SlCCD1A Enhances the Aroma Quality of Tomato Fruits by Promoting the Synthesis of Carotenoid-Derived Volatiles
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sampling Methods
2.3. Determination on Volatile Contents of Tomato Fruits
2.4. Sensory Evaluation on Aroma Quality of Tomato Fruits
2.5. Tissue-Specific Expression of SlCCD Genes in Tomato
2.6. Cloning of SlCCD1A and SlCCD1B from Tomato Fruits
2.7. Analysis of Nucleotide and Promoter Sequence and Transcription Factors of SlCCD1A and SlCCD1B
2.8. Analysis of Amino Acid Sequences of SlCCD1A and SlCCD1B Proteins
2.9. SlCCD1A Recombinant Vector Construction
2.9.1. The Overexpression Recombinant Vector Construction of SlCCD1A
2.9.2. The RNAi Recombinant Vector Construction of SlCCD1A
2.10. Genetic Transformation of SlCCD1A Recombinant Vectors
2.11. SlCCD1A Expression of Transgenic Tomato Plants
2.12. Volatile Contents and Evaluation of Aroma Quality of Transgenic Tomato Fruits
2.13. Data Statistics and Analysis
3. Results
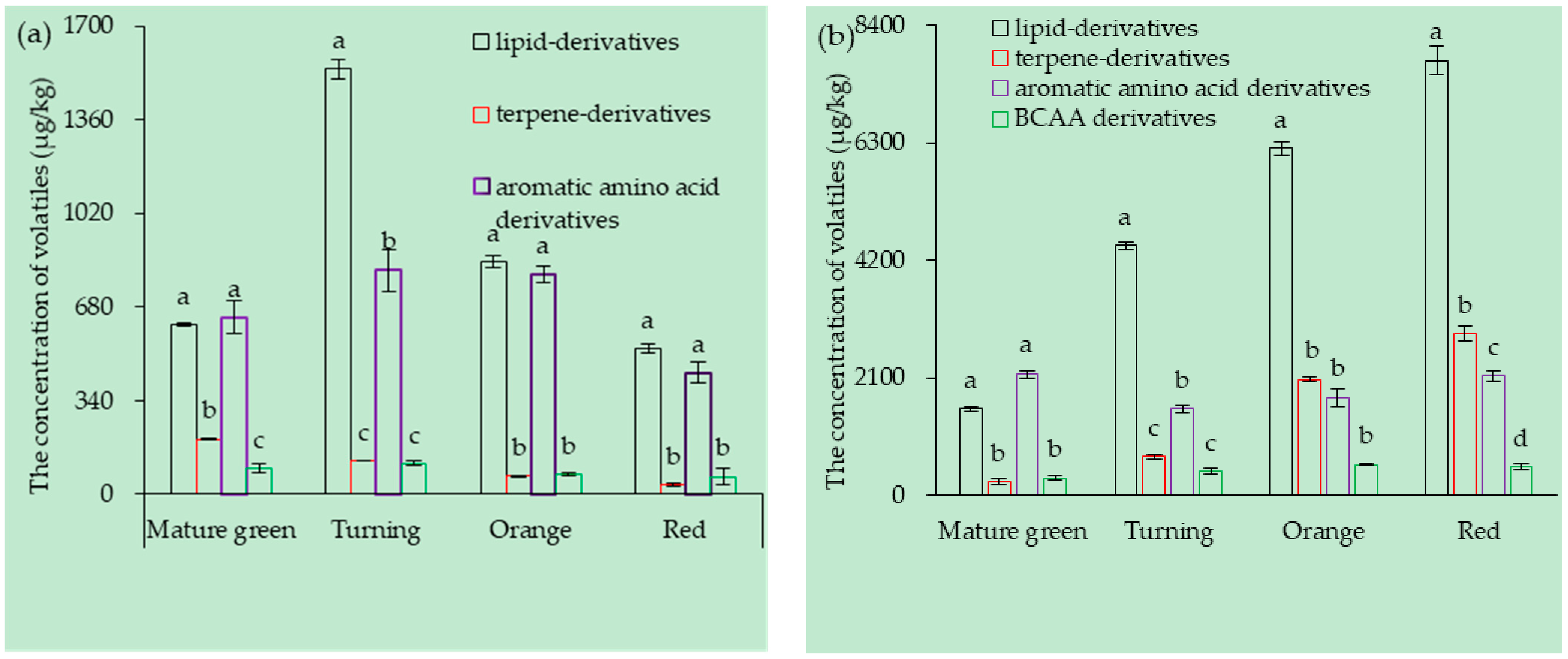
3.1. Tomato Fruit Volatiles Composition
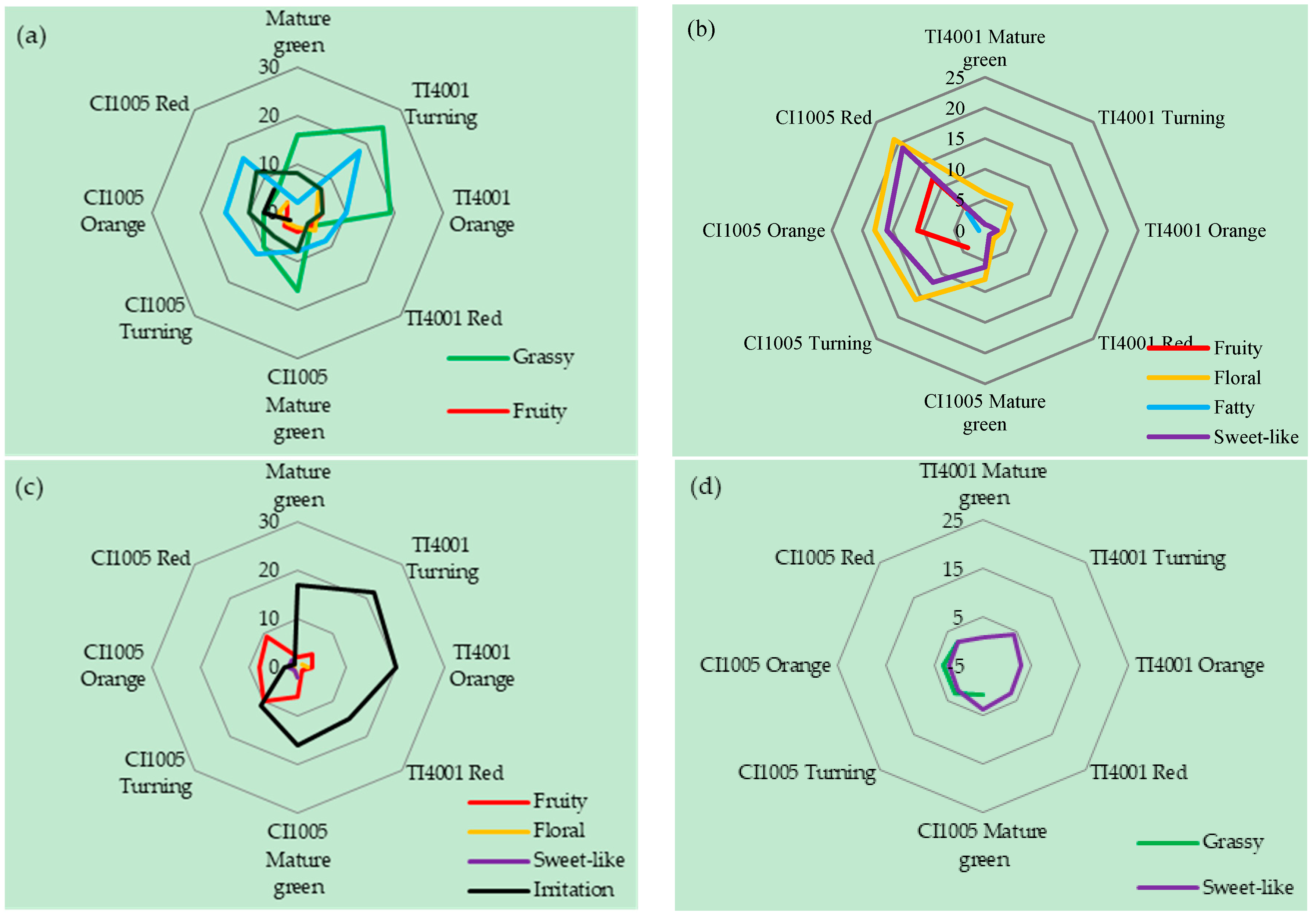
3.2. Aroma Characteristics of Tomato Fruits
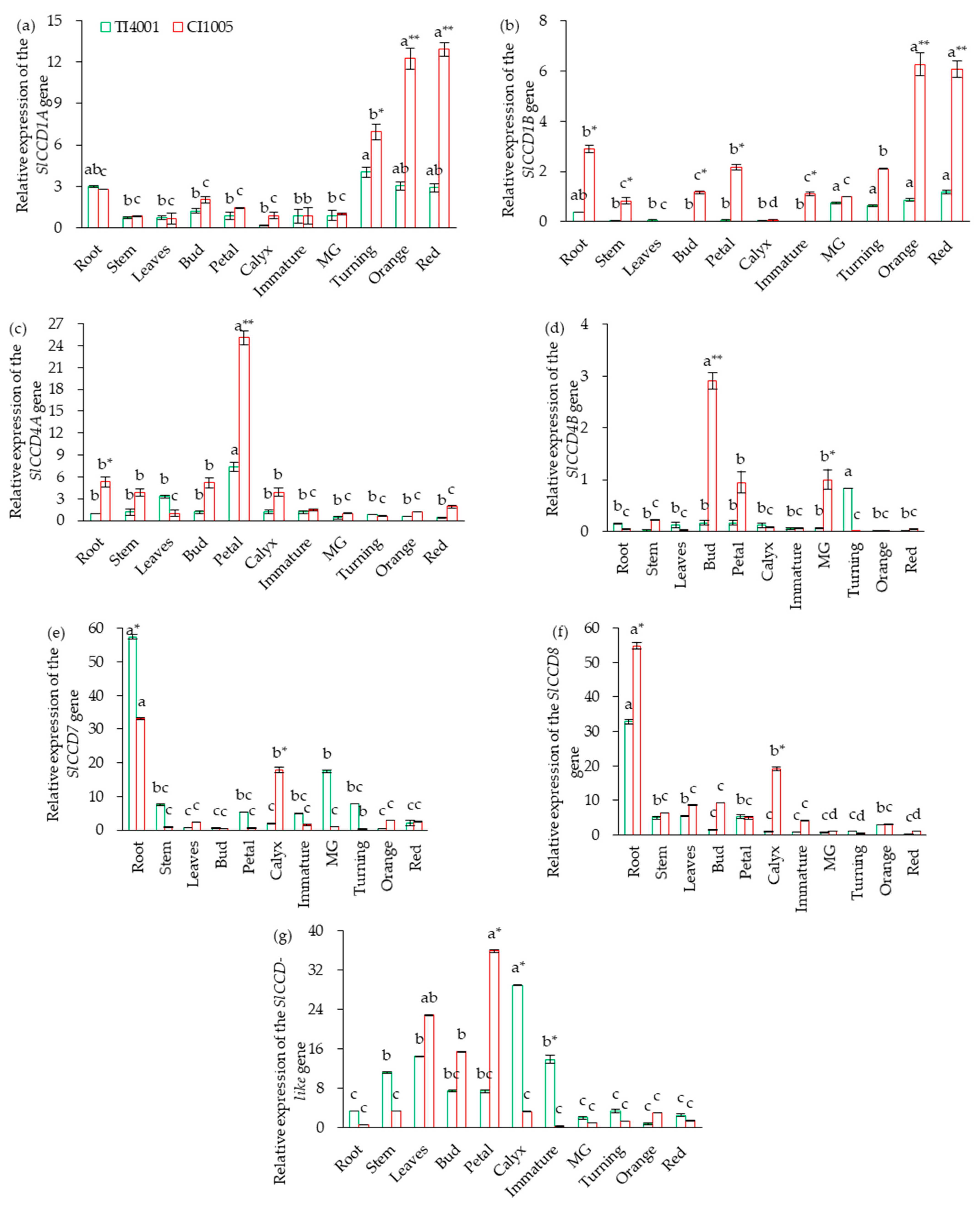
3.3. Tissue-Specific Expression of SlCCD in Tomatoes
3.4. Bioinformatics Analysis of SlCCD1A and SlCCD1B in Tomatoes
3.4.1. Sequence Analysis of tSlCCD1A and SlCCD1B
3.4.2. The Promoter Analysis of SlCCD1A and SlCCD1B in Tomatoes
3.4.3. Analysis of the SlCCD1A and SlCCD1B Transcription Factors in Tomatoes
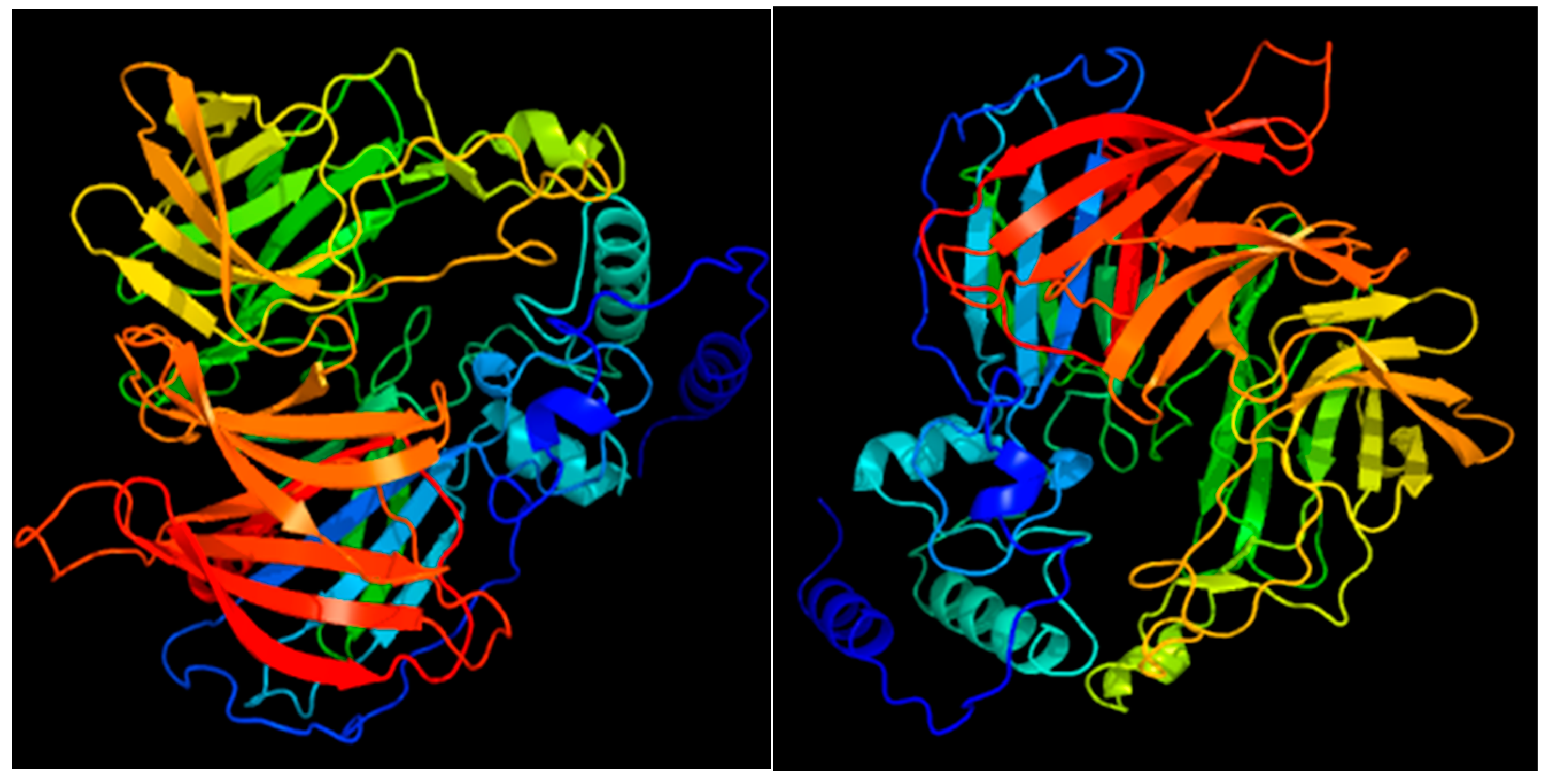
3.4.4. Structural Analysis of the SlCCD1A and SlCCD1B Proteins in Tomatoes
3.4.5. Conserved Structural Domains and Phylogenetic Analysis on the CCD1 Protein in Dicotyledonous Plants
