Low-Dimensional Compounds Containing Bioactive Ligands. Part XIX: Crystal Structures and Biological Properties of Copper Complexes with Halogen and Nitro Derivatives of 8-Hydroxyquinoline
Abstract
1. Introduction
2. Results and Discussion
2.1. Syntheses
2.2. Infrared Spectroscopy
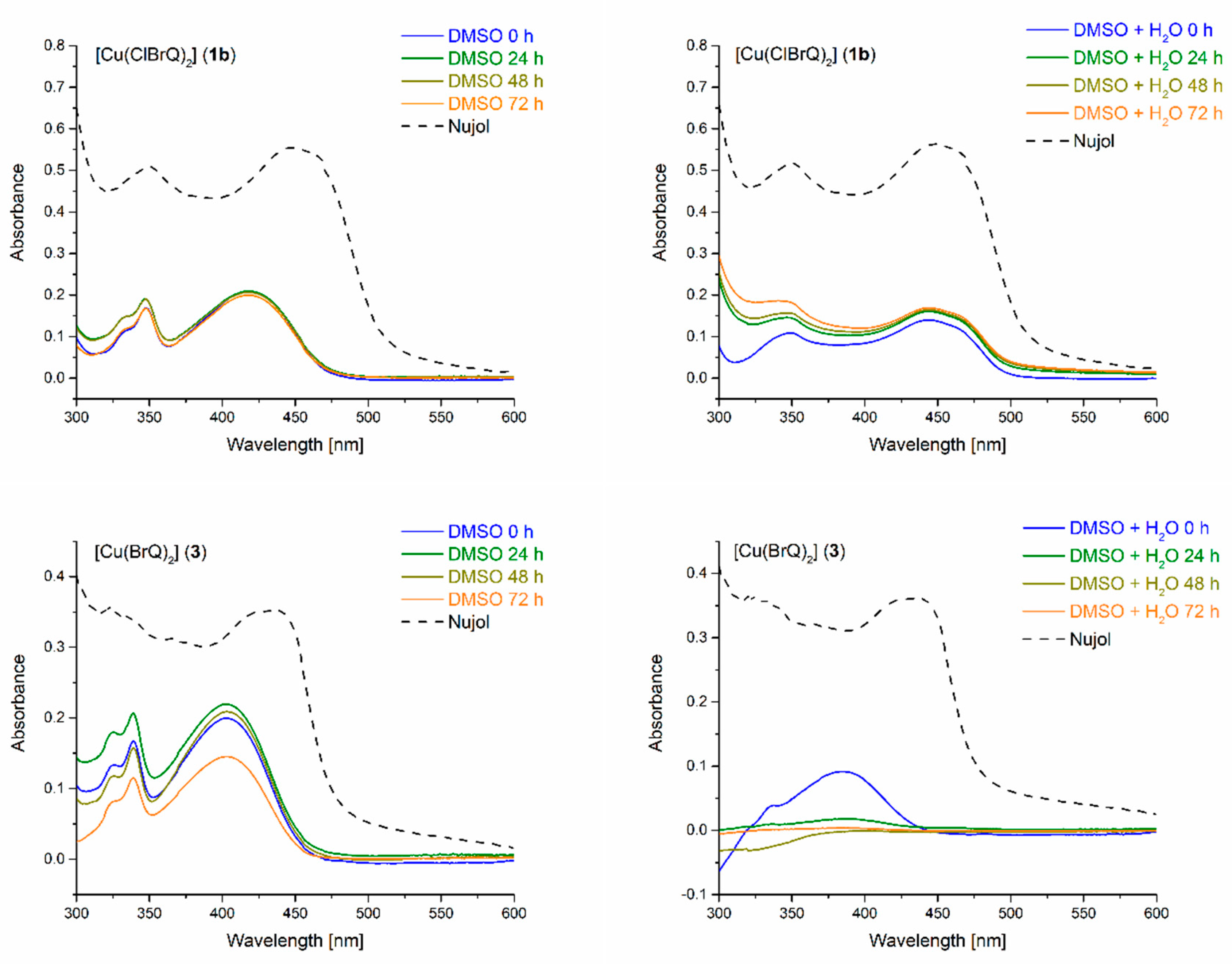
2.3. UV-Vis Spectroscopy
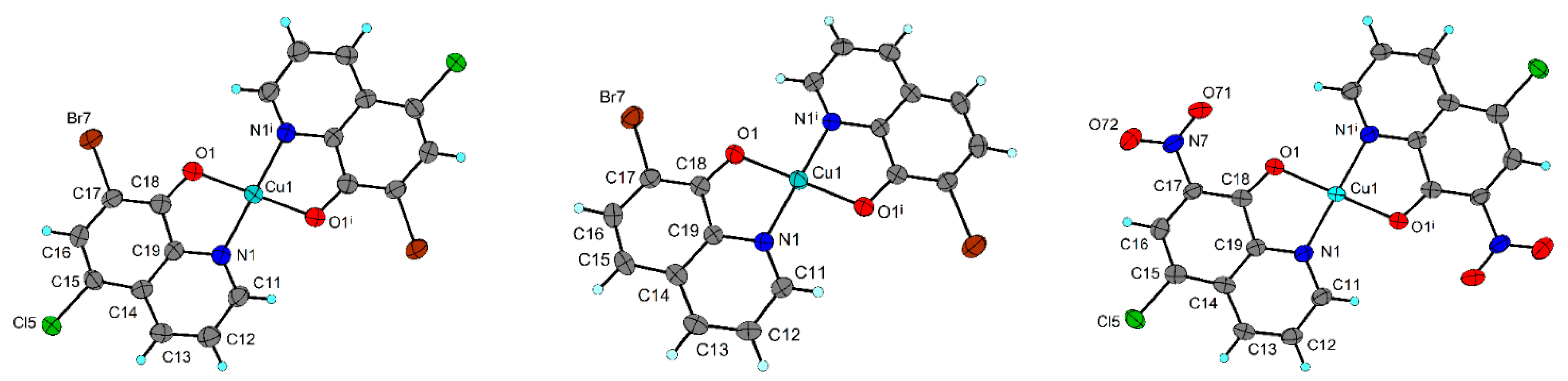
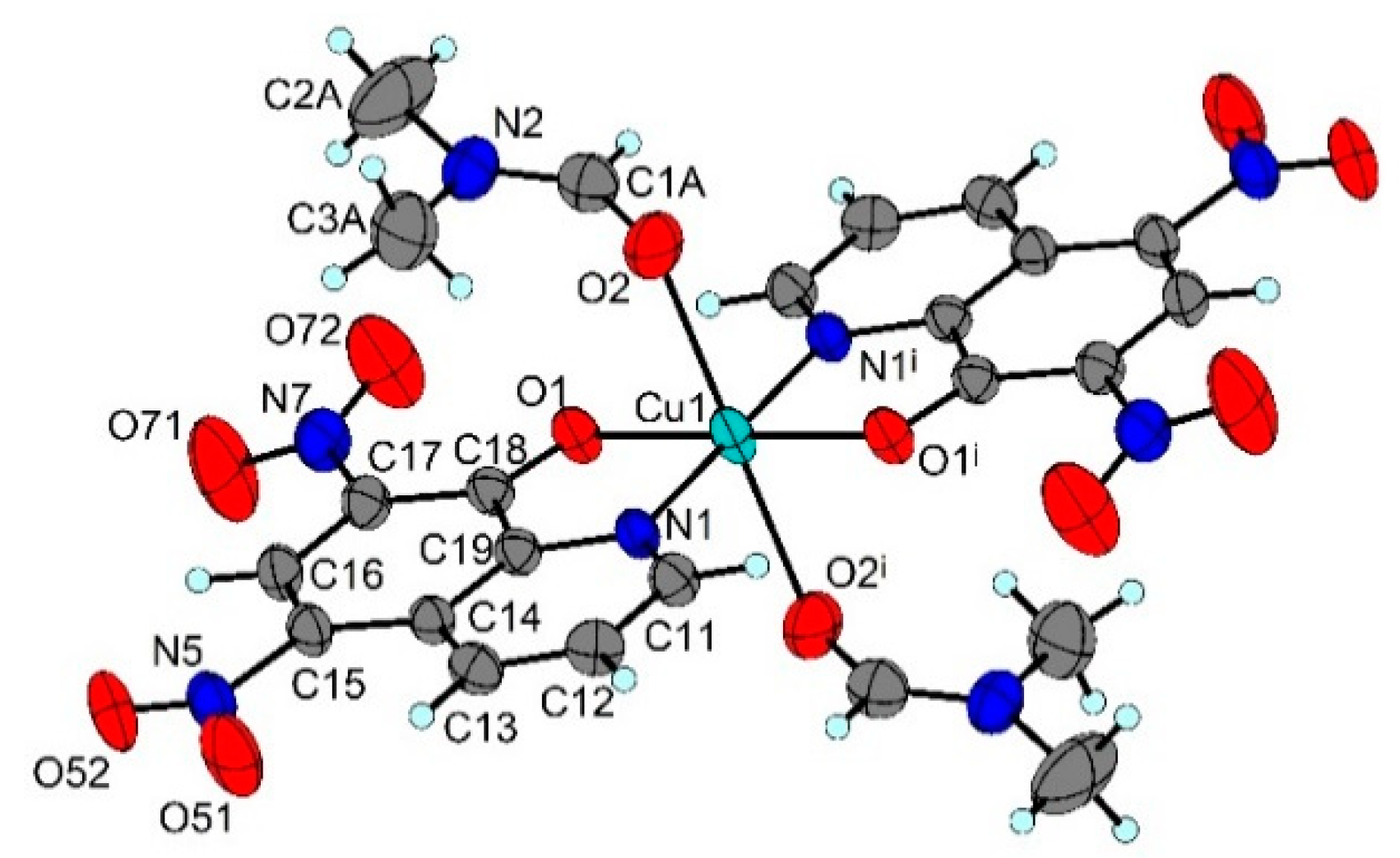
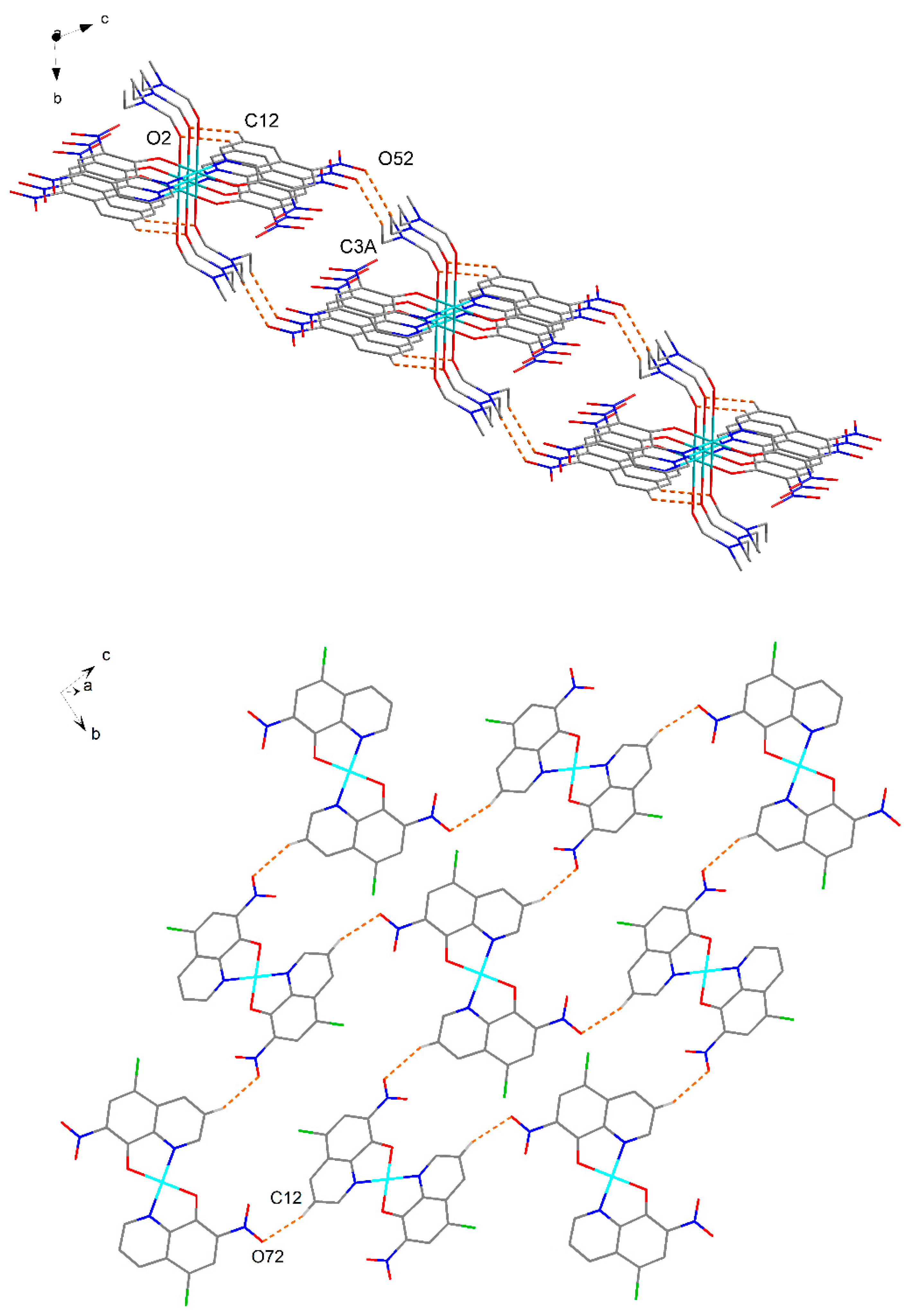
2.4. X-ray Structure Analysis
2.5. Antiproliferative Activity
2.6. Antibacterial Activity
2.7. Radical Scavenging Activity
3. Materials and Methods
3.1. Materials and Chemicals
3.2. Syntheses
3.2.1. Synthesis of [Cu(ClBrQ)2] (1a)
3.2.2. Synthesis of [Cu(ClBrQ)2] (1b)
3.2.3. Synthesis of [Cu(ClBrQ)2]·1/2 Diox (2)
3.2.4. Synthesis of [Cu(BrQ)2] (3)
3.2.5. Synthesis of [Cu(dNQ)2] (4)
3.2.6. Synthesis of [Cu(dNQ)2(DMF)2] (5)
3.2.7. Synthesis of [Cu(ClNQ)2] (6)
3.3. Physical Measurements
3.4. X-ray Structure Analysis
3.5. Cell Cultures
Screening of Antiproliferative/Cytotoxic Activity
3.6. Antibacterial Activity
3.6.1. Microorganisms Used
3.6.2. Agar Well-Diffusion Method
3.6.3. Determination of the Minimum Inhibitory Concentration (MIC) by the Microdilution Method
3.7. Radical Scavenging Experiments
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rosenberg, B.; van Camp, L.; Krigas, T. Platinum compounds: A new class of potent antitumour agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef]
- Peng, K.; Liang, B.B.; Liu, W.; Mao, Z.W. What blocks more anticancer platinum complexes from experiment to clinic: Major problems and potential strategies from drug design perspectives. Coord. Chem. Rev. 2021, 449, 214210. [Google Scholar] [CrossRef]
- Casini, A.; Vessières, A.; Meier-Menches, S.M. Metal-Based Anticancer Agents; Royal Society of Chemistry: Cambridge, UK, 2019; pp. 3–30. [Google Scholar]
