Anion Capture at the Open Core of a Geometrically Flexible Dicopper(II,II) Macrocycle Complex
Abstract
:1. Introduction
2. Results and Discussion
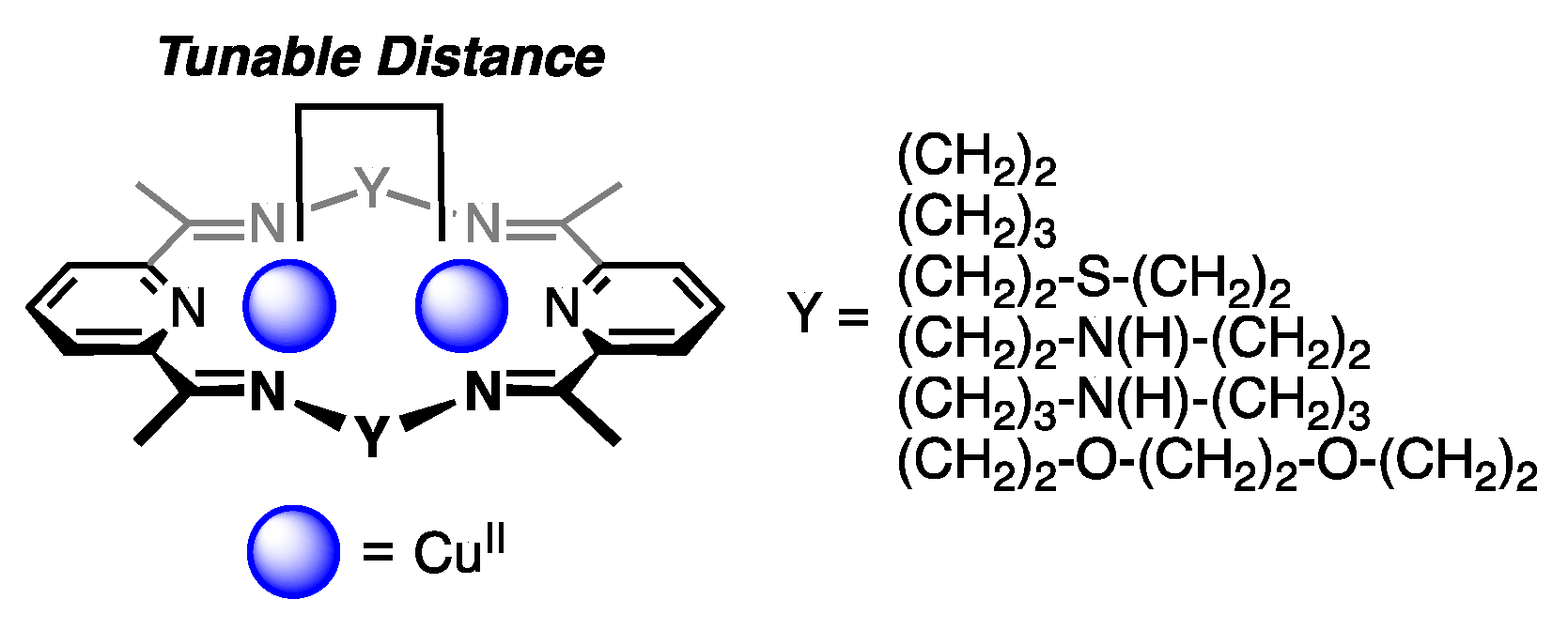
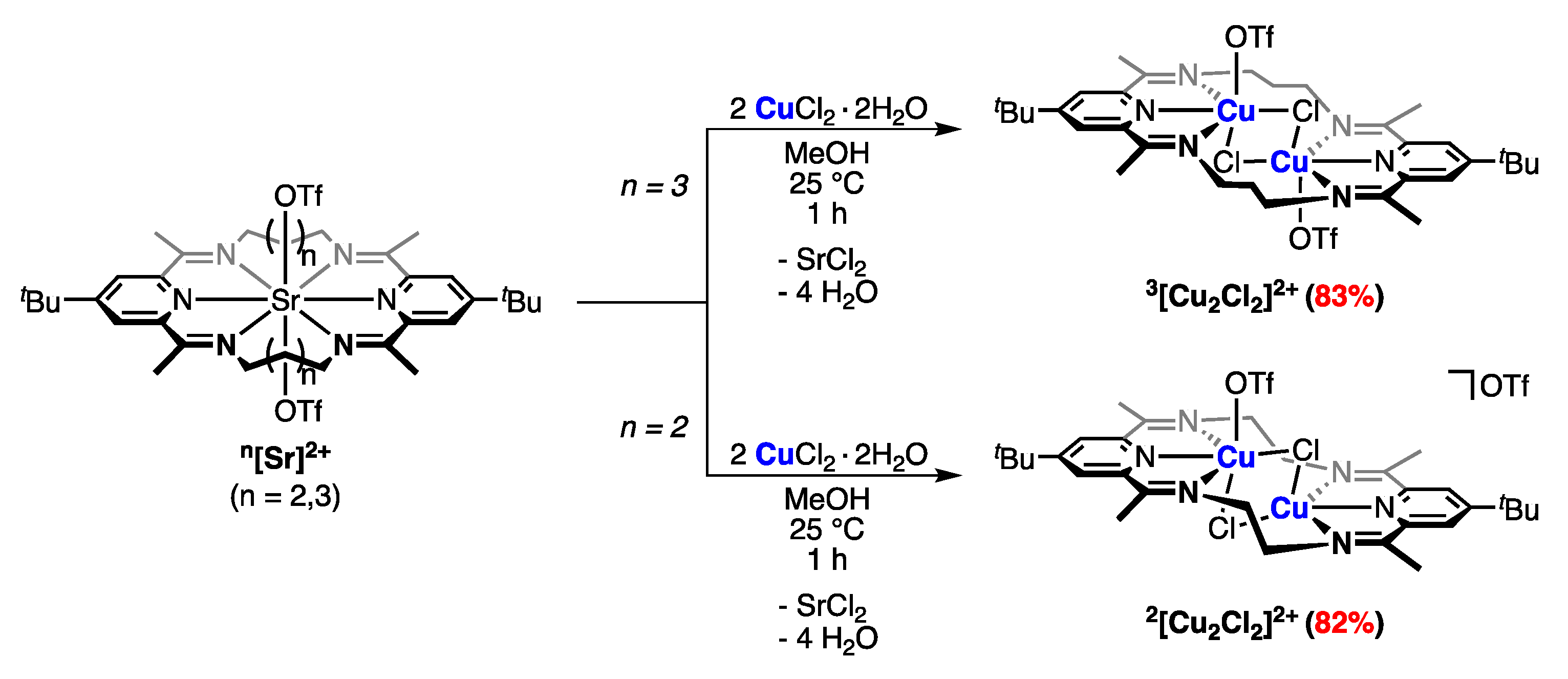
2.1. Ring-Size Modulated Structures of [Cu2Cl2]2+ Cluster Cores
2.2. Chloride Abstraction from 3[Cu2Cl2]2+
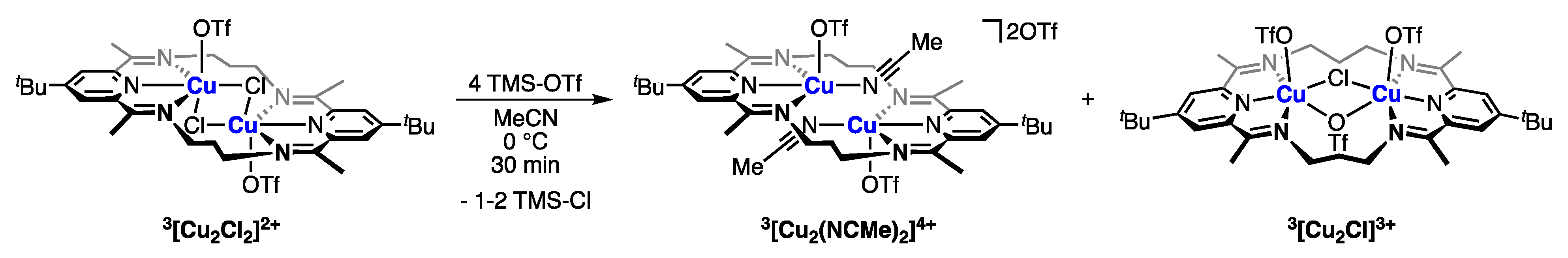
2.3. A Synthetic Route to an “Open-Core” Cu2(II,II) Macrocycle
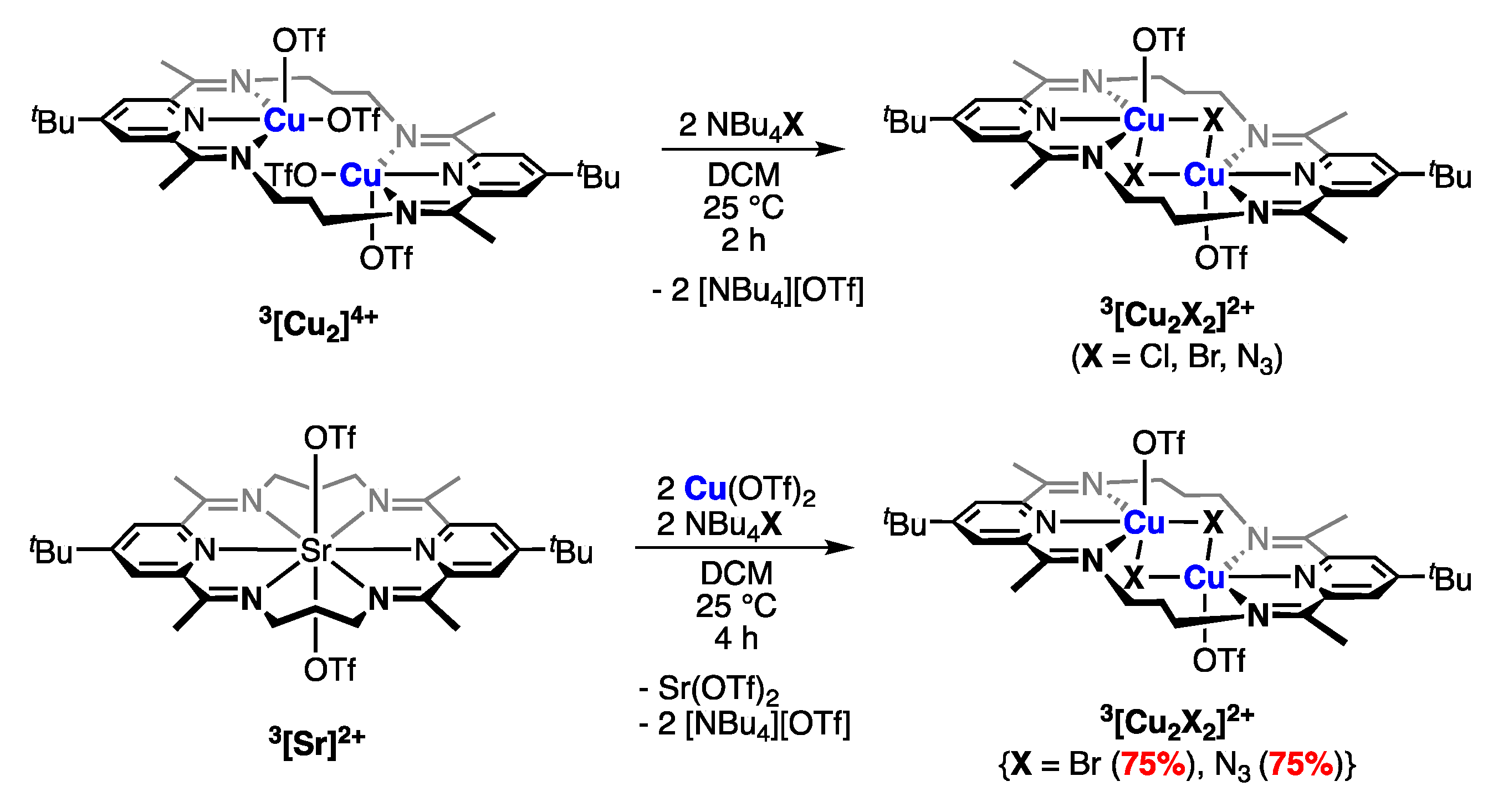
2.4. Anion Substitution Reactions with 3[Cu2]4+
2.4.1. Anion Substitution with Halides and Pseudohalides
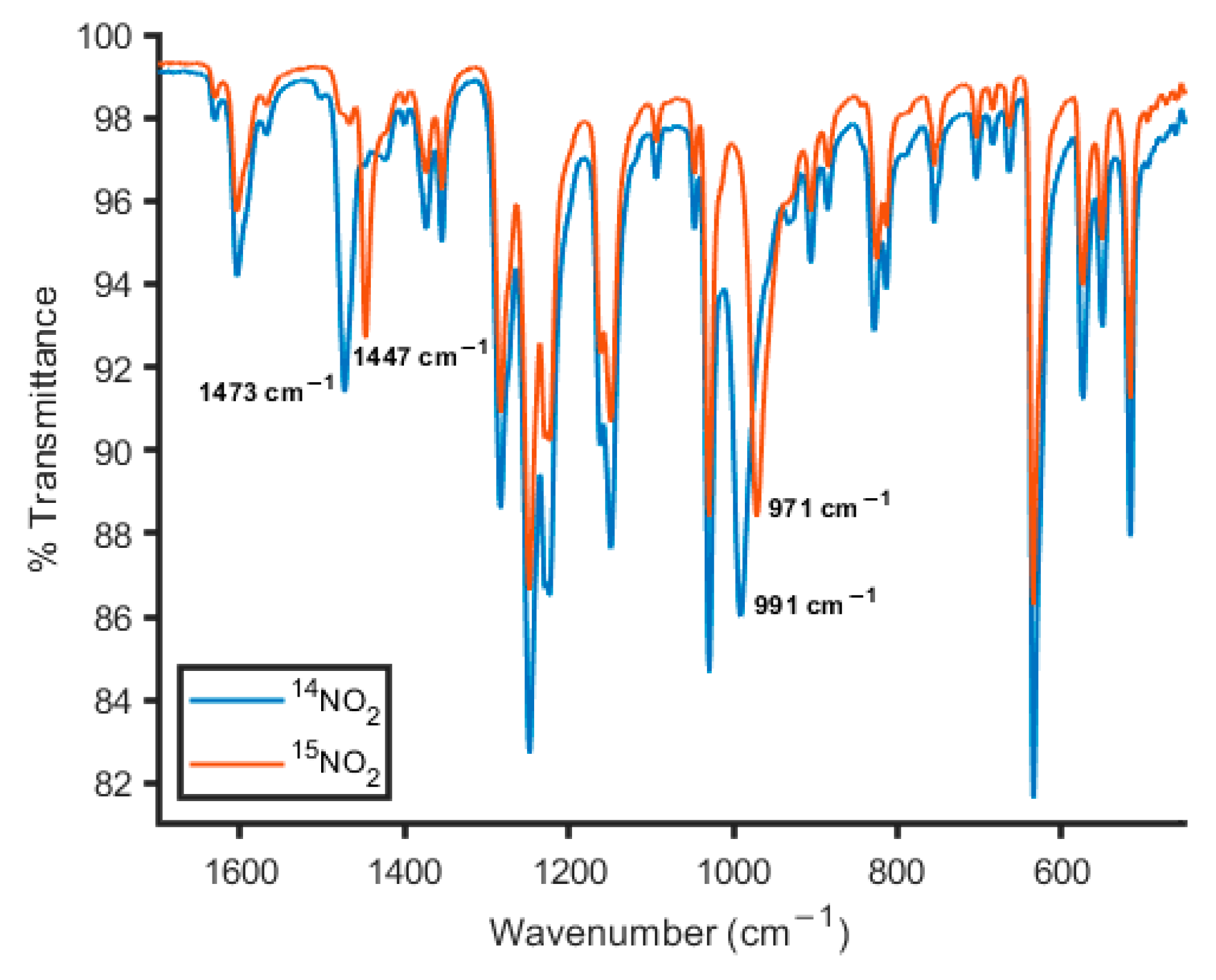
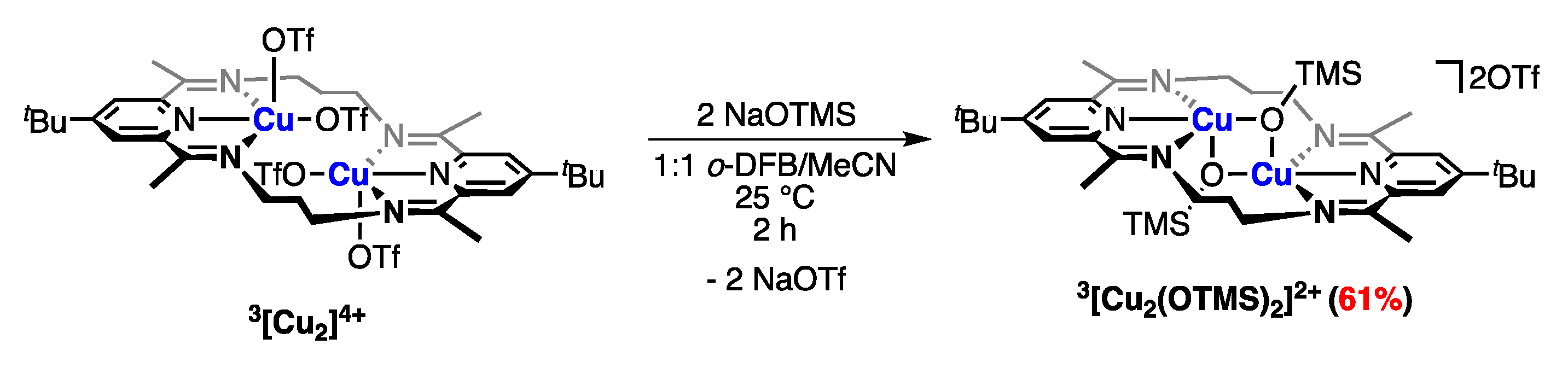
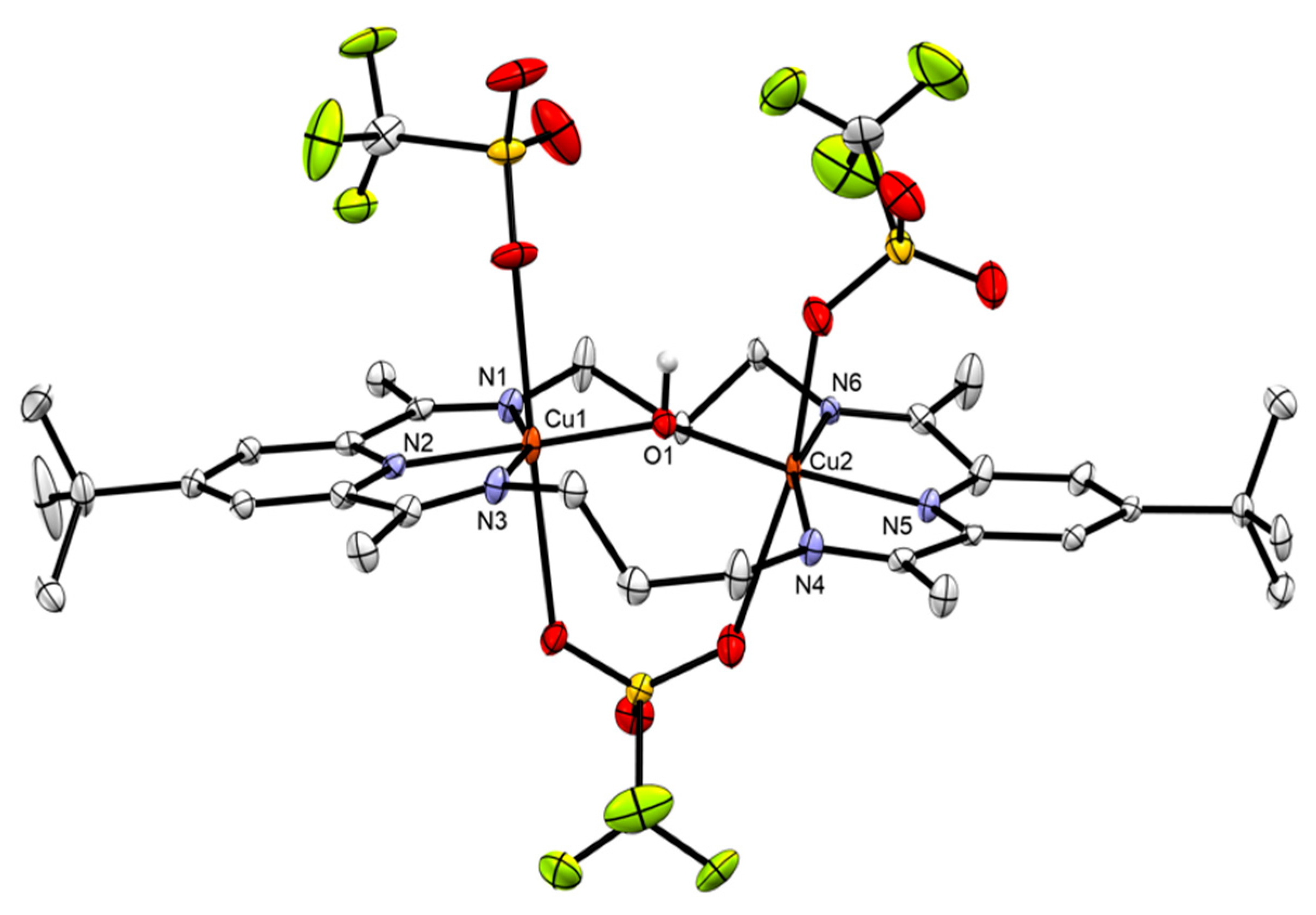
2.4.2. Anion Substitution with Nitrite and Trimethylsilanolate Anions
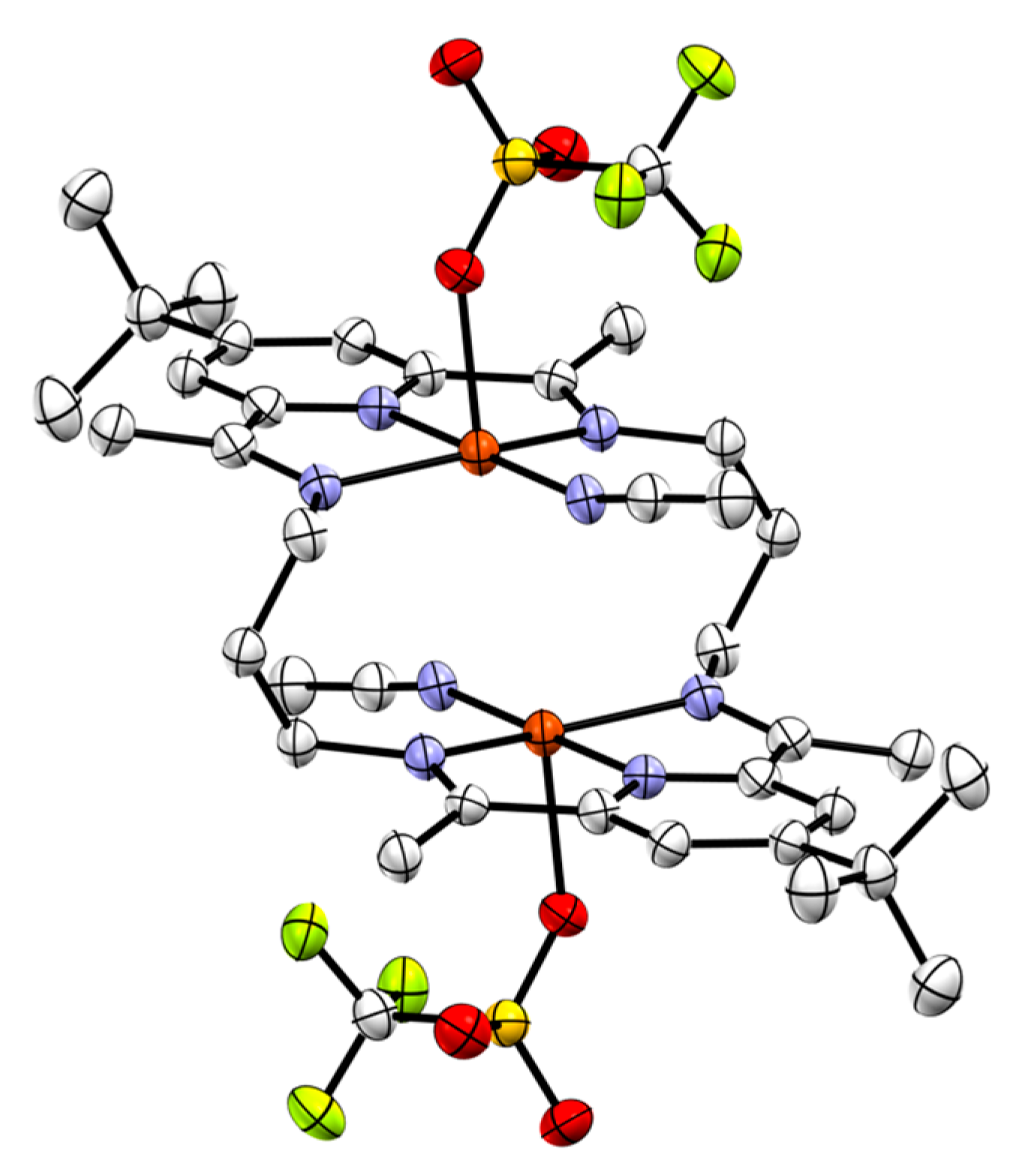
2.4.3. Synthesis of 3[Cu2OH]3+
3. Materials and Methods
3.1. General Considerations
3.2. Synthetic Procedures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tomkins, P.; Ranocchiari, M.; van Bokhoven, J.A. Direct Conversion of Methane to Methanol under Mild Conditions over Cu-Zeolites and beyond. Acc. Chem. Res. 2017, 50, 418–425. [Google Scholar]
- Snyder, B.E.R.; Bols, M.L.; Schoonheydt, R.A.; Sels, B.F.; Solomon, E.I. Iron and Copper Active Sites in Zeolites and Their Correlation to Metalloenzymes. Chem. Rev. 2018, 118, 2718–2768. [Google Scholar]
- Newton, M.A.; Knorpp, A.J.; Sushkevich, V.L.; Palagin, D.; van Bokhoven, J.A. Active sites and mechanisms in the direct conversion of methane to methanol using Cu in zeolitic hosts: A critical examination. Chem. Soc. Rev. 2020, 49, 1449–1486. [Google Scholar] [PubMed]
- Solomon, E.I.; Heppner, D.E.; Johnston, E.M.; Ginsbach, J.W.; Cirera, J.; Qayyum, M.; Kieber-Emmons, M.T.; Kjaergaard, C.H.; Hadt, R.G.; Tian, L. Copper Active Sites in Biology. Chem. Rev. 2014, 114, 3659–3853. [Google Scholar]
