[Pd(dach)Cl2] Complex Targets Proteins Involved in Ribosomal Biogenesis, and RNA Splicing in HeLa Cells
Abstract
1. Introduction
2. Results
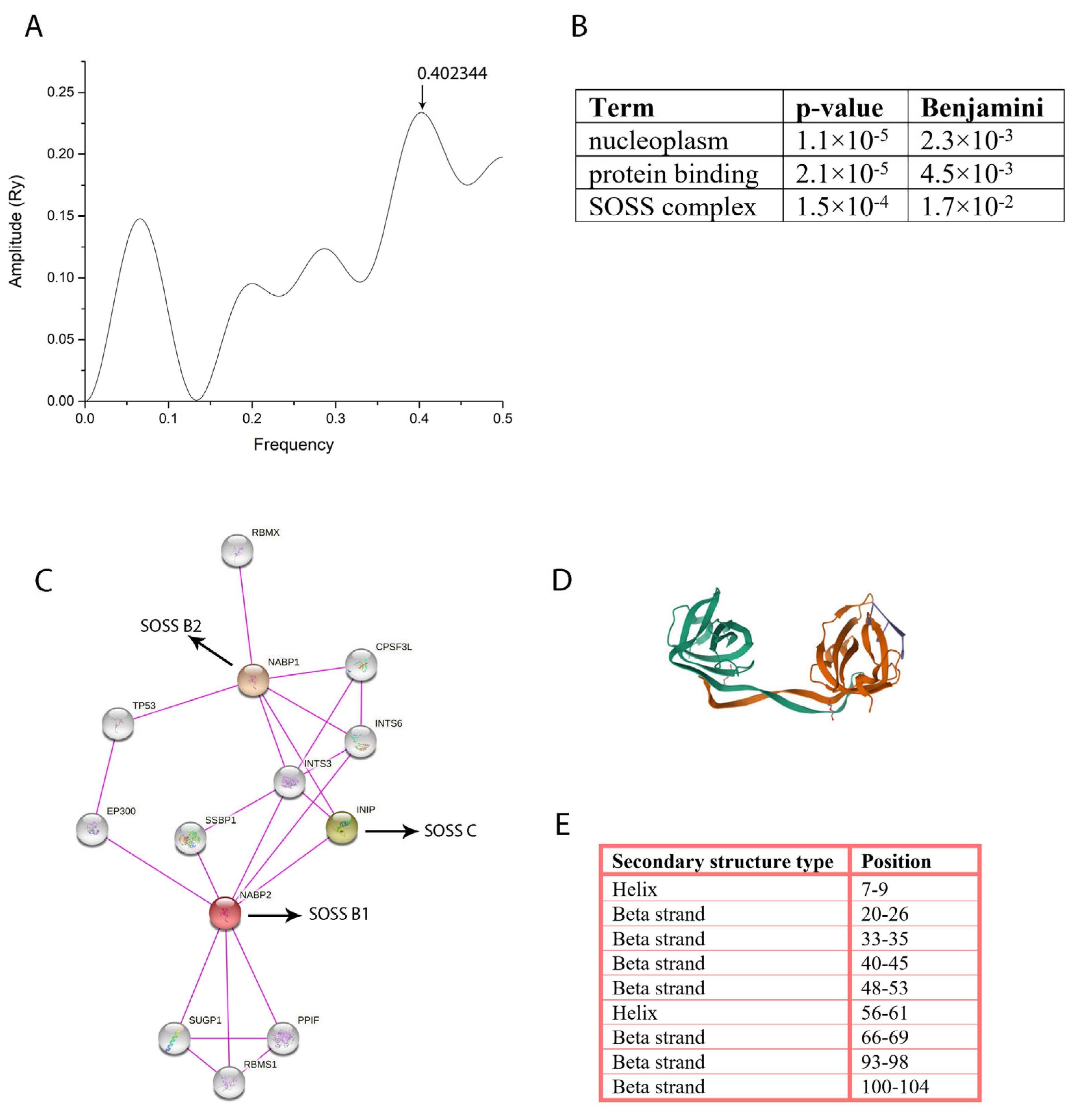
2.1. Predicting the Protein Targets for [Pd(dach)Cl2] by Computational Biology
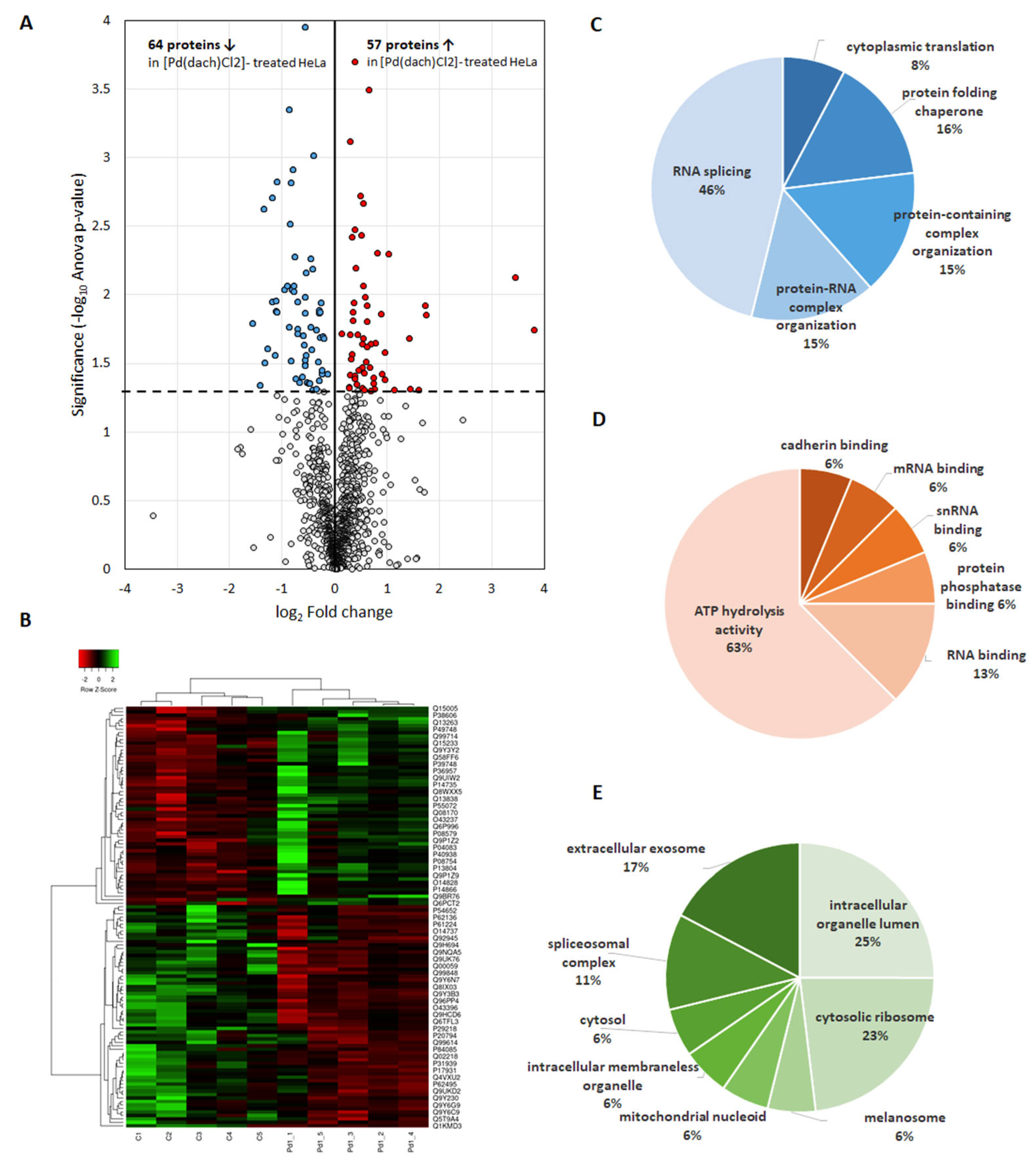
2.2. Differentially Abundant Proteins as a Result of [Pd(dach)Cl2] Treatment
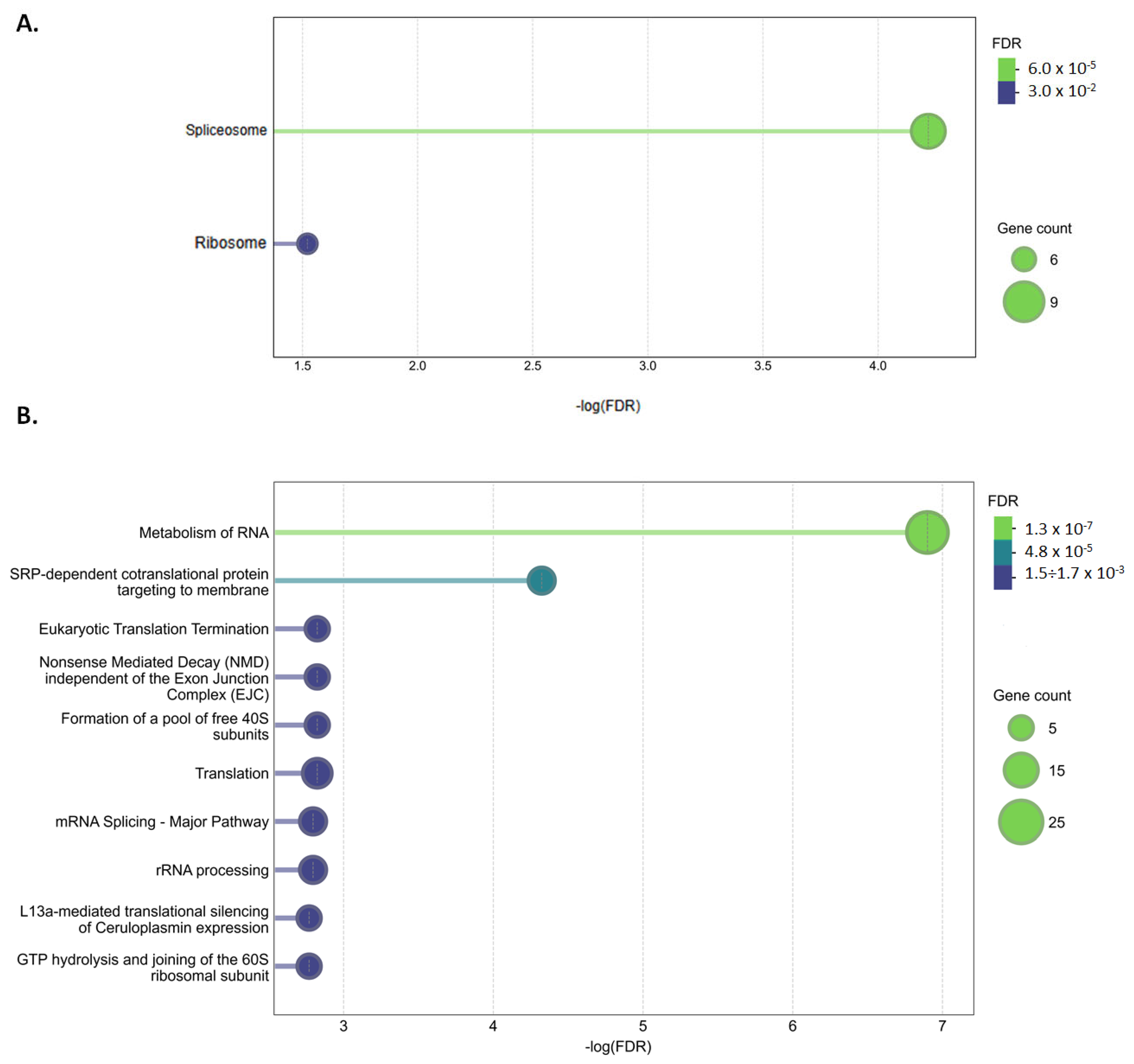
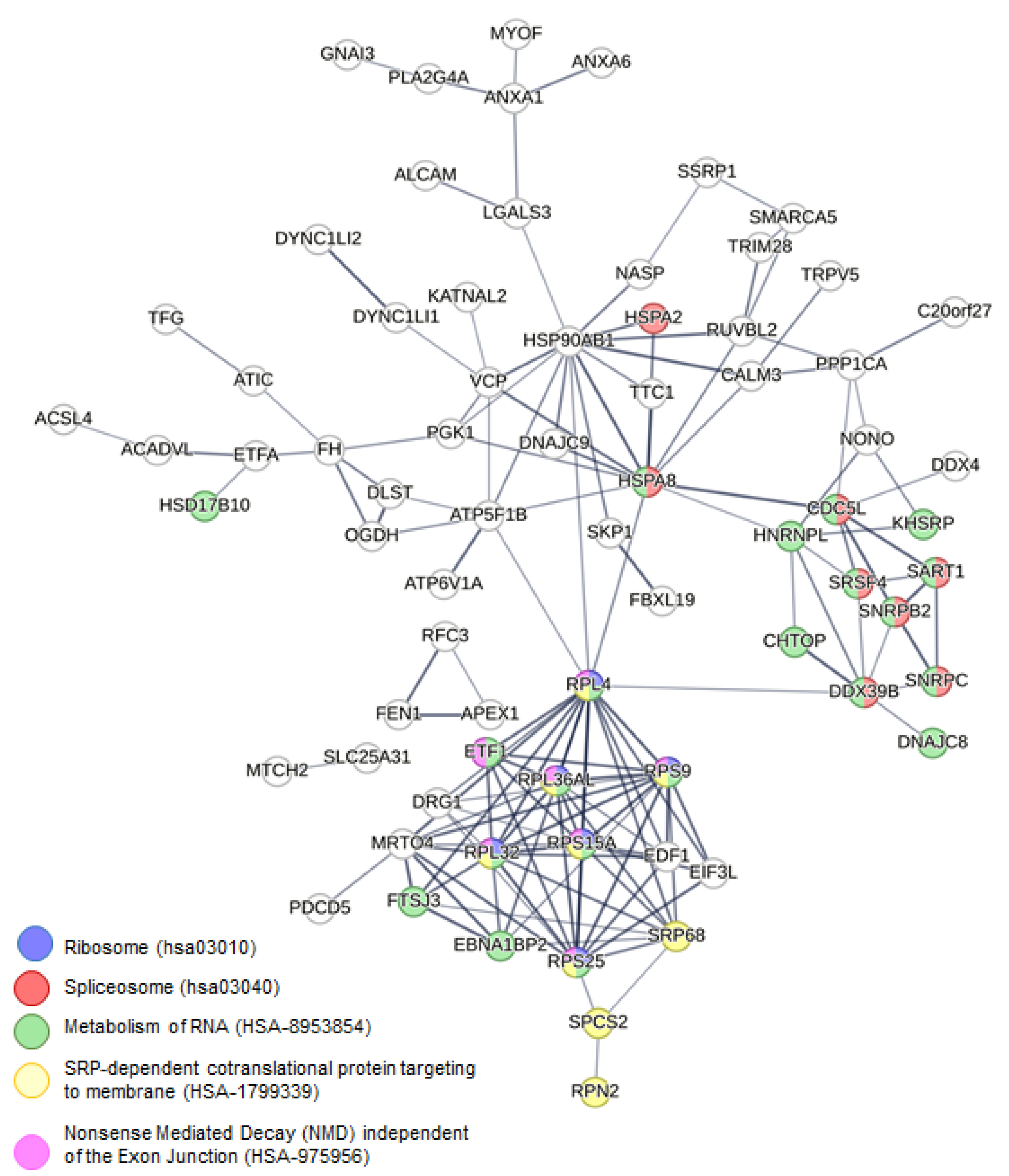
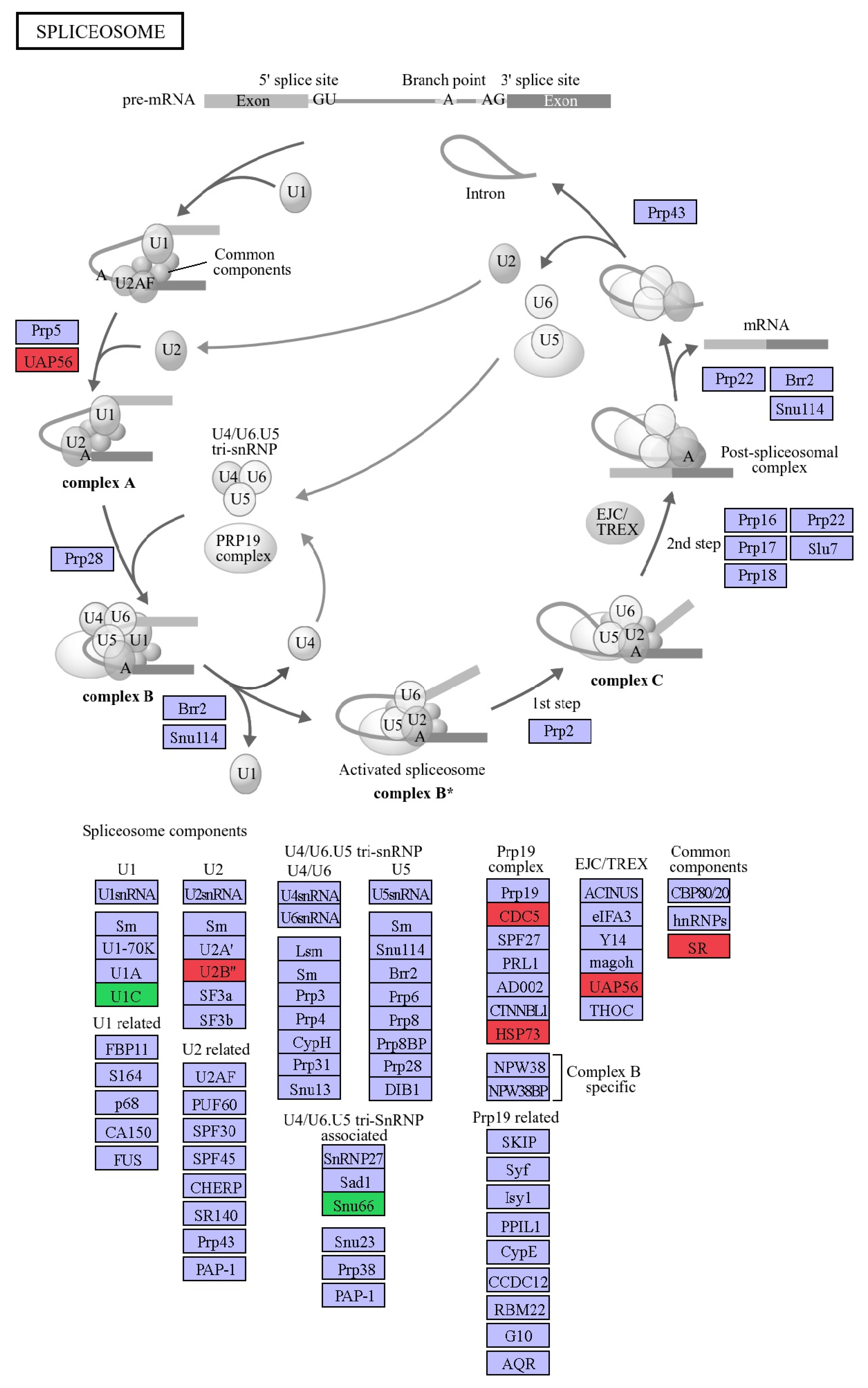
2.3. Altered Pathways in HeLa Cells Due to [Pd(dach)Cl2] Treatment
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Cell Culture
4.3. Sample Preparation
4.4. LC-MS/MS Acquisition
4.5. MS Data Processing and Identification
4.6. Proteomics Data Analysis
4.7. Computational Biology Analysis of the [Pd(dach)Cl2] Interaction with HeLa Cellular Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAPs | Differentially abundant proteins |
| DNA | Desoxyribonucleic acid |
| EJC | Exon Junction Complex |
| GO | Gene Ontology |