- Duan, Z.Y.; Liu, J.Q.; Yin, P.; Li, J.J.; Cai, G.Y.; Chen, X.M. Impact of aging on the risk of platinum-related renal toxicity: A systematic review and meta-analysis. Cancer Treat. Rev. 2018, 69, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Nedeljković, M.; Damjanović, A. Mechanisms of chemotherapy resistance in triple-negative breast cancer-how we can rise to the challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Luxami, V.; Paul, K. Insights of 8-hydroxyquinolines: A novel target in medicinal chemistry. Bioorganic Chem. 2021, 108, 104663. [Google Scholar] [CrossRef] [PubMed]
- Cherdtrakulkiat, R.; Boonpangrak, S.; Sinthupoom, N.; Prachayasittikul, S.; Ruchirawat, S.; Prachayasittikul, V. Derivatives (halogen, nitro and amino) of 8-hydroxyquinoline with highly potent antimicrobial and antioxidant activities. Biochem. Biophys. Rep. 2016, 6, 135–141. [Google Scholar] [CrossRef]
- Saadeh, H.A.; Sweidan, K.A.; Mubarak, M.S. Recent advances in the synthesis and biological activity of 8-hydroxyquinolines. Molecules 2020, 25, 4321. [Google Scholar] [CrossRef]
- Low, K.H.; Xu, Z.X.; Xiang, H.F.; Chui, S.S.Y.; Roy, V.A.L.; Che, C.M. Bis(5,7-dimethyl-8-hydroxyquinolinato)platinum(II) complex for efficient organic heterojunction solar cells. Chem. Asian J. 2011, 6, 3223–3229. [Google Scholar] [CrossRef]
- Vranec, P.; Potočňák, I. Low-dimensional compounds containing bioactive ligands. Part III: Palladium(II) complexes with halogenated quinolin-8-ol derivatives. J. Mol. Struct. 2013, 1041, 219–226. [Google Scholar] [CrossRef]
- Liu, Y.C.; Wei, J.H.; Chen, Z.F.; Liu, M.; Gu, Y.Q.; Huang, K.B.; Li, Z.Q.; Liang, H. The antitumor activity of zinc(II) and copper(II) complexes with 5,7-dihalo-substituted-8-quinolinoline. Eur. J. Med. Chem. 2013, 69, 554–563. [Google Scholar] [CrossRef]