- Hoshikawa, R.; Mitsuhashi, R.; Asato, E.; Liu, J.; Sakiyama, H. Structures of Dimer-of-Dimers Type Defect Cubane Tetranuclear Copper(II) Complexes with Novel Dinucleating Ligands. Molecules 2022, 27, 576. [Google Scholar] [PubMed]
- Mendy, J.S.; Saeed, M.A.; Fronczek, F.R.; Powell, D.R.; Hossain, M.A. Anion Recognition and Sensing by a New Macrocyclic Dinuclear Copper(II) Complex: A Selective Receptor for Iodide. Inorg. Chem. 2010, 49, 7223–7225. [Google Scholar]
- Rhaman, M.M.; Alamgir, A.; Wong, B.M.; Powell, D.R.; Hossain, M.A. A highly efficient dinuclear Cu(II) chemosensor for colorimetric and fluorescent detection of cyanide in water. RSC Adv. 2014, 4, 54263–54267. [Google Scholar]
- Mateus, P.; Lima, L.M.P.; Delgado, R. Di- and trinuclear copper(II) complexes of polyaza macrocycles and cryptands as anion receptors. Polyhedron 2013, 52, 25–42. [Google Scholar]
- Haack, P.; Kärgel, A.; Greco, C.; Dokic, J.; Braun, B.; Pfaff, F.F.; Mebs, S.; Ray, K.; Limberg, C. Access to a CuII–O–CuII Motif: Spectroscopic Properties, Solution Structure, and Reactivity. J. Am. Chem. Soc. 2013, 135, 16148–16160. [Google Scholar]
- Jurgeleit, R.; Grimm-Lebsanft, B.; Flöser, B.M.; Teubner, M.; Buchenau, S.; Senft, L.; Hoffmann, J.; Naumova, M.; Näther, C.; Ivanović-Burmazović, I.; et al. Catalytic Oxygenation of Hydrocarbons by Mono-μ-oxo Dicopper(II) Species Resulting from O−O Cleavage of Tetranuclear CuI/CuII Peroxo Complexes. Angew. Chem. Int. Ed. 2021, 60, 14154–14162. [Google Scholar]
- Carsch, K.M.; Lukens, J.T.; DiMucci, I.M.; Iovan, D.A.; Zheng, S.-L.; Lancaster, K.M.; Betley, T.A. Electronic Structures and Reactivity Profiles of Aryl Nitrenoid-Bridged Dicopper Complexes. J. Am. Chem. Soc. 2020, 142, 2264–2276. [Google Scholar] [CrossRef]
- Ziegler, M.S.; Lakshmi, K.V.; Tilley, T.D. Dicopper Cu(I)Cu(I) and Cu(I)Cu(II) Complexes in Copper-Catalyzed Azide–Alkyne Cycloaddition. J. Am. Chem. Soc. 2017, 139, 5378–5386. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, M.S.; Levine, D.S.; Lakshmi, K.V.; Tilley, T.D. Aryl Group Transfer from Tetraarylborato Anions to an Electrophilic Dicopper(I) Center and Mixed-Valence μ-Aryl Dicopper(I,II) Complexes. J. Am. Chem. Soc. 2016, 138, 6484–6491. [Google Scholar] [CrossRef]
