from soil. J. Appl. Biol. Biotechnol. 2018, 6, 17–23. [Google Scholar]
- Sindhu, R.; Manonmani, H.K. Expression and characterization of recombinant L-asparaginase from Pseudomonas fluorescens. Protein Expr. Purif. 2018, 143, 83–91. [Google Scholar] [CrossRef] [PubMed]
- El-Naggar, N.E.-A.; Deraz, S.F.; El-Ewasy, S.M.; Suddek, G.M. Purification, characterization and immunogenicity assessment of glutaminase free L-asparaginase from Streptomyces brollosae NEAE-115. BMC Pharmacol. Toxicol. 2018, 19, 51. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.W.M.; Farag, A.M.; Beltagy, E.A. Purification, characterization and anticancer activity of L-asparaginase produced by marine Aspergillus terreus. J. Pure Appl. Microbiol. 2018, 12, 1845–1854. [Google Scholar] [CrossRef]
- Alrumman, S.A.; Mostafa, Y.S.; Al-Izran, K.A.; Alfaifi, M.Y.; Taha, T.H.; Elbehairi, S.E. Production and anticancer activity of an L-asparaginase from Bacillus licheniformis isolated from the Red Sea, Saudi Arabia. Sci. Rep. 2019, 9, 3756. [Google Scholar] [CrossRef]
- Aly, N.; El-Ahwany, A.; Ataya, F.S.; Saeed, H. Bacillus sonorensis L. Asparaginase: Cloning, expression in E. coli and characterization. Protein J. 2020, 39, 717–729. [Google Scholar] [CrossRef]
- Saeed, H.; Hemida, A.; El-Nikhely, N.; Abdel-Fattah, M.; Shalaby, M.; Hussein, A.; Eldoksh, A.; Ataya, F.; Aly, N.; Labrou, N.; et al. Highly efficient Pyrococcus furiosus recombinant L-asparaginase with no glutaminase activity: Expression, purification, functional characterization, and cytotoxicity on THP-1, A549 and Caco-2 cell lines. Int. J. Biol. Macromol. 2020, 156, 812–828. [Google Scholar] [CrossRef]
- Prakash, P.; Singh, H.R.; Jha, S.K. Production, purification and kinetic characterization of glutaminase free anti-leukemic L-asparaginase with low endotoxin level from novel soil isolate. Prep. Biochem. Biotechnol. 2020, 50, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Chohan, S.M.; Sajed, M.; Naeem, S.U.; Rashid, N. Heterologous gene expression and characterization of TK2246, a highly active and thermostable plant type L-asparaginase from Thermococcus kodakarensis. Int. J. Biol. Macromol. 2020, 147, 131–137. [Google Scholar] [CrossRef]
- Dhevagi, P.; Poorani, E. Isolation and characterization of L-asparaginase from marine actinomycetes. Indian J. Biotechnol. 2006, 5, 514–520. [Google Scholar]
- Dash, C.; Mohapatra, S.B.; Maiti, P.K. Optimization, purification, and characterization of L-asparaginase from Actinomycetales bacterium BkSoiiA. Prep. Biochem. Biotechnol. 2016, 46, 1–7. [Google Scholar] [CrossRef]
- Dharmaraj, S. Study of L-asparaginase production by Streptomyces noursei MTCC 10469, isolated from marine sponge Callyspongia diffusa. Iran. J. Biotechnol. 2011, 9, 102–108. [Google Scholar]
- Farahat, M.G.; Amr, D.; Galal, A. Molecular cloning, structural modeling and characterization of a novel glutaminase-free L-asparaginase from Cobetia amphilecti AMI6. Int. J. Biol. Macromol. 2020, 143, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Meghavarnam, A.K.; Janakiraman, S. Purification and characterization of therapeutic enzyme L-asparaginase from a tropical soil fungal isolate Fusarium culmorum ASP-87. MOJ Proteom. Bioinform. 2015, 2, 171–175. [Google Scholar]

| Purification Steps | Total Activity (IU) | Total Protein (mg) | Specific Activity (IU mg−1) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Crude enzyme | 3110.86 | 200.04 | 15.55 | 1 | 100 |
| Nickel affinity purified GmASNase | 1328.57 | 2.73 | 486.65 | 31.29 | 42.71 |
| Metal Ions and Inhibitors | Relative Activity (%) |
|---|---|
| Control | 100 |
| Mn2+ | 147 ± 10 |
| Zn2+ | 103 ± 4 |
| K+ | 94 ± 3 |
| Na+ | 93 ± 1 |
| Al3+ | 84 ± 2 |
| Ca2+ | 89 ± 6 |
| Mg2+ | 193 ± 29 |
| Cu2+ | 77 ± 4 |
| Ni2+ | 160 ± 8 |
| Fe2+ | 72 ± 2 |
| Fe3+ | 80 ± 3 |
| SDS | 0 |
| EDTA | 84 ± 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, H.; Chen, M.; Jiao, L.; Lu, Z.; Bie, X.; Zhao, H.; Lu, F. Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential. Foods 2021, 10, 2819. https://doi.org/10.3390/foods10112819
Chi H, Chen M, Jiao L, Lu Z, Bie X, Zhao H, Lu F. Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential. Foods. 2021; 10(11):2819. https://doi.org/10.3390/foods10112819
Chicago/Turabian StyleChi, Huibing, Meirong Chen, Linshu Jiao, Zhaoxin Lu, Xiaomei Bie, Haizhen Zhao, and Fengxia Lu. 2021. "Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential" Foods 10, no. 11: 2819. https://doi.org/10.3390/foods10112819
APA StyleChi, H., Chen, M., Jiao, L., Lu, Z., Bie, X., Zhao, H., & Lu, F. (2021). Characterization of a Novel L-Asparaginase from Mycobacterium gordonae with Acrylamide Mitigation Potential. Foods, 10(11), 2819. https://doi.org/10.3390/foods10112819