- Zhang, H.R.; Liu, Y.C.; Meng, T.; Qin, Q.P.; Tang, S.F.; Chen, Z.F.; Zou, B.Q.; Liu, Y.N.; Liang, H. Cytotoxicity, DNA binding and cell apoptosis induction of a zinc(II) complex of HBrQ. MedChemComm 2015, 6, 2224–2231. [Google Scholar] [CrossRef]
- Valiahdi, S.M.; Heffeter, P.; Jakupec, M.A.; Marculescu, R.; Berger, W.; Rappersberger, K.; Keppler, B.K. The gallium complex KP46 exerts strong activity against primary explanted melanoma cells and induces apoptosis in melanoma cell lines. Melanoma Res. 2009, 19, 283–293. [Google Scholar] [CrossRef]
- Hreusova, M.; Novohradsky, V.; Markova, L.; Kostrhunova, H.; Potočňák, I.; Brabec, V.; Kasparkova, J. Gallium(III) Complex with Cloxyquin Ligands Induces Ferroptosis in Cancer Cells and Is a Potent Agent against Both Differentiated and Tumorigenic Cancer Stem Rhabdomyosarcoma Cells. Bioinorg. Chem. Appl. 2022, 2022, 3095749. [Google Scholar] [CrossRef] [PubMed]
- Kubista, B.; Schoefl, T.; Mayr, L.; van Schoonhoven, S.; Heffeter, P.; Windhager, R.; Keppler, B.K.; Berger, W. Distinct activity of the bone-targeted gallium compound KP46 against osteosarcoma cells—Synergism with autophagy inhibition. J. Exp. Clin. Cancer Res. 2017, 36, 5658–5668. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gobec, M.; Kljun, J.; Sosič, I.; Mlinarič-Raščan, I.; Uršič, M.; Gobec, S.; Turel, I. Structural characterization and biological evaluation of a clioquinol-ruthenium complex with copper-independent antileukaemic activity. Dalton Trans. 2014, 43, 9045–9051. [Google Scholar] [CrossRef]
- Kubanik, M.; Holtkamp, H.; Söhnel, T.; Jamieson, S.M.F.; Hartinger, C.G. Impact of the Halogen Substitution Pattern on the Biological Activity of Organoruthenium 8-Hydroxyquinoline Anticancer Agents. Organometallics 2015, 34, 5658–5668. [Google Scholar] [CrossRef]
- Chen, Z.F.; Gu, Y.Q.; Song, X.Y.; Liu, Y.C.; Peng, Y.; Liang, H. Synthesis, crystal structure, cytotoxicity and DNA interaction of 5,7-dichloro-8-quinolinolato-lanthanides. Eur. J. Med. Chem. 2013, 59, 194–202. [Google Scholar] [CrossRef]