- Drew, M.G.B.; McCann, M.; Nelson, S.M. Bi-copper(I) and bi-copper(II) complexes of a 30-membered macrocyclic ligand: The inclusion of substrate molecules and the crystal and molecular structures of a µ-hydroxo- and a µ-imidazolato-complex. J. Chem. Soc. Dalton Trans. 1981, 1868–1878. [Google Scholar] [CrossRef]
- Drew, M.G.B.; McCann, M.; Nelson, S.M. Binuclear macrocyclic copper(II) complexes as receptors for small bridging ligands: X-ray crystal and molecular structure of a µ-azido complex. J. Chem. Soc. Chem. Commun. 1979, 481–482. [Google Scholar] [CrossRef]
- Drew, M.G.B.; Nelson, J.; Esho, F.; McKee, V.; Nelson, S.M. Dicopper(II) complexes of a macrocyclic ligand containing single hydroxo-, methoxo-, or 1,1-azido-bridges: Synthesis, magnetic properties, electron spin resonance spectra, and the crystal and molecular structure of a µ-hydroxo-derivative. J. Chem. Soc. Dalton Trans. 1982, 1837–1843. [Google Scholar] [CrossRef]
- Nelson, S.M.; Esho, F.S.; Drew, M.G.B. Metal-ion controlled reactions of 2,6-diacetylpyridine with 1,2-di-aminoethane and 2,6-diformylpyridine with o-phenylenediamine and the crystal and molecular structure of a pentagonal pyramidal cadmium(II) complex containing unidentate o-phenylenediamine. J. Chem. Soc. Dalton Trans. 1982, 407–415. [Google Scholar] [CrossRef]
- Cui, P.; Wang, Q.; McCollom, S.P.; Manor, B.C.; Carroll, P.J.; Tomson, N.C. Ring-Size-Modulated Reactivity of Putative Dicobalt-Bridging Nitrides: C−H Activation versus Phosphinimide Formation. Angew. Chem. Int. Ed. 2017, 56, 15979–15983. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Gau, M.R.; Tomson, N.C. Mimicking the Constrained Geometry of a Nitrogen-Fixation Intermediate. J. Am. Chem. Soc. 2020, 142, 8142–8146. [Google Scholar] [CrossRef] [PubMed]