| HSP | Heat Shock Protein |
| ISM | Informational spectrum method |
| LC-MS/MS | Liquid Chromatography-Tandem mass spectrometry |
| MCC | Maximal Clique Centrality |
| NDM | Nonsense Mediated Decay |
| PPI | Protein–protein interaction |
| RNA | Ribonucleic Acid |
| snRNPs | Small Nuclear Ribonucleoproteins |
| SOSS DNA | Sensor of the single-stranded DNA |
| TP53 | Tumor Suppressor Protein p53 |
References
- Fowler, J.R.; Maani, E.V.; Dunton, C.J.; Gasalberti, D.P.; Jack, B.W. Cervical Cancer. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Arbyn, M.; Weiderpass, E.; Bruni, L.; De Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [PubMed]
- Ralić, V.; Nešić, M.D.; Dučić, T.; Stepić, M.; Korićanac, L.; Davalieva, K.; Petković, M. Analysis of Biomolecular Changes in HeLa Cervical Cancer Cell Line Induced by Interaction with [Pd(dach)Cl2]. Inorganics 2025, 13, 20. [Google Scholar] [CrossRef]
- Yodoshi, M.; Okabe, N. Structures and Interaction with DNA of Ternary Palladium(II) Complexes: [Pd(Gly)(X)] (Gly=Glycine; X=2,2′-Bipyridine, 1,10-Phenanthroline and 2,2′-Bi-pyridylamine). Chem. Pharm. Bull. 2008, 56, 908–914. [Google Scholar] [CrossRef][Green Version]
- Gao, E.; Liu, F.; Zhu, M.; Wang, L.; Huang, Y.; Liu, H.; Ma, S.; Shi, Q.; Wang, N. Synthesis, characterization, DNA interaction, and cytotoxicity of novel Pd(II) and Pt(II) complexes. J. Enzyme Inhib. Med. Chem. 2010, 25, 557–564. [Google Scholar] [CrossRef]
- Hobart, D.B.; Berg, M.A.G.; Merola, J.S. Bis-glycinato complexes of palladium(II): Synthesis, structural determination, and hydrogen bonding interactions. Inorganica Chim. Acta 2014, 423, 21–30. [Google Scholar] [CrossRef]
- Hobart, D.B.; Berg, M.A.G.; Rogers, H.M.; Merola, J.S. Synthesis, Characterization, and Non-Covalent Interactions of Palladium(II)-Amino Acid Complexes. Molecules 2021, 26, 4331. [Google Scholar] [CrossRef]
- Hobart, D.B.; Merola, J.S.; Rogers, H.M.; Sahgal, S.; Mitchell, J.; Florio, J.; Merola, J.W. Synthesis, Structure, and Catalytic Reactivity of Pd(II) Complexes of Proline and Proline Homologs. Catalysts 2019, 9, 515. [Google Scholar] [CrossRef]