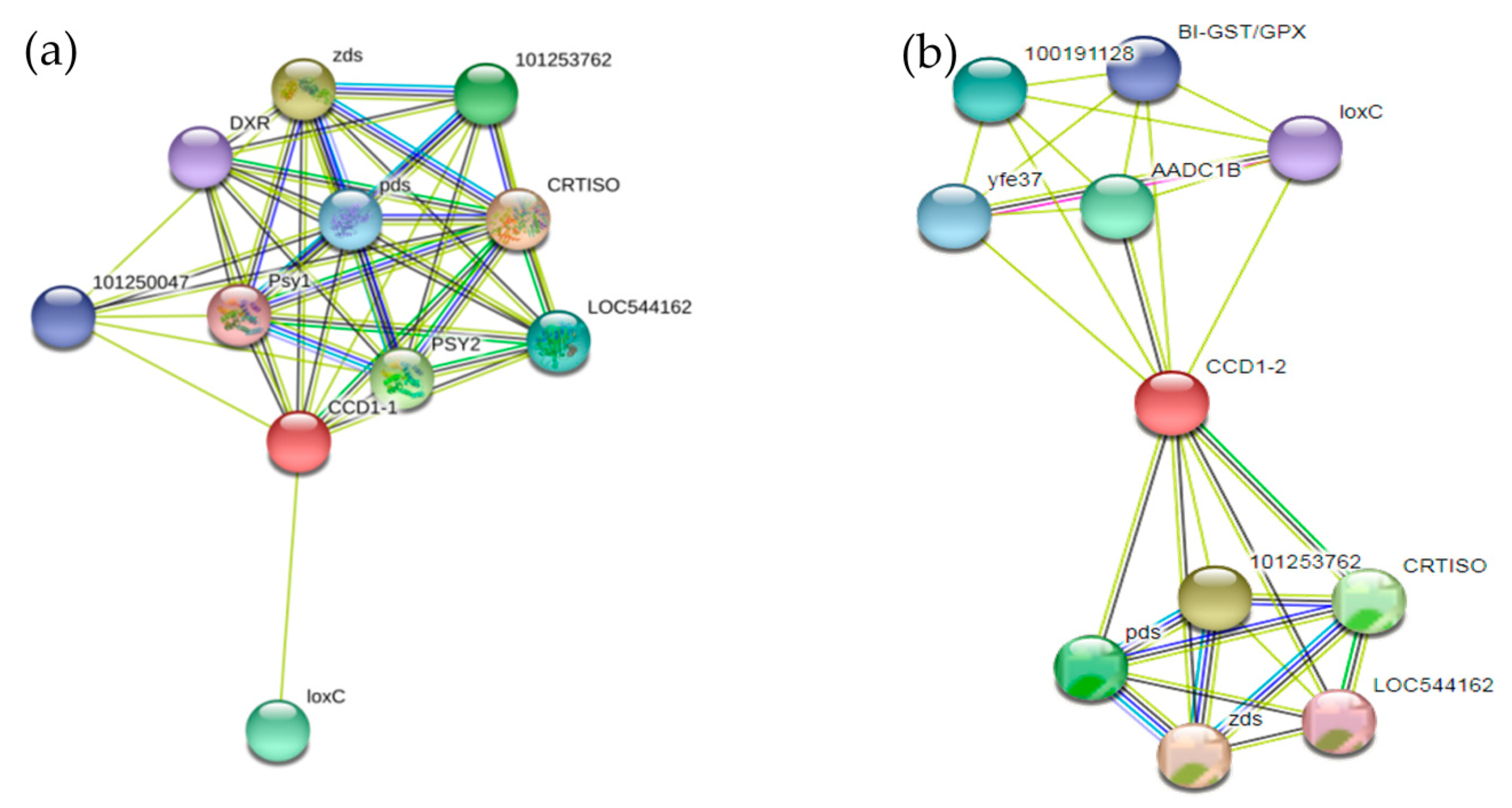
3.4.6. Regulatory Network Analysis of the SlCCD1A and SlCCD1B Proteins
3.5. Effect of SlCCD1A on Volatiles and Aroma in Tomato Fruits
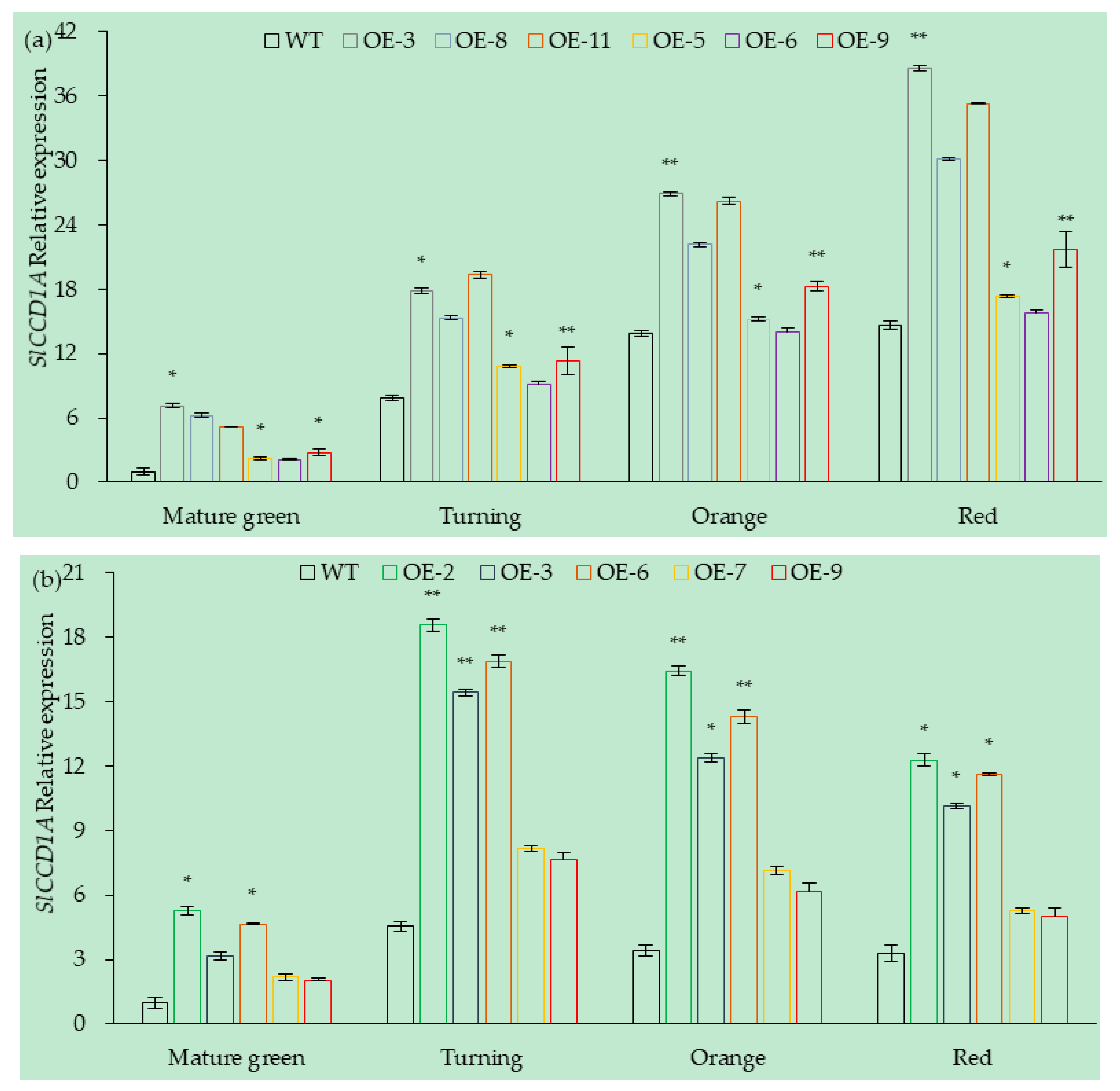
3.5.1. Analysis of SlCCD1A Expression in Transgenic Overexpressing Lines of Tomato Fruits
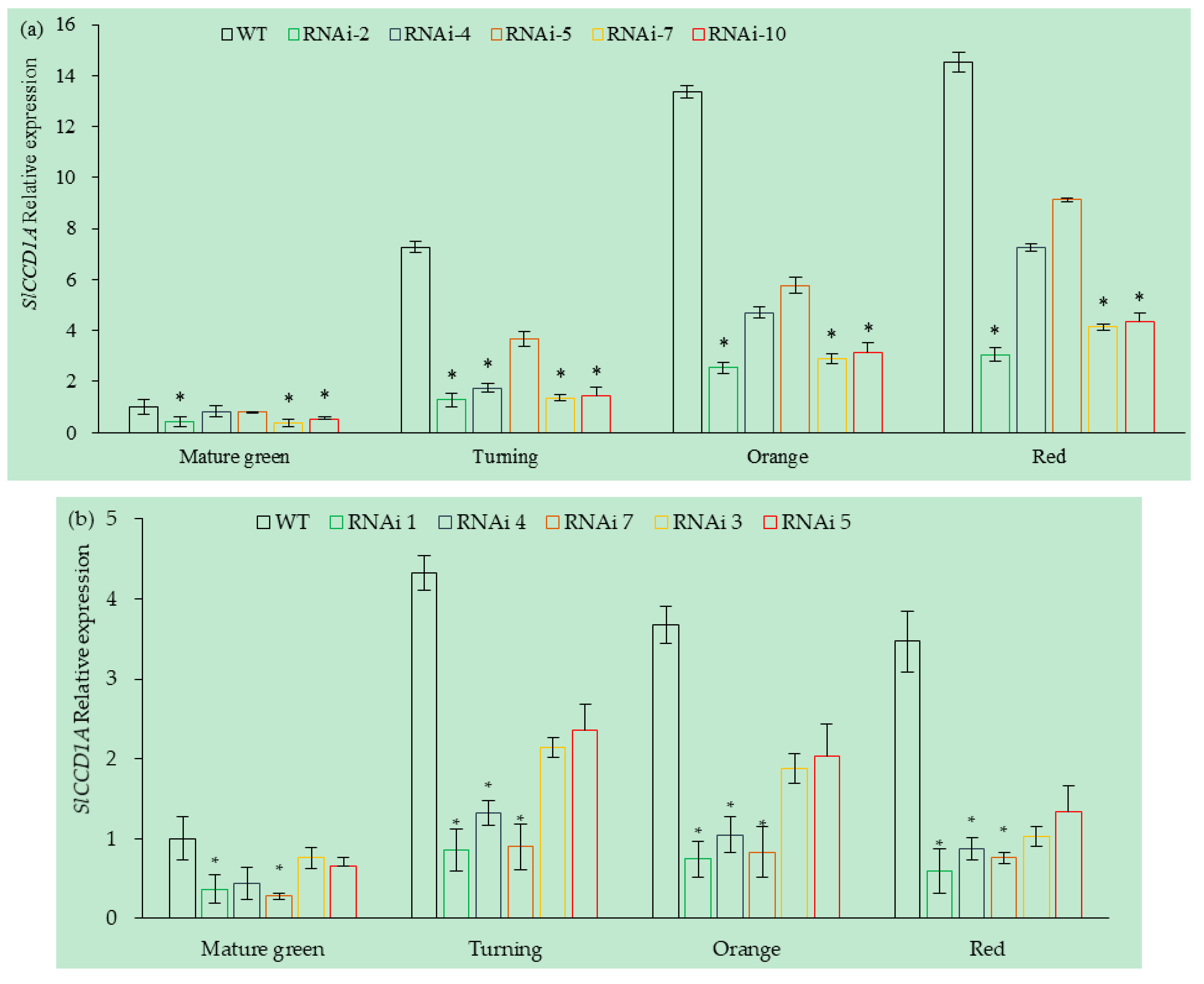
3.5.2. Analysis of SlCCD1A Expression in RNAi Transgenic Tomato Fruits
3.6. Effects of SlCCD1A on Volatiles in Tomato Fruits
3.6.1. Overexpression of SlCCD1A on Volatile Accumulation in Tomato Fruits
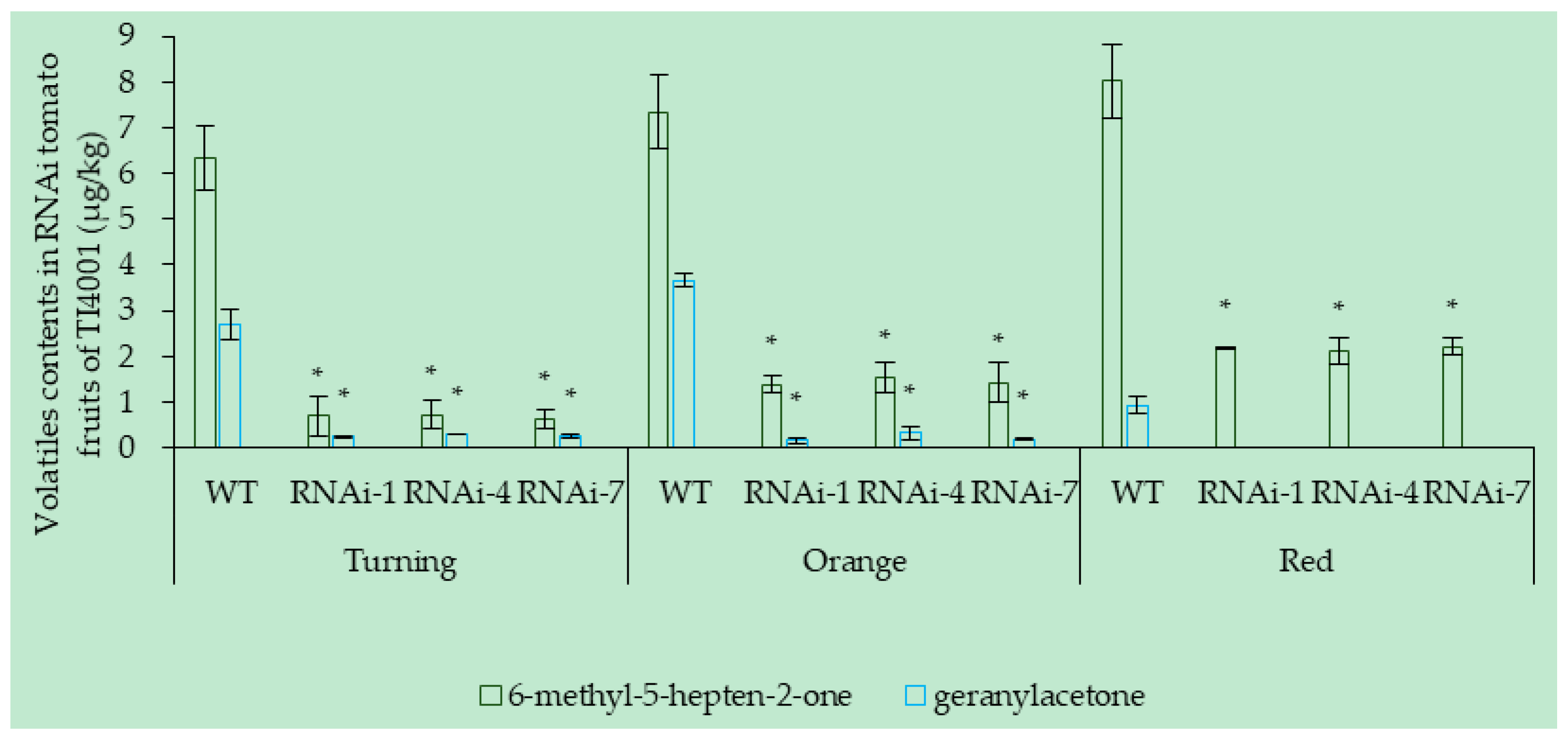
3.6.2. Effect of SlCCD1A RNAi on the Volatile Contents in Tomato Fruits
3.7. Effect of SlCCD1A on Aroma in Tomato Fruits
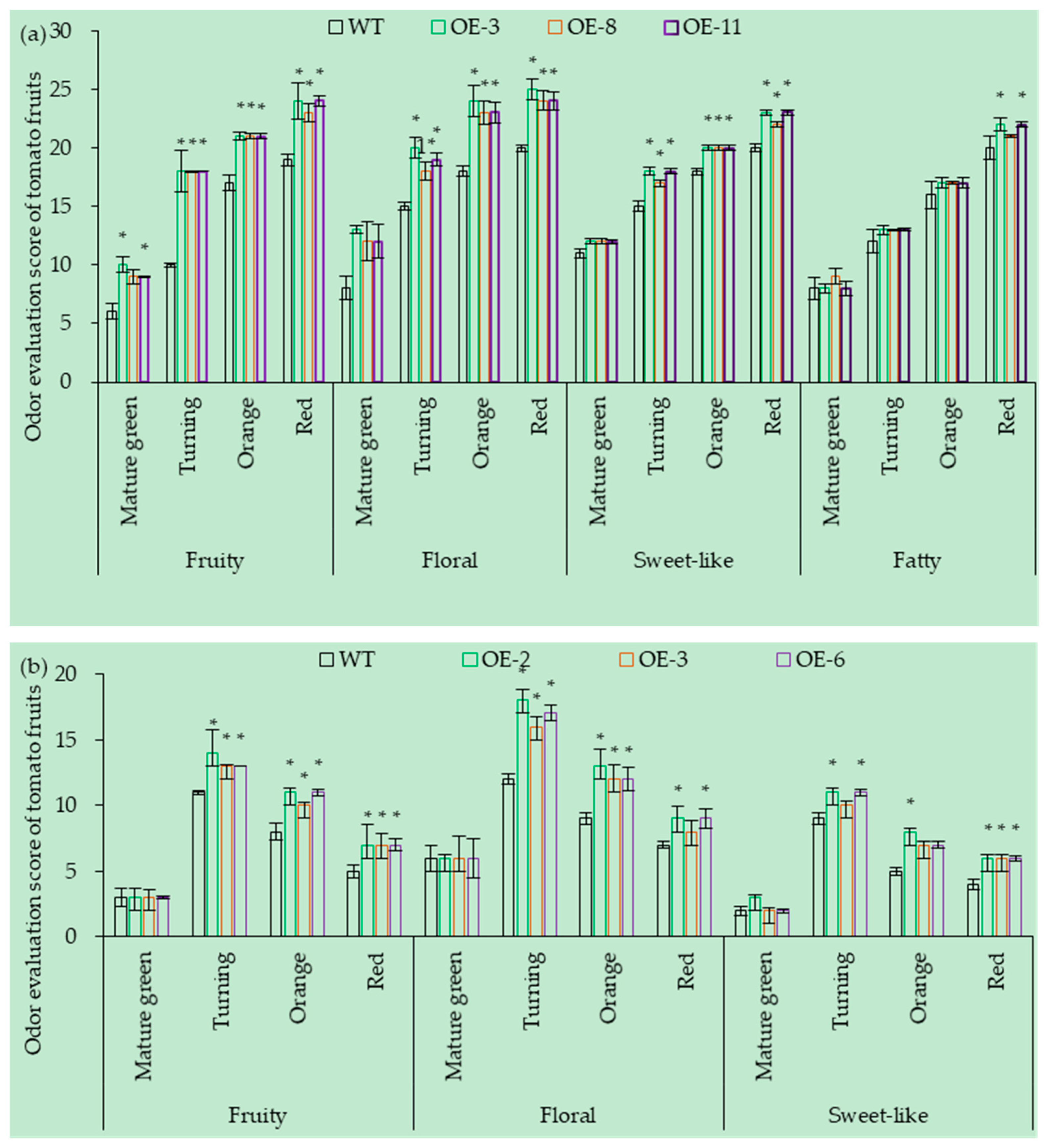
3.7.1. OE of SlCCD1A on the Aroma in Tomato Fruits
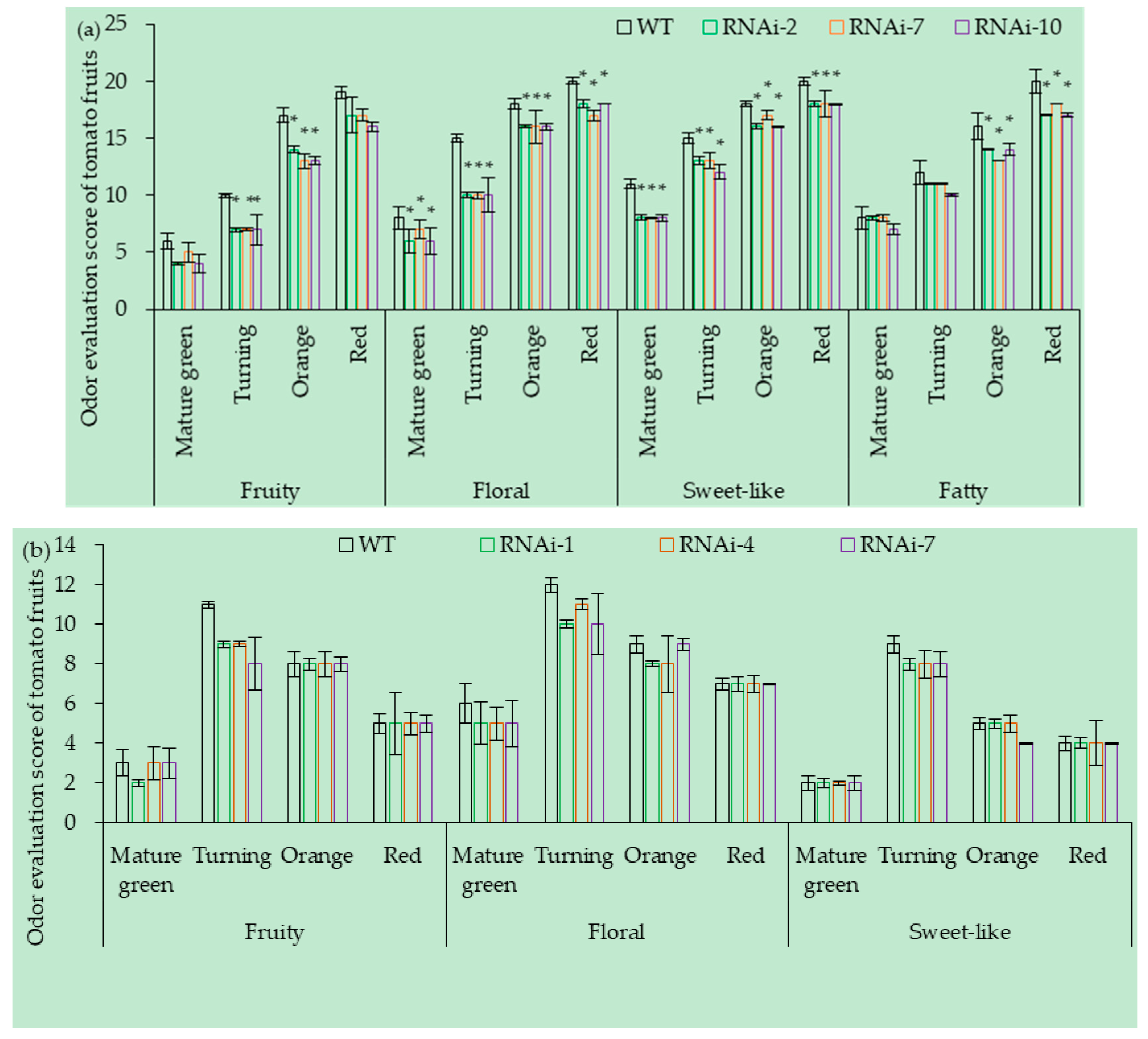
3.7.2. RNAi of SlCCD1A on the Aromas in Tomato Fruits
3.8. Effects of SlCCD1A on Carotenoid Contents in Tomato Fruits
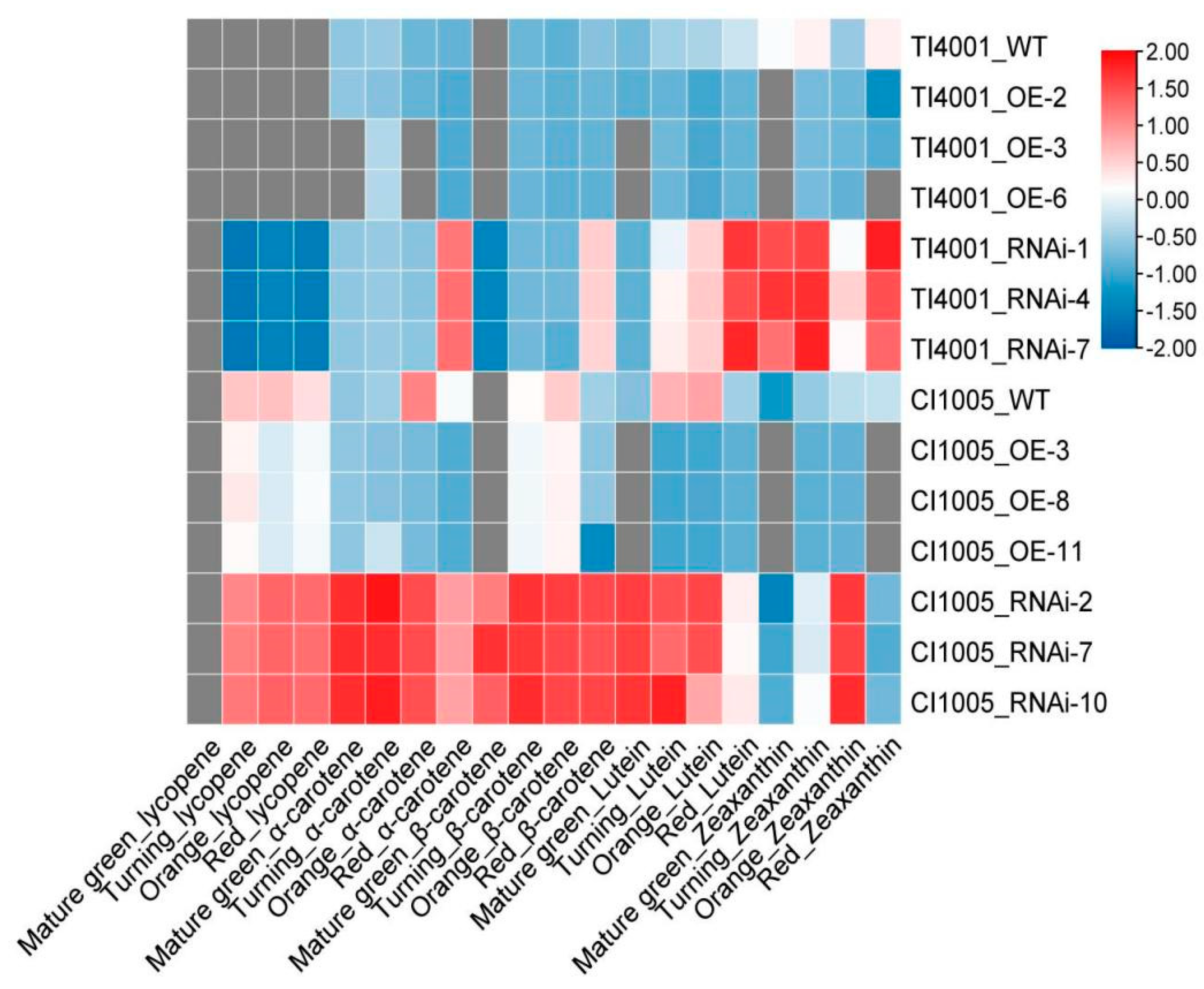
3.8.1. OE of SlCCD1A on Carotenoid Contents in Tomato Fruits
3.8.2. RNAi of SlCCD1A on Carotenoid Contents in Tomato Fruits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). Available online: http://www.fao.org/faostat/zh/?#data/QC/visualize (accessed on 22 December 2020).
- Giovannoni, J. Tomato multiomics reveals consequences of crop domestication and improvement. Cell 2018, 172, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.T.; Gou, J.B.; Klee, H.J.; Huang, S.W. Next-Gen Approaches to Flavor-Related Metabolism. In Annual Review of Plant Biology; Merchant, S.S., Ed.; Annual Reviews: Palo Alto, CA, USA, 2019; Volume 70, pp. 187–212. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.T.; Zhang, J.H.; Zhang, X.Y.; Yu, Q.H.; Zheng, Z.; Zhang, Z.H.; Lun, Y.Y.; Li, S.; Wang, X.X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Zhu, G.T.; Resende, J.R.M.F.R.; Lin, T.; Taylor, M.; Zhang, B.; Ikeda, H.; Liu, Z.Y.; Fisher, J.; Zemach, I.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Fulton, T.M.; Bucheli, P.; Voirol, E.; Lopez, J.; Petiard, V.; Tanksley, S.D. Quantitative trait loci (QTL) affecting sugar, organic acids and other biochemical properties possibly contributing to flavor, identified in four advanced backcross populations of tomato. Euphytica 2002, 127, 163–177. [Google Scholar] [CrossRef]
- Klee, H.J. Improving the flavor of fresh fruit: Genomics, biochemistry, and biotechnology. New Phytol. 2010, 187, 44–56. [Google Scholar] [CrossRef]
- Klee, H.J.; Tieman, D.M. The genetics of fruit flavour preferences. Nat. Rev. Genet. 2018, 19, 347–356. [Google Scholar] [CrossRef]
- Baldwin, E.A.; Goodner, K.; Plotto, A. Interaction of volatiles, sugars, and acids on perception of tomato aroma and flavor descriptors. J. Food Sci. 2008, 73, S294–S307. [Google Scholar] [CrossRef]
- Conrath, U.; Pieterse, C.M.J.; Mauch-Mani, B. Priming in plantpathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef]
- Tong, X.H.; Qi, J.F.; Zhu, X.D.; Mao, B.Z.; Zeng, L.J.; Wang, B.H.; Li, Q.; Zhou, G.X.; Xu, X.J.; Lou, Y.G.; et al. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J. 2012, 71, 763–775. [Google Scholar] [CrossRef]
- Hammerbacher, A.; Coutinho, T.A.; Gershenzon, J. Roles of plant volatiles in defence against microbial pathogens and microbial exploitation of volatiles. Plant Cell Environ. 2019, 42, 2827–2843. [Google Scholar] [CrossRef]