- Spentzos, A.Z.; Gau, M.R.; Carroll, P.J.; Tomson, N.C. Unusual cyanide and methyl binding modes at a dicobalt macrocycle following acetonitrile C–C bond activation. Chem. Commun. 2020, 56, 9675–9678. [Google Scholar] [CrossRef]
- Thierer, L.M.; Brooks, S.H.; Weberg, A.B.; Cui, P.; Zhang, S.; Gau, M.R.; Manor, B.C.; Carroll, P.J.; Tomson, N.C. Macrocycle-Induced Modulation of Internuclear Interactions in Homobimetallic Complexes. Inorg. Chem. 2022, 61, 6263–6280. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, S.; Cui, P.; Weberg, A.B.; Thierer, L.M.; Manor, B.C.; Gau, M.R.; Carroll, P.J.; Tomson, N.C. Interdependent Metal–Metal Bonding and Ligand Redox-Activity in a Series of Dinuclear Macrocyclic Complexes of Iron, Cobalt, and Nickel. Inorg. Chem. 2020, 59, 4200–4214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cui, P.; Liu, T.; Wang, Q.; Longo, T.J.; Thierer, L.M.; Manor, B.C.; Gau, M.R.; Carroll, P.J.; Papaefthymiou, G.C.; et al. N−H Bond Formation at a Diiron Bridging Nitride. Angew. Chem. Int. Ed. 2020, 59, 15215–15219. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Q.; Thierer, L.M.; Weberg, A.B.; Gau, M.R.; Carroll, P.J.; Tomson, N.C. Tuning Metal–Metal Interactions through Reversible Ligand Folding in a Series of Dinuclear Iron Complexes. Inorg. Chem. 2019, 58, 12234–12244. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Brooks, S.H.; Liu, T.; Tomson, N.C. Tuning metal–metal interactions for cooperative small molecule activation. Chem. Commun. 2021, 57, 2839–2853. [Google Scholar] [CrossRef]
- Römelt, C.; Weyhermüller, T.; Wieghardt, K. Structural characteristics of redox-active pyridine-1,6-diimine complexes: Electronic structures and ligand oxidation levels. Coord. Chem. Rev. 2019, 380, 287–317. [Google Scholar] [CrossRef]
- Addison, A.W.; Rao, T.N.; Reedijk, J.; van Rijn, J.; Verschoor, G.C. Synthesis; structure, and spectroscopic properties of copper(II) compounds containing nitrogen–sulphur donor ligands; the crystal and molecular structure of aqua[1,7-bis(N-methylbenzimidazol-2′-yl)-2,6-dithiaheptane]copper(II) perchlorate. J. Chem. Soc. Dalton Trans. 1984, 1349–1356. [Google Scholar] [CrossRef]