- Wang, Y.; Chiu, J.-F. Proteomic Approaches in Understanding Action Mechanisms of Metal-Based Anticancer Drugs. Met.-Based Drugs 2008, 2008, 716329. [Google Scholar] [CrossRef]
- Fotis, C.; Antoranz, A.; Hatziavramidis, D.; Sakellaropoulos, T.; Alexopoulos, L.G. Network-based technologies for early drug discovery. Drug Discov. Today 2018, 23, 626–635. [Google Scholar] [CrossRef]
- Nešić, M.D.; Dučić, T.; Gonçalves, M.; Stepić, M.; Algarra, M.; Soto, J.; Gemović, B.; Bandosz, T.J.; Petković, M. Biochemical changes in cancer cells induced by photoactive nanosystem based on carbon dots loaded with Ru-complex. Chem. Biol. Interact. 2022, 360, 109950. [Google Scholar] [CrossRef]
- Sencanski, M.; Perovic, V.; Pajovic, S.B.; Adzic, M.; Paessler, S.; Glisic, S. Drug Repurposing for Candidate SARS-CoV-2 Main Protease Inhibitors by a Novel In Silico Method. Molecules 2020, 25, 3830. [Google Scholar] [CrossRef] [PubMed]
- Veljkovic, N.; Glisic, S.; Prljic, J.; Perovic, V.; Botta, M.; Veljkovic, V. Discovery of New Therapeutic Targets by the Informational Spectrum Method. Curr. Protein Pept. Sci. 2008, 9, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Veljkovic, V. A Theoretical Approach to the Preselection of Carcinogens and Chemical Carcinogenesis; Gordon and Breach Science Publishers: New York, NY, USA, 1980. [Google Scholar]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Huang, J.; Gong, Z.; Ghosal, G.; Chen, J. SOSS Complexes Participate in the Maintenance of Genomic Stability. Mol. Cell 2009, 35, 384–393. [Google Scholar] [CrossRef]
- Ren, W.; Chen, H.; Sun, Q.; Tang, X.; Lim, S.C.; Huang, J.; Song, H. Structural Basis of SOSS1 Complex Assembly and Recognition of ssDNA. Cell Rep. 2014, 6, 982–991. [Google Scholar] [CrossRef]
- Li, Y.H.; Gao, Z.Q.; Dong, Y.H. Crystal structure of SSB from homo sapiens: 5d8e. 2016. Available online: https://www.rcsb.org/structure/5D8E (accessed on 1 June 2025).