- Czerny, M.; Christlbauer, M.; Christlbauer, M.; Fischer, A.; Granvogl, M.; Hammer, M.; Hartl, C.; Hernandez, N.; Schieberle, P. Re-investigation on odour thresholds of key food aroma compounds and development of an aroma language based on odour qualities of defined aqueous odorant solutions. Eur. Food Res. Technol. 2008, 228, 265–273. [Google Scholar] [CrossRef]
- Klee, H.J.; Tieman, D.M. Genetic challenges of flavor improvement in tomato. Trends Genet. 2013, 29, 257–262. [Google Scholar] [CrossRef] [PubMed]
- Kreissl, J.; Schieberle, P. Characterization of odor-active compounds in Italian tomatoes with emphasis on new odorants. J. Agr. Food Chem. 2017, 65, 5198–5208. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Bliss, P.; McIntyre, L.M.; Blandon-Ubeda, A.; Bies, D.; Klee, H.J. The chemical interactions underlying tomato flavor preferences. Curr. Biol. 2012, 22, 1035–1039. [Google Scholar] [CrossRef]
- Piombino, P.; Sinesio, F.; Moneta, E.; Cammareri, M.; Genovese, A.; Lisanti, M.T.; Mogno, M.R.; Peparaio, M.; Termolino, P.; Moio, L.; et al. Investigating physicochemical, volatile and sensory parameters playing a positive or a negative role on tomato liking. Food Res. Int. 2013, 50, 409–419. [Google Scholar] [CrossRef]
- Cheng, G.T.; Chang, P.P.; Shen, Y.B.; Wu, L.T.; El-Sappah, A.H.; Zhang, F.; Liang, Y. Comparing the flavor characteristics of 71 tomato (Solanum lycopersicum) accessions in central Shaanxi. Front. Plant Sci. 2020, 11, 586834. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Bradbury, L.M.; Wurtzel, E.T. The carotenoid dioxygenase gene family in maize, sorghum and rice. Arch. Biochem. Biophys. 2010, 504, 104–111. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, Q.; Li, P.; Zhang, S.; Liu, C.; Jin, J.; Cao, P.; Yang, Y. Carotenoid cleavage dioxygenases: Identification, expression, and evolutionary analysis of this gene family in tobacco. Int. J. Mol. Sci. 2019, 20, 5796. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.K.; Hans, J.; Walter, M.H.; Matusova, R.; Beekwilder, J.; Verstappen, F.W.A.; Ming, Z.; Echtelt, E.V.; Strack, D.; Bisseling, T.; et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 2008, 228, 789–801. [Google Scholar] [CrossRef]
- Vogel, J.T.; Tan, B.C.; Mccarty, D.R.; Klee, H.J. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef]
- Ilg, A.; Beyer, P.; Al-Babili, S. Characterization of the rice carotenoid cleavage dioxygenase 1 reveals a novel route for geranial biosynthesis. FEBS J. 2009, 276, 736–747. [Google Scholar] [CrossRef] [PubMed]
- El Hadi, M.A.; Zhang, F.J.; Wu, F.F.; Zhou, C.H.; Tao, J. Advances in fruit aroma volatile research. Molecules 2013, 18, 8200–8229. [Google Scholar] [CrossRef]
- Ilg, A.; Bruno, M.; Beyer, P.; Al-Babili, S. Tomato carotenoid cleavage dioxygenases 1A and 1B: Relaxed double bond specificity leads to a plenitude of dialdehydes, mono-apocarotenoid and isoprenoid volatiles. FEBS Open Bio 2014, 4, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Baldwin, E.A.; Bai, J. Recent advance in aromatic volatile compounds research in tomato fruit: The metabolisms and regulations. Food Bioprocess Technol. 2016, 9, 203–216. [Google Scholar] [CrossRef]
- United States Standards for Grades of Fresh Tomatoes. United States: Department of Agriculture. Available online: https://www.ams.usda.Gov/grades-standards/tomato-grades-and-standards (accessed on 5 February 2018).