- Alvarez, S. A cartography of the van der Waals territories. Dalton Trans. 2013, 42, 8617–8636. [Google Scholar] [CrossRef] [PubMed]
- McKee, V.; Smith, J. Synthesis and X-ray structure of a bicopper(II) Schiff base macrocyclic complex having a single endogenous alkoxy bridge. J. Chem. Soc. Chem. Commun. 1983, 1465–1467. [Google Scholar] [CrossRef]
- Cheung, P.M.; Berger, R.F.; Zakharov, L.N.; Gilbertson, J.D. Square planar Cu(I) stabilized by a pyridinediimine ligand. Chem. Commun. 2016, 52, 4156–4159. [Google Scholar] [CrossRef]
- Bower, J.K.; Cypcar, A.D.; Henriquez, B.; Stieber, S.C.E.; Zhang, S. C(sp3)–H Fluorination with a Copper(II)/(III) Redox Couple. J. Am. Chem. Soc. 2020, 142, 8514–8521. [Google Scholar] [CrossRef] [PubMed]
- Bower, J.K.; Cypcar, A.D.; Henriquez, B.; Stieber, S.C.E.; Zhang, S. Correction to “C(sp3)–H Fluorination with a Copper(II)/(III) Redox Couple”. J. Am. Chem. Soc. 2022, 144, 6118–6119. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Ray, M.S.; Drew, M.G.B.; Figuerola, A.; Diaz, C.; Ghosh, A. Facile synthesis of Cu(II) complexes of monocondensed N,N,N donor Schiff base ligands: Crystal structure, spectroscopic and magnetic properties. Polyhedron 2006, 25, 2241–2253. [Google Scholar] [CrossRef]
- Adhikary, C.; Mal, D.; Sen, R.; Bhattacharjee, A.; Gütlich, P.; Chaudhuri, S.; Koner, S. Synthesis, X-ray crystal structure and magnetic study of a novel μ2-1,1-azido bridged dimeric copper(II) complex. Polyhedron 2007, 26, 1658–1662. [Google Scholar] [CrossRef]
- Rahaman, S.H.; Bose, D.; Ghosh, R.; Mostafa, G.; Fun, H.-K.; Ghosh, B.K. Synthesis, crystal and molecular structures of 1D and 2D supramers of copper(II) with N,N′-{bis(pyridin-2-yl) benzylidene}butane-1,4-diamine. Struct. Chem. 2007, 18, 237–244. [Google Scholar] [CrossRef]