- Zidovska, A. The self-stirred genome: Large-scale chromatin dynamics, its biophysical origins and implications. Curr. Opin. Genet. Dev. 2020, 61, 83–90. [Google Scholar] [CrossRef]
- Hanel, W.; Moll, U.M. Links between mutant p53 and genomic instability. J. Cell Biochem. 2012, 113, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Salz, T.; Zajac-Kaye, M.; Liao, D.; Huang, S.; Qiu, Y. Overexpression of histone deacetylases in cancer cells is controlled by interplay of transcription factors and epigenetic modulators. FASEB J. 2014, 28, 4265–4279. [Google Scholar] [CrossRef]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef]
- Bi, Y.; Kong, P.; Zhang, L.; Cui, H.; Xu, X.; Chang, F.; Yan, T.; Li, J.; Cheng, C.; Song, B.; et al. EP300 as an oncogene correlates with poor prognosis in esophageal squamous carcinoma. J. Cancer 2019, 10, 5413–5426. [Google Scholar] [CrossRef]
- Richard, D.J.; Bolderson, E.; Cubeddu, L.; Wadsworth, R.I.M.; Savage, K.; Sharma, G.G.; Nicolette, M.L.; Tsvetanov, S.; McIlwraith, M.J.; Pandita, R.K.; et al. Single-stranded DNA-binding protein hSSB1 is critical for genomic stability. Nature 2008, 453, 677–681. [Google Scholar] [CrossRef]
- Long, Q.; Sebesta, M.; Sedova, K.; Haluza, V.; Alagia, A.; Liu, Z.; Stefl, R.; Gullerova, M. The phosphorylated trimeric SOSS1 complex and RNA polymerase II trigger liquid-liquid phase separation at double-strand breaks. Cell Rep. 2023, 42, 113489. [Google Scholar] [CrossRef] [PubMed]
- Venables, J.P.; Klinck, R.; Koh, C.; Gervais-Bird, J.; Bramard, A.; Inkel, L.; Durand, M.; Couture, S.; Froehlich, U.; Lapointe, E.; et al. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 2009, 16, 670–676. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.Y.-T.; Simon, L.M.; Neill, N.J.; Marcotte, R.; Sayad, A.; Bland, C.S.; Echeverria, G.V.; Sun, T.; Kurley, S.J.; Tyagi, S.; et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015, 525, 384–388. [Google Scholar] [CrossRef]
- Higareda-Almaraz, J.C.; Valtierra-Gutiérrez, I.A.; Hernandez-Ortiz, M.; Contreras, S.; Hernandez, E.; Encarnacion, S. Analysis and Prediction of Pathways in HeLa Cells by Integrating Biological Levels of Organization with Systems-Biology Approaches. PLoS ONE 2013, 8, e65433. [Google Scholar] [CrossRef]
- Zhang, Y.; Shan, N.; Zhou, W.; Zhang, S. Identification of HSPA8 as a candidate biomarker for endometrial carcinoma by using iTRAQ-based proteomic analysis. OncoTargets Ther. 2016, 2016, 2169–2179. [Google Scholar] [CrossRef]
- Li, B.; Ming, H.; Qin, S.; Zhou, L.; Huang, Z.; Jin, P.; Peng, L.; Luo, M.; Zhang, T.; Wang, K.; et al. HSPA8 Activates Wnt/β-Catenin Signaling to Facilitate BRAF V600E Colorectal Cancer Progression by CMA-Mediated CAV1 Degradation. Adv. Sci. 2024, 11, 2306535. [Google Scholar] [CrossRef]
- Wu, J.; Lu, F.; Yu, B.; Wang, W.; Ye, X. The oncogenic role of SNRPB in human tumors: A pan-cancer analysis. Front. Mol. Biosci. 2022, 9, 994440. [Google Scholar] [CrossRef]
- Szymura, S.J.; Bernal, G.M.; Wu, L.; Zhang, Z.; Crawley, C.D.; Voce, D.J.; Campbell, P.-A.; Ranoa, D.E.; Weichselbaum, R.R.; Yamini, B. DDX39B interacts with the pattern recognition receptor pathway to inhibit NF-κB and sensitize to alkylating chemotherapy. BMC Biol. 2020, 18, 32. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Su, B.; Yu, P.; He, J.; Meng, L.; Xiao, Q.; Sun, J.; Zhou, K.; Xue, Y.; et al. Transcriptome-wide analysis and modelling of prognostic alternative splicing signatures in invasive breast cancer: A prospective clinical study. Sci. Rep. 2020, 10, 16504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; He, C.; Guo, X.; Fang, Y.; Lai, Q.; Wang, X.; Pan, X.; Li, H.; Qin, K.; Li, A.; et al. DDX39B contributes to the proliferation of colorectal cancer through direct binding to CDK6/CCND1. Cell Death Discov. 2022, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Nakata, D.; Nakao, S.; Nakayama, K.; Araki, S.; Nakayama, Y.; Aparicio, S.; Hara, T.; Nakanishi, A. The RNA helicase DDX39B and its paralog DDX39A regulate androgen receptor splice variant AR-V7 generation. Biochem. Biophys. Res. Commun. 2017, 483, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Meng, T.; Huang, R.; Zeng, Z.; Huang, Z.; Yin, H.; Jiao, C.; Yan, P.; Hu, P.; Zhu, X.; Li, Z.; et al. Identification of Prognostic and Metastatic Alternative Splicing Signatures in Kidney Renal Clear Cell Carcinoma. Front. Bioeng. Biotechnol. 2019, 7, 270. [Google Scholar] [CrossRef]