- Cheng, G.T.; Chang, P.P.; Wang, X.Y.; Lou, Q.Q.; El-Sappah, A.H.; Zhang, F.; Liang, Y. Effect of fruit color and ripeness on volatiles profiles in cherry tomato (Solanum Lycopersicum var. cerasiforme) fruit. Food Sci. 2021, 42, 173–183. [Google Scholar] [CrossRef]
- Bai, J.; Baldwin, E.A.; McCollum, G.; Plotto, A.; Manthey, J.A.; Widmer, W.W.; Luzio, G.; Cameron, R. Changes in volatile and non-volatile flavor chemicals of “Valencia” orange juice over the harvest seasons. Foods 2016, 5, 4. [Google Scholar] [CrossRef]
- Liu, T.; Zhu, W.; Huang, J.; Chen, H.H.; Nie, R.Z.; Li, C.M. Comparison of the nutritional as well as the volatile composition of inseason and off season Hezuo 903 tomato at the red stage. Eur. Food Res. Technol. 2017, 243, 203–214. [Google Scholar] [CrossRef]
- Zhang, B.; Tiemana, D.M.; Jiao, C.; Xu, Y.M.; Chen, K.S.; Fei, Z.J.; Giovannoni, J.J.; Klee, H.J. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. Proc. Natl. Acad. Sci. USA 2016, 113, 12580–12585. [Google Scholar] [CrossRef]
- Zhu, Y.Z.; Sims, C.A.; Klee, H.J.; Sarnoski, P.J. Sensory and flavor characteristics of tomato juice from garden gem and roma tomatoes with comparison to commercial tomato juice. J. Food Sci. 2018, 83, 153–161. [Google Scholar] [CrossRef]
- Zhang, K.; Yang, J.; Qiao, Z.; Cao, X.; Luo, Q.; Zhao, J.; Wang, F.; Zhang, W. Assessment of beta-glucans, phenols, flavor and volatile profiles of hulless barley wine originating from highland areas of China. Food Chem. 2019, 293, 32–40. [Google Scholar] [CrossRef]
- Liu, R.M.; Du, Z.H.; Zhang, Y.; Shi, Y.Y.; Chen, X.B.; Lin, L.; Xiong, Y.M.; Chen, M.J. Volatile component quantification in combination with putative gene expression analysis reveal key players in aroma formation during fruit ripening in Carica papaya cv ‘Hong fei’. Postharvest Biol. Tec. 2019, 158, 110987. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Hou, Y.L.; Meng, K.; Ye, H.; Ban, Q.Y.; Wang, B.; Suo, J.T.; Lv, J.Y.; Rao, J.P. The persimmon 9-lipoxygenase gene DkLOX3 plays positive roles in both promoting senescence and enhancing tolerance to abiotic stress. Front. Plant Sci. 2015, 6, 01073. [Google Scholar] [CrossRef]
- Yasmeen, A.; Mirza, B.; Inayatullah, S.; Safdar, N.; Jamil, M.; Ali, S.; Choudhry, M.F. In planta transformation of tomato. Plant Mol. Biol. Rep. 2009, 27, 20–28. [Google Scholar] [CrossRef]
- Li, C.; Yan, J.M.; Li, Y.Z.; Zhang, Z.C.; Wang, Q.L.; Liang, Y. Silencing the SpMPK1, SpMPK2, and SpMPK3 genes in tomato reduces abscisic acid—mediated drought tolerance. Int. J. Mol. Sci. 2013, 14, 21983–21996. [Google Scholar] [CrossRef]
- Yang, T.X.; Deng, L.; Zhao, W.; Zhang, R.X.; Jiang, H.L.; Ye, Z.B.; Li, C.B.; Li, C.Y. Rapid breeding of pink-fruited tomato hybrids using the CRISPR/Cas9 system. J. Genet. Genom. 2019, 46, 505–508. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, S.; Fan, W.; Duan, M.; Han, Y.; Li, H. Identification of volatile compounds and odour activity values in quinoa porridge by gas chromatography-mass spectrometry. J. Sci. Food. Agr. 2019, 99, 3957–3966. [Google Scholar] [CrossRef]
- Ronningen, I.; Miller, M.; Xia, Y.; Peterson, D.G. Identification and validation of sensory-active compounds from data-driven research: A flavoromics approach. J. Sci. Food. Agr. 2018, 66, 2473–2479. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.T.; Tran, T.L.H.; Hertog, M.; Picha, D.; Nicolai, B. Quality changes of tomato during fruit development and climacteric ripening. Eur. J. Hortic. Sci. 2017, 82, 319–325. [Google Scholar] [CrossRef]
- Casals, J.; Rivera, A.; Sabate, J.; del Castillo, R.R.; Simo, J. Cherry and fresh market tomatoes: Differences in chemical, morphological, and sensory traits and their implications for consumer acceptance. Agronomy 2019, 9, 9. [Google Scholar] [CrossRef]
- Mele, M.A.; Kang, H.M.; Lee, Y.T.; Islam, M.Z. Grape terpenoids: Flavor importance, genetic regulation, and future potential. Crit. Rev. Food Sci. 2020, 61, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Simkin, A.J.; Schwartz, S.H.; Auldridge, M.; Taylor, M.G.; Klee, H.J. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 2004, 40, 882–892. [Google Scholar] [CrossRef]
- Ilahy, R.; Tlili, I.; Siddiqui, M.W.; Hdider, C.; Lenucci, M.S. Inside and beyond color: Comparative overview of functional quality of tomato and watermelon fruits. Front. Plant Sci. 2019, 10, 769. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Zorrilla-Lopez, U.; Farre, G.; Zhu, C.F.; Sandmann, G.; Twyman, R.M.; Capell, T.; Christou, P. Nutritionally important carotenoid as consumer products. Phytochem. Rev. 2015, 4, 727–743. [Google Scholar] [CrossRef]
- Wei, Y.P.; Wan, H.J.; Wu, Z.M.; Wang, R.Q.; Ruan, M.Y.; Ye, Q.J.; Li, Z.M.; Zhou, G.Z.; Yao, Z.P.; Yang, Y.J. A comprehensive analysis of carotenoid cleavage dioxygenases. Plant Mol. Biol. Rep. 2016, 34, 512–523. [Google Scholar] [CrossRef]
- Cruz, A.B.; Bianchetti, R.E.; Alves, F.R.R.; Purgatto, E.; Peres, L.E.P.; Rossi, M.; Freschi, L. Light, ethylene and auxin signaling interaction regulates carotenoid biosynthesis during tomato fruits ripening. Front. Plant Sci. 2018, 9, 1370. [Google Scholar] [CrossRef]