- Nandy, M.; Shit, S.; Garribba, E.; Gómez-García, C.J.; Mitra, S. Double azido/cyanato bridged copper(II) dimers incorporating tridentate nitrogen donors Schiff base: Structure, EPR and magnetic studies. Polyhedron 2015, 102, 137–146. [Google Scholar] [CrossRef]
- Camus, A.; Marsich, N.; Lanfredi, A.M.M.; Ugozzoli, F.; Massera, C. Copper(II)nitrito complexes with 2,2′-dipyridylamine. Crystal structures of the [(acetato)(2,2′-dipyridylamine)(nitrito-O,O′)copper(II)] and [(2,2′-dipyridylamine)(nitrito-O,O′)(μ-nitrito-O)copper(II)]2·2(acetonitrile). Inorg. Chim. Acta 2000, 309, 1–9. [Google Scholar] [CrossRef]
- Arnold, P.J.; Davies, S.C.; Durrant, M.C.; Griffiths, D.V.; Hughes, D.L.; Sharpe, P.C. Copper(II) nitrite complexes of tripodal ligands derived from 1,1,1-tris(2-pyridyl)methylamine. Inorg. Chim. Acta 2003, 348, 143–149. [Google Scholar] [CrossRef]
- Zhu, R.-Q. Di-µ-nitrito-κ4O:O-bis[bis(1-ethyl-1H-imidazole-κN3)(nitrito-κO)copper(II)]. Acta Crystallogr. E 2011, 67, m869. [Google Scholar] [CrossRef]
- Zhu, R.-Q. Bis(µ-nitrito-κ2O:O)bis[bis(1-methyl-1H-imidazole-κN3)(nitrito-κO)copper(II)]. Acta Crystallogr. E 2012, 68, m398. [Google Scholar] [CrossRef]
- McGeary, M.J.; Wedlich, R.C.; Coan, P.S.; Folting, K.; Caulton, K.G. Synthesis and thermal decomposition of copper(I) silyloxide complexes. X-ray crystal structures of [Cu(OSiPh3)]4 and [Cu(OSiPh3)(PMe2Ph)]2. Polyhedron 1992, 11, 2459–2473. [Google Scholar] [CrossRef]
- Dai, X.; Warren, T.H. Dioxygen activation by a neutral β-diketiminato copper(I) ethylene complex. Chem. Commun. 2001, 19, 1998–1999. [Google Scholar] [CrossRef]
- Kitajima, N.; Koda, T.; Iwata, Y.; Morooka, Y. Reaction aspects of a µ-peroxo binuclear copper(II) complex. J. Am. Chem. Soc. 1990, 112, 8833–8839. [Google Scholar] [CrossRef]
- Kitajima, N.; Koda, T.; Hashimoto, S.; Kitagawa, T.; Morooka, Y. Synthesis and characterization of the dinuclear copper(II) complexes [Cu(HB(3,5-Me2pz)3)]2X (X = O2−, (OH)22−, CO32−, O22−). J. Am. Chem. Soc. 1991, 113, 5664–5671. [Google Scholar] [CrossRef]