- Wang, R.; Zou, B.Q.; Qin, Q.P.; Wang, Z.F.; Tan, M.X.; Liang, H. Synthesis, characterization and the anticancer activity of six lanthanides(III) complexes with 5,7-dihalogenated-8-quinolinol and 2,2′-bipyridine derivatives. Transit. Met. Chem. 2020, 45, 477–483. [Google Scholar] [CrossRef]
- Meng, T.; Qin, Q.P.; Chen, Z.L.; Zou, H.H.; Wang, K.; Liang, F.P. High in vitro and in vivo antitumor activities of Ln(III) complexes with mixed 5,7-dichloro-2-methyl-8-quinolinol and 4,4′-dimethyl-2,2′-bipyridyl chelating ligands. Eur. J. Med. Chem. 2019, 169, 103–110. [Google Scholar] [CrossRef]
- Liu, Y.C.; Chen, Z.F.; Song, X.Y.; Peng, Y.; Qin, Q.P.; Liang, H. Synthesis, crystal structure, cytotoxicity and DNA interaction of 5,7-dibromo-8-quinolinolato-lanthanides. Eur. J. Med. Chem. 2013, 59, 168–175. [Google Scholar] [CrossRef]
- Tardito, S.; Barilli, A.; Bassanetti, I.; Tegoni, M.; Bussolati, O.; Franchi-Gazzola, R.; Mucchino, C.; Marchiò, L. Copper-dependent cytotoxicity of 8-hydroxyquinoline derivatives correlates with their hydrophobicity and does not require caspase activation. J. Med. Chem. 2012, 55, 10448–10459. [Google Scholar] [CrossRef]
- Kuchárová, V.; Kuchár, J.; Zaric, M.; Canovic, P.; Arsenijevic, N.; Volarevic, V.; Misirkic, M.; Trajkovic, V.; Radojević, I.D.; Čomić, L.R.; et al. Low-dimensional compounds containing bioactive ligands. Part XI: Synthesis, structures, spectra, in vitro anti-tumor and antimicrobial activities of 3d metal complexes with 8-hydroxyquinoline-5-sulfonic acid. Inorg. Chim. Acta 2019, 497, 119062. [Google Scholar] [CrossRef]
- Lüköová, A.; Baran, P.; Volarevic, V.; Ilic, A.; Vilková, M.; Litecká, M.; Harmošová, M.; Potočňák, I. Low-dimensional compounds containing bioactive ligands. Part XVI: Halogenated derivatives of 8-quinolinol N-oxides and their copper(II) complexes. J. Mol. Struct. 2021, 1246, 131144. [Google Scholar] [CrossRef]