- Wu, Q.; Tao, X.; Ai, X.; Luo, Z.S.; Mao, L.C.; Ying, T.J.; Li, L. Effect of exogenous auxin on aroma volatiles of cherry tomato (Solanum lycopersicum L.) fruit during postharvest ripening. Postharvest Biol. Technol. 2018, 146, 108–116. [Google Scholar] [CrossRef]
- Han, Y.; Wang, H.; Wang, X.; Li, K.; Dong, M.; Li, Y.; Zhu, Q.; Shang, F.D. Mechanism of floral scent production in osmanthus fragrans and the production and regulation of its key floral constituents, β-ionone and linalool. Hortic. Res. 2019, 6, 106. [Google Scholar] [CrossRef]
- Scherzinger, D.; Scheffer, E.; Bar, C.; Ernst, H.; Al-Babili, S. The Mycobacterium tuberculosis ORF Rv0654 encodes a carotenoid oxygenase mediating central and excentric cleavage of conventional and aromatic carotenoid. FEBS J. 2010, 277, 4662–4673. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.D.; McCarty, D.R. VP14 of maize catalyzes the carotenoid cleavage reaction of abscisic acid biosynthesis. Plant Physiol. 1997, 114, S798. [Google Scholar]
- Priya, R.; Sneha, P.; Dass, J.F.P.; Doss, C.G.; Manickavasagam, M.; Siva, R. Exploring the codon patterns between CCD and NCED genes among different plant species. Comput. Biol. Med. 2019, 114, 103449. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Mccarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant Physiol. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney, S.C.; Mila, I.; Bouzayen, M.; Magallanes, L.M.; Dellapenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Bertoni, G. Maize viviparous14: Structure meets function. Plant Cell 2010, 22, 2925. [Google Scholar] [CrossRef][Green Version]
- Messing, S.A.; Gabelli, S.B.; Echeverria, I.; Vogel, J.T.; Guan, J.C.; Tan, B.C.; Klee, H.J.; McCarty, D.R.; Amzel, L.M. Structural insights into maize viviparous14, a key enzyme in the biosynthesis of the phytohormone abscisic acid. Plant Cell 2010, 22, 2970–2980. [Google Scholar] [CrossRef]
- Priya, R.; Dass, J.F.P.; Siva, R. Gene expression prediction and hierarchical clustering analysis of plant CCD genes. Plant Mol. Biol. Rep. 2016, 34, 618–627. [Google Scholar] [CrossRef]
- Sharma, D.; Koul, A.; Kaul, S.; Dhar, M.K. Tissue-specific transcriptional regulation and metabolite accumulation in tomato (Solanum lycopersicum L.). Protoplasma 2020, 257, 1093–1108. [Google Scholar] [CrossRef]
- Nawade, B.; Shaltiel-Harpaz, L.; Yahyaa, M.; Bosamia, T.C.; Kabaha, A.; Kedoshim, R.; Zohar, M.; Isaacson, T.; Ibdah, M. Analysis of apocarotenoid volatiles during the development of Ficus carica fruits and characterization of carotenoid cleavage dioxygenase genes. Plant Sci. 2020, 290, 110292. [Google Scholar] [CrossRef]


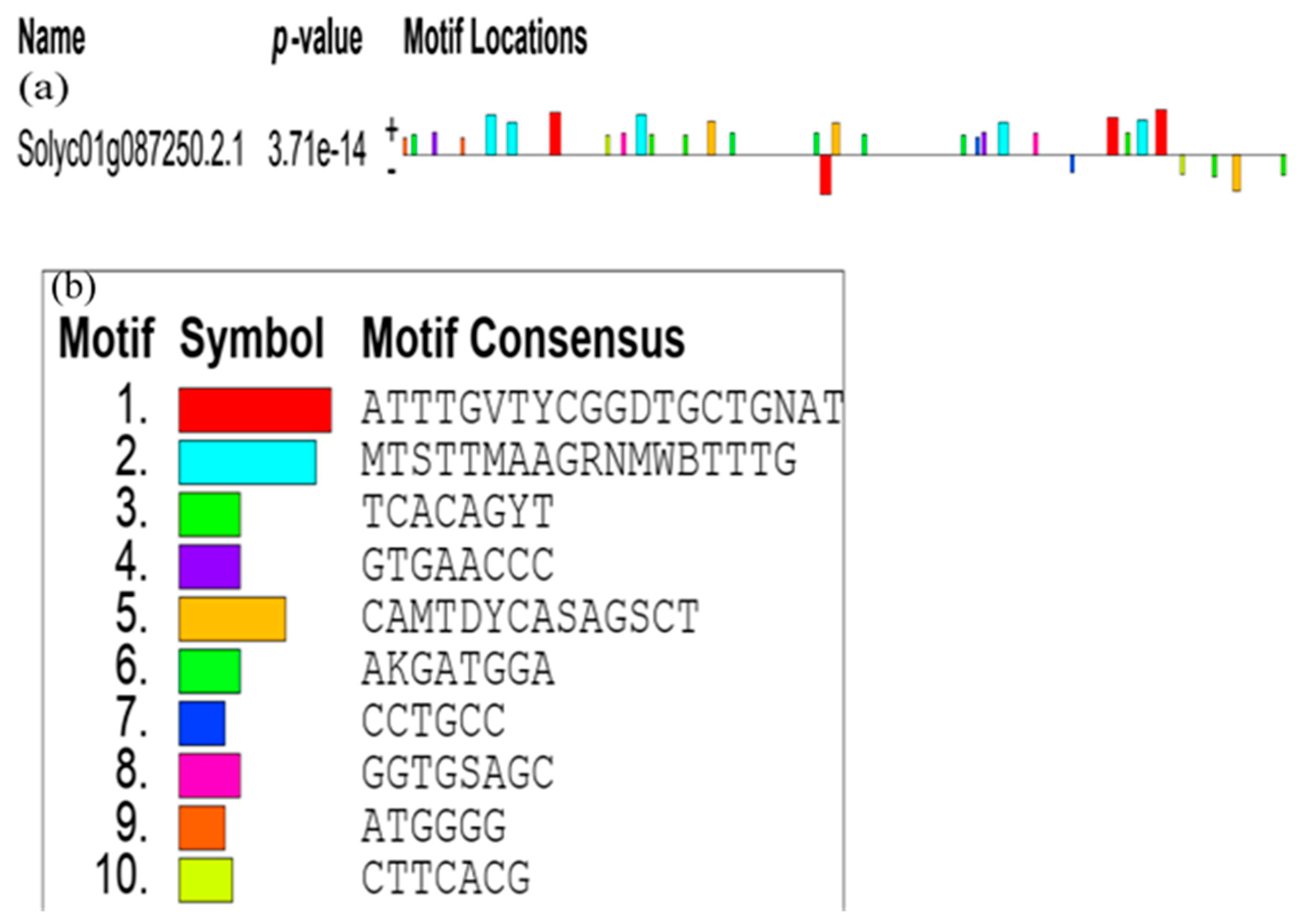

| Matrix ID | Name | Family | Score | Relative Score | Strand | Predicted Sequence | |
|---|---|---|---|---|---|---|---|
| SlCCD1A and SlCCD1B | MA0020.1 | Dof2 | Dof-type | 8.76 | 1 | − a | aaagca |
| MA0053.1 | MNB1A | Dof-type | 8.11 | 1 | − | aaagc | |
| MA0063.1 | Nkx2-5 | NK-related factors | 9 | 0.99 | − | ataattg | |
| MA0067.1 | Pax2 | Paired domain only | 8.36 | 0.97 | − | tgtcatgc | |
| MA0095.1 | YY1 | More than 3 adjacent | 7.39 | 0.95 | − | tccatc | |
| zinc finger factors | |||||||
| MA0102.3 | CEBPA | C/EBP-related factors | 12.06 | 0.94 | − | atttcatcaca | |
| MA0049.1 | hb | Factors with multiple | 10.71 | 0.93 | − | caacaaaaaa | |
| dispersed zinc fingers | |||||||
| MA0046.2 | HNF1A | POU domain factors | 14.6 | 0.91 | + b | aactaataatttaca | |
| MA0108.1 | TBP | TBP-related factors | 10.96 | 0.9 | + | gtataaaattgggag | |
| MA0100.2 | Myb | Myb/SANT domain factors | 7.75 | 0.88 | - | atagctgaca | |
| SlCCD1A | MA0035.1 | Gata1 | GATA-type zinc fingers | 6.92 | 0.99 | + | agatgg |
| SlCCD1B | MA0036.1 | GATA2 | GATA-type zinc fingers | 6.65 | 1 | − | ggata |
| MA0064.1 | PBF | Dof-type | 8.06 | 1 | − | aaagc | |
| MA0024.2 | E2F1 | E2F-related factors | 7.29 | 0.88 | − | cgagcgggaat | |
| MA0050.1 | IRF1 | Interferon-regulatory factors | 11.77 | 0.88 | − | taaaatgaaact |
| Volatiles | Mature Green | Turning | Orange | Red | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | OE-3 | OE-8 | OE-11 | WT | OE-3 | OE-8 | OE-11 | WT | OE-3 | OE-8 | OE-11 | WT | OE-3 | OE-8 | OE-11 | |
| (E)-citral | — | — | — | — | 6.31 | 22.14 * | 21.35 * | 21.66 * | 42.32 | 84.44 * | 70.12 * | 78.55 * | 33.09 | 105.36 * | 95.3 * | 106.34 * |
| β-cyclocitral | — | — | — | — | 7.28 | 23.16 * | 20.18 * | 22.03 * | 13.25 | 46.38 * | 34.65 * | 44.38 * | 9.14 | 66.13 * | 55.24 * | 58.72 * |
| 6-methyl-5-hepten-2-ol | 1.77 | 4.11 * | 3.86 * | 3.94 * | 1.46 | 16.98 * | 15.32 * | 15.69 * | 5.68 | 33.17 * | 28.17 * | 30.57 * | 6.35 | 44.28 * | 38.17 * | 40.42 * |
| 6-methyl-5-hepten-2-one | 8.71 | 10.69 | 9.25 | 9.87 | 45.86 | 108.14 * | 101.6 * | 103.12 * | 139.47 | 357.13 * | 348.19 * | 348.78 * | 179.15 | 502.32 ** | 478.37 ** | 489.32 ** |
| geranylacetone | 3.06 | 8.24 * | 7.54 * | 8.03 * | 15.53 | 64.16 * | 62.18 * | 63.94 * | 59.08 | 138.29 * | 112.13 * | 115.18 * | 77.54 | 213.47 ** | 190.45 ** | 203.69 ** |
| (E)-á-ionone | — | — | — | — | 18.68 | 45.76 * | 42.17 * | 42.66 * | 20.36 | 63.19 * | 56.88 * | 59.11 * | 32.48 | 112.94 * | 96.42 * | 110.35 * |
| β-ionone | — | — | — | — | 4.61 | 38.19 * | 36.23 * | 37.03 * | 5.4 | 67.65 ** | 50.65 ** | 56.73 ** | 11.23 | 135.54 * | 130.23 * | 130.69 * |