- Chaudhuri, P.; Ventur, D.C.; Wieghardt, K.; Peters, E.M.; Peters, K. Preparation, Magnetism, and Crystal Structures of the Tautomers [LCu(μ2-OH)2CuL](ClO4)2 (Blue) and [LCu(μ2-OH2)(μ2-O)CuL](ClOμ4)μ2 (Green): μ-Aqua-μ-oxo vs. Di-μ-hydroxo Linkage. Angew. Chem. Int. Ed. Eng. 1985, 24, 57–59. [Google Scholar] [CrossRef]
- Thierer, L.M.; Wang, Q.; Brooks, S.H.; Cui, P.; Qi, J.; Gau, M.R.; Manor, B.C.; Carroll, P.J.; Tomson, N.C. Pyridyldiimine macrocyclic ligands: Influences of template ion, linker length and imine substitution on ligand synthesis, structure and redox properties. Polyhedron 2021, 198, 115044. [Google Scholar] [CrossRef] [PubMed]
- Rosenau, C.P.; Jelier, B.J.; Gossert, A.D.; Togni, A. Exposing the Origins of Irreproducibility in Fluorine NMR Spectroscopy. Angew. Chem. Int. Ed. 2018, 57, 9528–9533. [Google Scholar] [CrossRef] [PubMed]



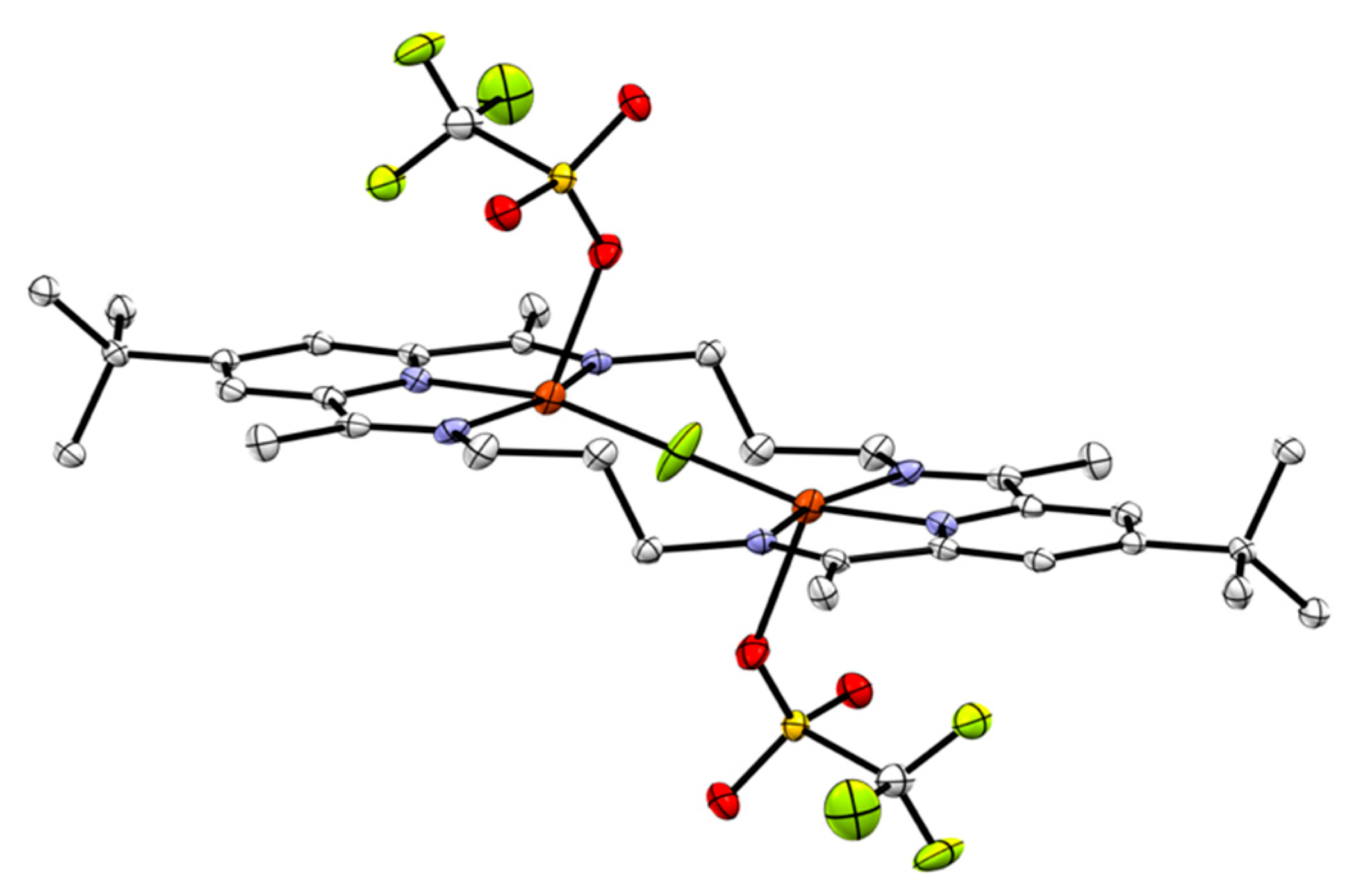

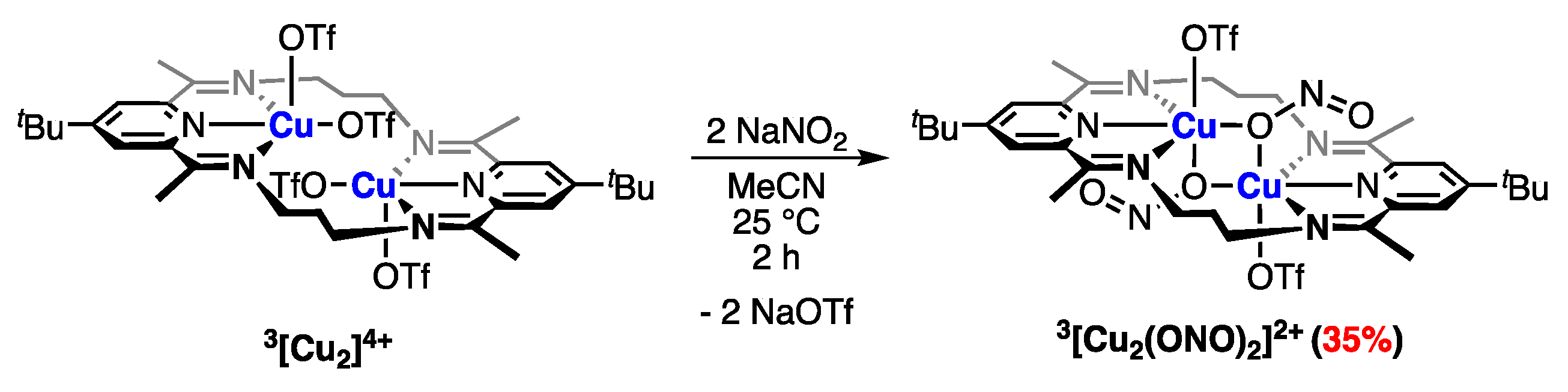
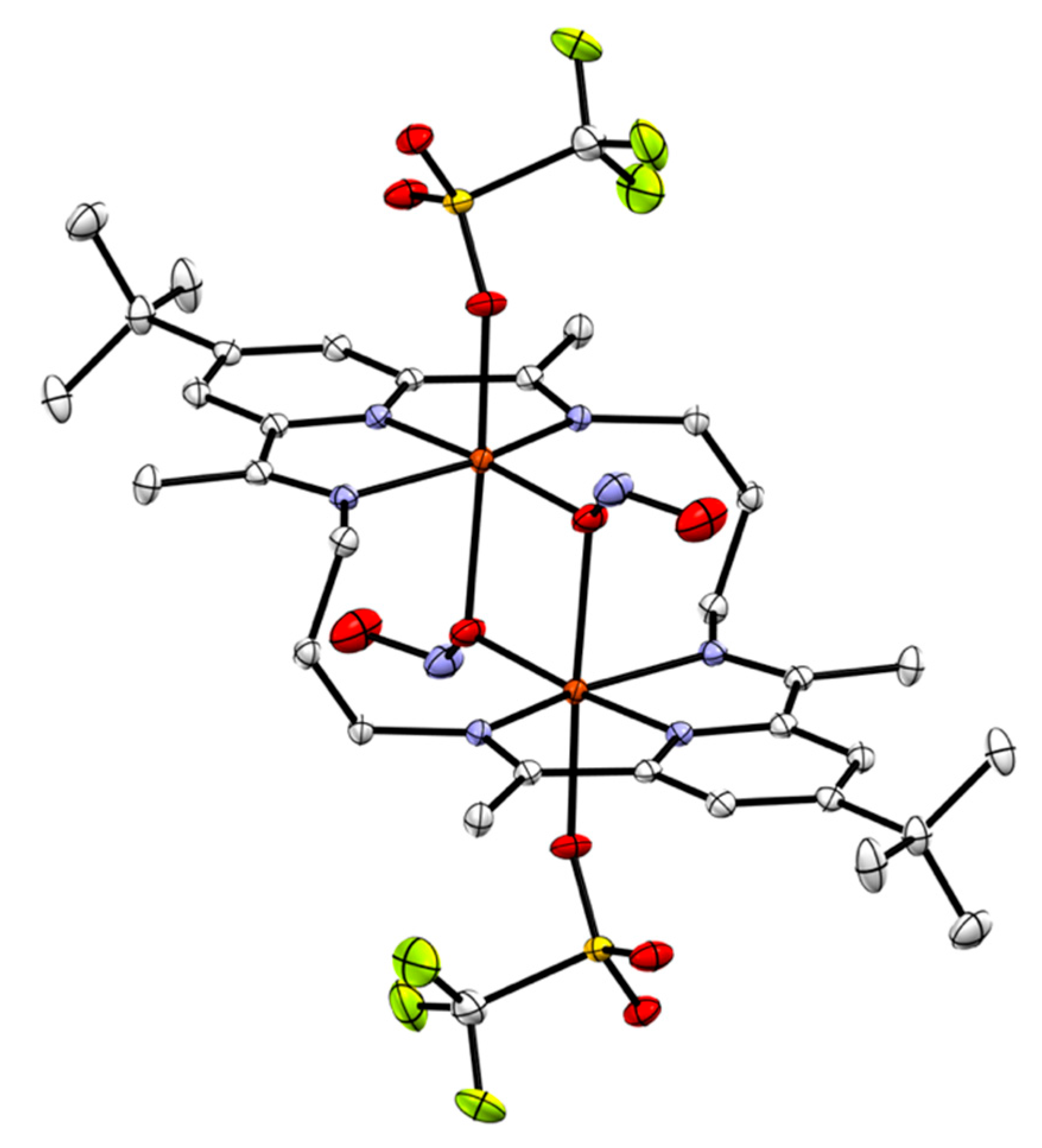


| 3[Cu2Cl2]2+a | 2[Cu2Cl2]2+b | [(dippPDI)CuCl][PF6] | |
|---|---|---|---|
| Cu(1)—N(1)/Å | 2.056(4) | 2.057 | 2.070(2) |
| Cu(1)—N(2)/Å | 1.902(3) | 1.903 | 1.922(2) |
| Cu(1)—N(3)/Å | 2.047(3) | 2.044 | 2.062(2) |
| Cu(1)—Cl(1)/Å | 2.1860(11) | 2.2056 | 2.1450(5) |
| Cu(1)—Cl(2)/Å | 2.873(1) | 3.021 | - |
| Cu(1)—O(1)/Å | 2.710(1) | 2.4015 | - |
| Cu(1)—Cu(2)/Å | 3.129(1) | 2.9922 | - |
| Cu(2)—N(4)/Å | - | 2.041 | - |
| Cu(2)—N(5)/Å | - | 1.898 | - |
| Cu(2)—N(6)/Å | - | 2.042 | - |
| Cu(2)—Cl(1)/Å | - | 2.7273 | - |
| Cu(2)—Cl(2)/Å | - | 2.2043 | - |
| ∠Cu(1)-Cl(1)-Cu(2)/° | 75.05(4) | 73.83 | - |
| ∠Cu(1)-Cl(2)-Cu(2)/° | - | 67.80 | - |
| ∆/Å | 0.187 | 0.182 | 0.178 |
| 3[Cu2OH]3+ | [(EtPDI2)Cu2OH]3+ b,c | [(OctPDI2)Cu2OH]3+ c | |
|---|---|---|---|
| Cu(1)—N(1)/Å | 2.097(2) | 2.066(12) | 2.039(11) |
| Cu(1)—N(2)/Å | 1.912(2) | 1.916(13) | 1.907(16) |
| Cu(1)—N(3)/Å | 2.040(3) | 2.061(13) | 2.060(9) |