- Potočňák, I.; Vranec, P.; Farkasová, V.; Sabolová, D.; Vataščinová, M.; Kudláčová, J.; Radojević, I.D.; Čomić, L.R.; Markovic, B.S.; Volarevic, V.; et al. Low-dimensional compounds containing bioactive ligands. Part VI: Synthesis, structures, in vitro DNA binding, antimicrobial and anticancer properties of first row transition metal complexes with 5-chloro-quinolin-8-ol. J. Inorg. Biochem. 2016, 154, 67–77. [Google Scholar] [CrossRef]
- Nikolaides, N.; Bogdan, S.E.; Szalma, J.S. Preparation of 8-alkyl 7-(2-imidazolinylamino)quinolines via palladium mediated alkylations. Synth. Commun. 2002, 32, 2027–2033. [Google Scholar] [CrossRef]
- Clavier, S.; Rist, Ø.; Hansen, S.; Gerlach, L.O.; Högberg, T.; Bergman, J. Preparation and evaluation of sulfur-containing metal chelators. Org. Biomol. Chem. 2003, 1, 4248–4253. [Google Scholar] [CrossRef] [PubMed]
- Basak, A.; Abouelhassan, Y.; Norwood, V.M.; Bai, F.; Nguyen, M.T.; Jin, S.; Huigens, R.W. Synthetically Tuning the 2-Position of Halogenated Quinolines: Optimizing Antibacterial and Biofilm Eradication Activities via Alkylation and Reductive Amination Pathways. Chem. A Eur. J. 2016, 22, 9181–9189. [Google Scholar] [CrossRef]
- Bhatt, S.; Nayak, S.K. Copper(II) bromide: A simple and selective monobromination reagent for electron-rich aromatic compounds. Synth. Commun. 2007, 37, 1381–1388. [Google Scholar] [CrossRef]
- Hao, W.; Liu, Y. C-H bond halogenation catalyzed or mediated by copper: An overview. Beilstein J. Org. Chem. 2015, 11, 2132–2144. [Google Scholar] [CrossRef]
- Arjunan, V.; Mohan, S.; Ravindran, P.; Mythili, C.V. Vibrational spectroscopic investigations, ab initio and DFT studies on 7-bromo-5-chloro-8-hydroxyquinoline. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2009, 72, 783–788. [Google Scholar] [CrossRef]