| geraniol | — | — | — | — | 12.04 | 35.63 * | 33.19 * | 33.69 * | 13.16 | 67.18 * | 65.89 * | 63 * | 32.07 | 87.27 * | 80.38 * | 85.33 * |
| 3,7-dimethyl-6-octen-1-ol | — | 4.36 | 3.18 | 3.24 | 9.28 | 35.36 * | 30.79 * | 32.11 * | 17.24 | 52.17 * | 47.88 * | 53.24 * | ||||
| neral | 2.1 | 3.6 | 3.2 | 3.3 | 7.16 | 35.66 * | 25.63 * | 31.18 * | 20.17 | 65.47 * | 56.24 * | 58.65 * | 34.15 | 92.88 * | 87.36 * | 90.43 * |
| acrylacetaldehyde | — | — | — | — | 4.28 | 5.69 * | 5.23 * | 5.44 * | 18.03 | 37.63 * | 26.69 * | 28.63 * | 18.01 | 82.14 * | 76.22 * | 79.38 * |
| (E)-farnesal | — | — | — | — | — | 5.63 | 3.16 | 5.01 | 9.42 | 54.16 * | 50.98 * | 52.19 * | 22.67 | 83.31 * | 79.44 * | 81.34 * |
| hexahydropseudoionone | — | — | — | — | — | 14.46 | 12.01 | 13.06 | — | 28.17 | 20.18 | 25.77 | 20.15 | 55.42 * | 50.16 * | 51.07 * |
| pseudoionone | — | — | — | — | 3.17 | 27.15 * | 23.05 * | 23.96 * | 18.06 | 64.69 * | 53.84 * | 56.69 * | 34.77 | 45.86 * | 46.12 * | 45.25 * |
| farnesyl acetone | 0.47 | 3.69 * | 3.44 * | 3.58 * | 5.27 | 48.36 * | 42.36 * | 45.23 * | 34.17 | 62.65 * | 54.89 * | 60.45 * | 48.94 | 75.17 * | 73.99 * | 76.45 * |
| Volatiles | Turning | Orange | Red | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | OE-2 | OE-3 | OE-6 | WT | OE-2 | OE-3 | OE-6 | WT | OE-2 | OE-3 | OE-6 | |
| 6-methyl-5-hepten-2-one | 6.45 | 28.36 * | 24.18 * | 27.48 * | 7.27 | 23.44 * | 18.16 * | 22.16 * | 7.7 | 30.11 * | 20.43 * | 32.08 * |
| geranylacetone | 2.82 | 15.33 * | 12.16 * | 14.36 * | 3.64 | 26.63 * | 20.14 * | 25.63 * | 0.86 | 21.14 * | 18.55 * | 20.47 * |
| (E)-á-ionone | — | 12.36 | 9.16 | 11.98 | 2.06 | 10.25 * | 8.23 * | 9.65 * | — | 9.36 | 6.23 | 8.96 |
| β-ionone | — | 7.24 | 5.62 | 6.89 | — | 9.43 | 7.93 | 8.74 | — | — | — | — |
| geraniol | — | 2.44 | 1.36 | 2.14 | — | 1.12 | 0.71 | 0.97 | — | 0.69 | 0.55 | 0.72 |
| neral | — | 1.84 | 1.56 | 1.74 | — | 0.93 | 0.63 | 0.86 | — | — | — | — |
| pseudoionone | — | 2.18 | 1.69 | 2.03 | — | 1.33 | 0.86 | 1.23 | — | — | — | — |
| Volatiles | Mature Green | Turning | Orange | Red | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | RNAi-2 | RNAi-7 | RNAi-10 | WT | RNAi-2 | RNAi-7 | RNAi-10 | WT | RNAi-2 | RNAi-7 | RNAi-10 | WT | RNAi-2 | RNAi-7 | RNAi-10 | |
| (E)-citral | — | — | — | — | 6.27 | 2.74 | 2.88 | 3.21 | 43.16 | 34.69 | 36.12 | 35.01 | 32.54 | 19.06 * | 18.98 * | 20.15 * |
| β-cyclocitral | — | — | — | — | 7.32 | 2.89 | 3.02 | 3.16 | 12.44 | 3.87 * | 4.02 * | 4.29 * | 8.16 | 2.78 | 3.22 | 3.18 |
| 6-methyl-5-hepten-2-ol | 1.56 | — | — | — | 1.48 | — | — | — | 5.35 | 1.3 | 1.24 | 1.33 | 6.77 | 3.04 | 2.15 | 2.47 |
| 6-methyl-5-hepten-2-one | 8.63 | 2.88 * | 3.05 * | 3.17 * | 46.23 | 8.97 * | 9.23 * | 10.65 * | 137.66 | 31.77 * | 32.17 * | 33.17 * | 176.22 | 43.18 ** | 44.72 ** | 48.5 * |
| geranylacetone | 3.28 | — | — | — | 15.34 | 2.77 * | 2.87 * | 3.14 * | 58.44 | 11.26 * | 11.57 * | 12.33 * | 76.12 | 16.45 * | 17.35 * | 18.31 * |
| (E)-á-ionone | — | — | — | — | 18.92 | 4.57 * | 4.64 * | 4.26 * | 22.17 | 2.55 * | 2.65 * | 2.88 * | 30.09 | 5.15 * | 5.7 * | 6.1 * |
| β-ionone | — | — | — | — | 4.94 | 1.63 | 1.88 | 2.1 | 5.61 | — | — | — | 13.05 | 2.99 * | 3.04 * | 3.17 * |
| geraniol | — | — | — | — | 12.88 | — | — | 1.47 * | 12.47 | 2.04 | 1.97 | 2.18 | 31.56 | 5.02 * | 5.34 * | 5.39 * |
| 3,7-dimethyl-6-octen-1-ol | — | — | — | — | — | — | — | — | 9.56 | 2.06 | 2.04 | 2.22c | 16.88 | 3.14 | 3.22 | 3.16 |
| neral | 2.4 | — | — | — | 7.39 | 1.68 | 1.96 | 2.16 | 19.16 | 3.19 | 3.18 | 3.24c | 33.22 | 7.35 | 7.88 | 7.96 |
| acrylacetaldehyde | — | — | — | — | 5.26 | — | — | — | 18.23 | 3.14 | 3.22 | 3.18c | 17.66 | 8.04 | 8.35 | 9.2 |
| (E)-farnesal | — | — | — | — | — | — | — | — | 9.68 | 7.73 | 7.88 | 8.16b | 21.35 | 2.35 | 2.98 | 3.6c |
| hexahydropseudoionone | — | — | — | — | 0.69 | — | — | — | — | — | — | — | 18.44 | 4.5 | 4.65 | 4.26 |
| pseudoionone | — | — | — | — | 3.41 | 1.75 | 1.86 | 2.19 | 17.54 | 6.8 | 7.1 | 7.06 | 33.83 | 9.56 * | 9.77 * | 10.19 * |
| farnesyl acetone | 0.55 | — | — | — | 6.18 | 1.98 | 2.04 | 2.36 | 33.18 | 8.33 * | 8.54 * | 8.63 * | 45.14 | 5.34 * | 5.57 * | 5.83 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, G.-T.; Li, Y.-S.; Qi, S.-M.; Wang, J.; Zhao, P.; Lou, Q.-Q.; Wang, Y.-F.; Zhang, X.-Q.; Liang, Y. SlCCD1A Enhances the Aroma Quality of Tomato Fruits by Promoting the Synthesis of Carotenoid-Derived Volatiles. Foods 2021, 10, 2678. https://doi.org/10.3390/foods10112678
Cheng G-T, Li Y-S, Qi S-M, Wang J, Zhao P, Lou Q-Q, Wang Y-F, Zhang X-Q, Liang Y. SlCCD1A Enhances the Aroma Quality of Tomato Fruits by Promoting the Synthesis of Carotenoid-Derived Volatiles. Foods. 2021; 10(11):2678. https://doi.org/10.3390/foods10112678
Chicago/Turabian StyleCheng, Guo-Ting, Yu-Shun Li, Shi-Ming Qi, Jin Wang, Pan Zhao, Qian-Qi Lou, Yan-Feng Wang, Xiang-Qian Zhang, and Yan Liang. 2021. "SlCCD1A Enhances the Aroma Quality of Tomato Fruits by Promoting the Synthesis of Carotenoid-Derived Volatiles" Foods 10, no. 11: 2678. https://doi.org/10.3390/foods10112678
APA StyleCheng, G.-T., Li, Y.-S., Qi, S.-M., Wang, J., Zhao, P., Lou, Q.-Q., Wang, Y.-F., Zhang, X.-Q., & Liang, Y. (2021). SlCCD1A Enhances the Aroma Quality of Tomato Fruits by Promoting the Synthesis of Carotenoid-Derived Volatiles. Foods, 10(11), 2678. https://doi.org/10.3390/foods10112678