- Walbrecq, G.; Lecha, O.; Gaigneaux, A.; Fougeras, M.R.; Philippidou, D.; Margue, C.; Tetsi Nomigni, M.; Bernardin, F.; Dittmar, G.; Behrmann, I.; et al. Hypoxia-Induced Adaptations of miRNomes and Proteomes in Melanoma Cells and Their Secreted Extracellular Vesicles. Cancers 2020, 12, 692. [Google Scholar] [CrossRef]
- Turunen, J.J.; Niemelä, E.H.; Verma, B.; Frilander, M.J. The significant other: Splicing by the minor spliceosome. WIREs RNA 2013, 4, 61–76. [Google Scholar] [CrossRef]
- Alioto, T.S. U12DB: A database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007, 35, D110–D115. [Google Scholar] [CrossRef]
- Ye, Z.; Bing, A.; Zhao, S.; Yi, S.; Zhan, X. Comprehensive analysis of spliceosome genes and their mutants across 27 cancer types in 9070 patients: Clinically relevant outcomes in the context of 3P medicine. EPMA J. 2022, 13, 335–350. [Google Scholar] [CrossRef]
- Peng, Q.; Zhou, Y.; Oyang, L.; Wu, N.; Tang, Y.; Su, M.; Luo, X.; Wang, Y.; Sheng, X.; Ma, J.; et al. Impacts and mechanisms of alternative mRNA splicing in cancer metabolism, immune response, and therapeutics. Mol. Ther. 2022, 30, 1018–1035. [Google Scholar] [CrossRef]
- Kitamura, K.; Nimura, K. Regulation of RNA Splicing: Aberrant Splicing Regulation and Therapeutic Targets in Cancer. Cells 2021, 10, 923. [Google Scholar] [CrossRef]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Zisi, A.; Bartek, J.; Lindström, M.S. Targeting Ribosome Biogenesis in Cancer: Lessons Learned and Way Forward. Cancers 2022, 14, 2126. [Google Scholar] [CrossRef]
- Ramalho, S.; Dopler, A.; Faller, W.J. Ribosome specialization in cancer: A spotlight on ribosomal proteins. NAR Cancer 2024, 6, zcae029. [Google Scholar] [CrossRef] [PubMed]
- Trifa, Y.; Privat, I.; Gagnon, J.; Baeza, L.; Lerbs-Mache, S. The Nuclear RPL4 Gene Encodes a Chloroplast Protein That Co-purifies with the T7-like Transcription Complex as Well as Plastid Ribosomes. J. Biol. Chem. 1998, 273, 3980–3985. [Google Scholar] [CrossRef]
- He, X.; Li, Y.; Dai, M.-S.; Sun, X.-X. Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget 2016, 7, 16217–16226. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, W.; Wang, H.; Wang, M.-H.; Xu, W.; Zhang, R. Identification of ribosomal protein S25 (RPS25)–MDM2–p53 regulatory feedback loop. Oncogene 2013, 32, 2782–2791. [Google Scholar] [CrossRef]
- Xie, J.; Zhang, W.; Liang, X.; Shuai, C.; Zhou, Y.; Pan, H.; Yang, Y.; Han, W. RPL32 Promotes Lung Cancer Progression by Facilitating p53 Degradation. Mol. Ther.—Nucleic Acids 2020, 21, 75–85. [Google Scholar] [CrossRef]
- Xu, L.; Wang, L.; Jiang, C.; Zhu, Q.; Chen, R.; Wang, J.; Wang, S. Biological effect of ribosomal protein L32 on human breast cancer cell behavior. Mol. Med. Rep. 2020, 22, 2478–2486. [Google Scholar] [CrossRef]
- Xu, M.; Wang, Y.; Chen, L.; Pan, B.; Chen, F.; Fang, Y.; Yu, Z.; Chen, G. Down-regulation of ribosomal protein S15A mRNA with a short hairpin RNA inhibits human hepatic cancer cell growth in vitro. Gene 2014, 536, 84–89. [Google Scholar] [CrossRef]
- Chen, J.; Wei, Y.; Feng, Q.; Ren, L.; He, G.; Chang, W.; Zhu, D.; Yi, T.; Lin, Q.; Tang, W.; et al. Ribosomal protein S15A promotes malignant transformation and predicts poor outcome in colorectal cancer through misregulation of p53 signaling pathway. Int. J. Oncol. 2016, 48, 1628–1638. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Liu, J. RPS15a Silencing Suppresses Cell Proliferation and Migration of Gastric Cancer. Yonsei Med. J. 2018, 59, 1166. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Shuangshuang, D.; Liang, X.; Zhou, X. RPS9 promotes the progression of NSCLC via activation Stat3 and Erk signaling pathways. J. Cancer 2022, 13, 1346–1355. [Google Scholar] [CrossRef]
- Cheng, D.; Zhu, B.; Li, S.; Yuan, T.; Yang, Q.; Fan, C. Down-regulation of RPS9 Inhibits Osteosarcoma Cell Growth through Inactivation of MAPK Signaling Pathway. J. Cancer 2017, 8, 2720–2728. [Google Scholar] [CrossRef]