- Krishnakumar, V.; Ramasamy, R. DFT studies and vibrational spectra of isoquinoline and 8-hydroxyquinoline. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.C.; Calvo, S.; Torre, M.H.; Baran, E.J. Vibrational spectra of clioquinol and its Cu(II) complex. J. Raman Spectrosc. 2007, 38, 373–376. [Google Scholar] [CrossRef]
- Vranec, P.; Potočňák, I.; Sabolová, D.; Farkasová, V.; Ipóthová, Z.; Pisarčíková, J.; Paulíková, H. Low-dimensional compounds containing bioactive ligands. V: Synthesis and characterization of novel anticancer Pd(II) ionic compounds with quinolin-8-ol halogen derivatives. J. Inorg. Biochem. 2014, 131, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Bahgat, K.; Ragheb, A.G. Analysis of vibratinal spectra of 8-hydroxyquinoline and its 5,7-dichloro, 5,7-dibromo, 5,7-diiodo and 5,7-dinitro derivatives based on density functional theory calculations. Cent. Eur. J. Chem. 2007, 5, 201–220. [Google Scholar] [CrossRef]
- Malherbe, F.E.; Bernstein, H.J. The infrared and Raman spectra of p-dioxane. J. Am. Chem. Soc. 1952, 74, 4408–4410. [Google Scholar] [CrossRef]
- Alver, Ö.; Parlak, C. Spectroscopic investigations of 1,4-dioxane adsorbed on bentonite from anatolia. Anadolu Univ. J. Sci. Technol. A Appl. Sci. Eng. 2016, 17, 273–277. [Google Scholar] [CrossRef]
- Borowski, P.; Gac, W.; Pulay, P.; Woliński, K. The vibrational spectrum of 1,4-dioxane in aqueous solution-theory and experiment. New J. Chem. 2016, 40, 7663–7670. [Google Scholar] [CrossRef]
- Shastri, A.; Das, A.K.; Krishnakumar, S.; Singh, P.J.; Raja Sekhar, B.N. Spectroscopy of N, N-dimethylformamide in the VUV and IR regions: Experimental and computational studies. J. Chem. Phys. 2017, 147, 224305. [Google Scholar] [CrossRef]
- Wang, C.; Niu, J.; Li, J.; Ma, X. Synthesis, characterization, structures and Suzuki coupling reaction of Cu(II) complexes derived from N and O-containing organic ligand. Inorg. Chim. Acta 2017, 464, 81–87. [Google Scholar] [CrossRef]