| Cu(2)—N(4)/Å | 2.096 a | - | 2.076(4) |
| Cu(2)—N(5)/Å | 1.926 a | - | 1.925(12) |
| Cu(2)—N(6)/Å | 2.038 a | - | 2.059(14) |
| Cu(1)—O(1)/Å | 1.872(2) | 1.916(9) | 1.866(13) |
| Cu(2)—O(1)/Å | 1.908 a | - | 1.912(10) |
| Cu(1)—Cu(2)/Å | 3.603 a | 3.145(4) | 3.57 |
| ∠Cu(1)-O(1)-Cu(2)/° | 145.09 a | 110.3(7) | 141.7(7) |
| ∆/Å | 0.196 a | 0.230 | 0.147 |
| Conformation | Cu-Cu/Å | Cu-Xequatorial/Å a | Cu-X-Cu/° a | |
|---|---|---|---|---|
| 3[Cu2Cl]2+ | Unfolded | 3.129(1) | 2.1860 | 75.05 |
| 2[Cu2Cl]2+ | Unfolded | 2.9922 a | 2.2056 | 70.82 |
| 3[Cu2(NCMe)2]4+ | Unfolded | 3.3479(7) | 1.973 | 77.64 |
| 3[Cu2Cl]3+ | Arched | 3.7045(7) | 2.2823 | 108.50 |
| 3[Cu2F]3+ | Unfolded | 3.7240(4) | 1.8620 | 180.00 |
| 3[Cu2(N3)2]2+ | Unfolded | 3.1362(6) | 1.936 | 88.02 |
| 3[Cu2(ONO)2]2+ | Unfolded | 3.2553(6) | 1.904 | 94.18 |
| 3[Cu2(OTMS)2]2+ | Unfolded | 2.8639(4) | 1.8969 | 86.65 |
| 3[Cu2OH]3+ | Planar | 3.603 a | 1.890 | 145.09 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brooks, S.H.; Richards, C.A.; Carroll, P.J.; Gau, M.R.; Tomson, N.C. Anion Capture at the Open Core of a Geometrically Flexible Dicopper(II,II) Macrocycle Complex. Inorganics 2023, 11, 348. https://doi.org/10.3390/inorganics11090348
Brooks SH, Richards CA, Carroll PJ, Gau MR, Tomson NC. Anion Capture at the Open Core of a Geometrically Flexible Dicopper(II,II) Macrocycle Complex. Inorganics. 2023; 11(9):348. https://doi.org/10.3390/inorganics11090348
Chicago/Turabian StyleBrooks, Sam H., Corey A. Richards, Patrick J. Carroll, Michael R. Gau, and Neil C. Tomson. 2023. "Anion Capture at the Open Core of a Geometrically Flexible Dicopper(II,II) Macrocycle Complex" Inorganics 11, no. 9: 348. https://doi.org/10.3390/inorganics11090348
APA StyleBrooks, S. H., Richards, C. A., Carroll, P. J., Gau, M. R., & Tomson, N. C. (2023). Anion Capture at the Open Core of a Geometrically Flexible Dicopper(II,II) Macrocycle Complex. Inorganics, 11(9), 348. https://doi.org/10.3390/inorganics11090348