- Shu, G.; Chen, M.; Liao, W.; Fu, L.; Lin, M.; Gui, C.; Cen, J.; Lu, J.; Chen, Z.; Wei, J.; et al. PABPC1L Induces IDO1 to Promote Tryptophan Metabolism and Immune Suppression in Renal Cell Carcinoma. Cancer Res. 2024, 84, 1659–1679. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-Q.; Ju, C.-L.J.; Wang, B.-J.; Wang, R.-G. PABPC1L depletion inhibits proliferation and migration via blockage of AKT pathway in human colorectal cancer cells. Oncol. Lett. 2019, 17, 3439–3445. [Google Scholar] [CrossRef]
- An, T.; Deng, L.; Wang, Y.; Yang, Z.; Chai, C.; Ouyang, J.; Lu, X.; Zhang, C. The Prognostic Impacts of PABPC1 Expression on Gastric Cancer Patients. Future Oncol. 2021, 17, 4471–4479. [Google Scholar] [CrossRef]
- Castro, M.E.; Ferrer, I.; Cascon, A.; Guijarro, M.V.; Lleonart, M.; Cajal, S.R.Y.; Leal, J.F.M.; Robledo, M.; Carnero, A. PPP1CA contributes to the senescence program induced by oncogenic Ras. Carcinogenesis 2007, 29, 491–499. [Google Scholar] [CrossRef]
- Hu, A.; Hong, F.; Li, D.; Xie, Q.; Chen, K.; Zhu, L.; He, H. KDM3B-ETF1 fusion gene downregulates LMO2 via the WNT/β-catenin signaling pathway, promoting metastasis of invasive ductal carcinoma. Cancer Gene Ther. 2022, 29, 215–224. [Google Scholar] [CrossRef]
- Stoddart, A.; Qian, Z.; Fernald, A.A.; Bergerson, R.J.; Wang, J.; Karrison, T.; Anastasi, J.; Bartom, E.T.; Sarver, A.L.; McNerney, M.E.; et al. Retroviral insertional mutagenesis identifies the del(5q) genes, CXXC5, TIFAB and ETF1, as well as the Wnt pathway, as potential targets in del(5q) myeloid neoplasms. Haematologica 2016, 101, e232–e236. [Google Scholar] [CrossRef]
- Petrović, B.; Bugarčić, Ž.D.; Van Eldik, R. Kinetic studies on the reactions of [Pd(dach)(X–Y)] complexes with some DNA constituents. Dalton Trans. 2008, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Davalieva, K.; Rusevski, A.; Velkov, M.; Noveski, P.; Kubelka-Sabit, K.; Filipovski, V.; Plaseski, T.; Dimovski, A.; Plaseska-Karanfilska, D. Comparative proteomics analysis of human FFPE testicular tissues reveals new candidate biomarkers for distinction among azoospermia types and subtypes. J. Proteomics 2022, 267, 104686. [Google Scholar] [CrossRef] [PubMed]
- Distler, U.; Kuharev, J.; Navarro, P.; Levin, Y.; Schild, H.; Tenzer, S. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 2014, 11, 167–170. [Google Scholar] [CrossRef]
- Davalieva, K.; Kiprijanovska, S.; Dimovski, A.; Rosoklija, G.; Dwork, A.J. Comparative evaluation of two methods for LC-MS/MS proteomic analysis of formalin fixed and paraffin embedded tissues. J. Proteomics 2021, 235, 104117. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Chin, C.-H.; Chen, S.-H.; Wu, H.-H.; Ho, C.-W.; Ko, M.-T.; Lin, C.-Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Papatheodorou, I.; Moreno, P.; Manning, J.; Fuentes, A.M.-P.; George, N.; Fexova, S.; Fonseca, N.A.; Füllgrabe, A.; Green, M.; Huang, N.; et al. Expression Atlas update: From tissues to single cells. Nucleic Acids Res. 2019, 48, D77–D83. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ralić, V.; Davalieva, K.; Gemović, B.; Senćanski, M.; Nešić, M.D.; Žakula, J.; Stepić, M.; Petković, M. [Pd(dach)Cl2] Complex Targets Proteins Involved in Ribosomal Biogenesis, and RNA Splicing in HeLa Cells. Inorganics 2025, 13, 215. https://doi.org/10.3390/inorganics13070215
Ralić V, Davalieva K, Gemović B, Senćanski M, Nešić MD, Žakula J, Stepić M, Petković M. [Pd(dach)Cl2] Complex Targets Proteins Involved in Ribosomal Biogenesis, and RNA Splicing in HeLa Cells. Inorganics. 2025; 13(7):215. https://doi.org/10.3390/inorganics13070215
Chicago/Turabian StyleRalić, Vanja, Katarina Davalieva, Branislava Gemović, Milan Senćanski, Maja D. Nešić, Jelena Žakula, Milutin Stepić, and Marijana Petković. 2025. "[Pd(dach)Cl2] Complex Targets Proteins Involved in Ribosomal Biogenesis, and RNA Splicing in HeLa Cells" Inorganics 13, no. 7: 215. https://doi.org/10.3390/inorganics13070215
APA StyleRalić, V., Davalieva, K., Gemović, B., Senćanski, M., Nešić, M. D., Žakula, J., Stepić, M., & Petković, M. (2025). [Pd(dach)Cl2] Complex Targets Proteins Involved in Ribosomal Biogenesis, and RNA Splicing in HeLa Cells. Inorganics, 13(7), 215. https://doi.org/10.3390/inorganics13070215