- di Vaira, M.; Bazzicalupi, C.; Orioli, P.; Messori, L.; Bruni, B.; Zatta, P. Clioquinol, a drug for Alzheimer’s disease specifically interfering with brain metal metabolism: Structural characterization of its zinc(II) and copper(II) complexes. Inorg. Chem. 2004, 43, 3795–3797. [Google Scholar] [CrossRef]
- Denoyer, D.; Clatworthy, S.A.S.; Cater, M.A. Copper Complexes in Cancer Therapy. Met. Ions Life Sci. 2018, 18, 469–506. [Google Scholar] [CrossRef]
- Shen, W.Y.; Jia, C.P.; Mo, A.N.; Liang, H.; Chen, Z.F. Chemodynamic therapy agents Cu(II) complexes of quinoline derivatives induced ER stress and mitochondria-mediated apoptosis in SK-OV-3 cells. Eur. J. Med. Chem. 2021, 223, 113636. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, K.; Khamrang, T.; Sambantham, K.; Sali, V.K.; Chitgupi, U.; Lovell, J.F.; Mohammad, A.A.; Venugopal, R. Identification of cytotoxic copper(II) complexes with phenanthroline and quinoline, quinoxaline or quinazoline-derived mixed ligands. Polyhedron 2021, 194, 114886. [Google Scholar] [CrossRef]
- Hu, K.; Liu, C.; Li, J.; Liang, F. Copper(II) complexes based on quinoline-derived Schiff-base ligands: Synthesis, characterization, HSA/DNA binding ability, and anticancer activity. MedChemComm 2018, 9, 1663–1672. [Google Scholar] [CrossRef]
- Firmino, G.S.S.; de Souza, M.V.N.; Pessoa, C.; Lourenco, M.C.S.; Resende, J.A.L.C.; Lessa, J.A. Synthesis and evaluation of copper(II) complexes with isoniazid-derived hydrazones as anticancer and antitubercular agents. BioMetals 2016, 29, 953–963. [Google Scholar] [CrossRef]
- Hindo, S.S.; Frezza, M.; Tomco, D.; Heeg, M.J.; Hryhorczuk, L.; McGarvey, B.R.; Dou, Q.P.; Verani, C.N. Metals in anticancer therapy: Copper(II) complexes as inhibitors of the 20S proteasome. Eur. J. Med. Chem. 2009, 44, 4353–4361. [Google Scholar] [CrossRef]
- Martínez Medina, J.J.; Naso, L.G.; Pérez, A.L.; Rizzi, A.; Okulik, N.B.; Valcarcel, M.; Salado, C.; Ferrer, E.G.; Williams, P.A.M. Synthesis, characterization, theoretical studies and biological (antioxidant, anticancer, toxicity and neuroprotective) determinations of a copper(II) complex with 5-hydroxytryptophan. Biomed. Pharmacother. 2019, 111, 414–426. [Google Scholar] [CrossRef]
- Rossi, A.; Poverini, R.; di Lullo, G.; Modesti, A.; Modicat, A.; Scarinos, M.L. Heavy metal toxicity following apical and basolateral exposure in the human intestinal cell line Caco-2. Toxicol. Vitr. 1996, 10, 27–36. [Google Scholar] [CrossRef]
- Panjehpour, M.; Taher, M.-A.; Bayesteh, M. The growth inhibitory effects of cadmium and copper on the MDA-MB468 human breast cancer cells. J. Res. Med. Sci. 2010, 15, 279–286. [Google Scholar]
- Peña, Q.; Sciortino, G.; Maréchal, J.D.; Bertaina, S.; Simaan, A.J.; Lorenzo, J.; Capdevila, M.; Bayón, P.; Iranzo, O.; Palacios, Ò. Copper(II) N, N, O-chelating complexes as potential anticancer agents. Inorg. Chem. 2021, 60, 2939–2952. [Google Scholar] [CrossRef]
- Beeton, M.L.; Aldrich-Wright, J.R.; Bolhuis, A. The antimicrobial and antibiofilm activities of copper(II) complexes. J. Inorg. Biochem. 2014, 140, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Litecká, M.; Hreusová, M.; Kašpárková, J.; Gyepes, R.; Smolková, R.; Obuch, J.; David, T.; Potočňák, I. Low-dimensional compounds containing bioactive ligands. Part XIV: High selective antiproliferative activity of tris(5-chloro-8-quinolinolato)gallium(III) complex against human cancer cell lines. Bioorganic Med. Chem. Lett. 2020, 30, 127206. [Google Scholar] [CrossRef] [PubMed]
- Lüköová, A.; Drweesh, E.A.; Volarevic, V.; Miloradovic, D.; Markovic, B.S.; Smolková, R.; Samoľová, E.; Kuchár, J.; Vilková, M.; Potočňák, I. Low-dimensional compounds containing bioactive ligands. Part XIII: Square planar anti-cancer Pd(II) complexes with halogenderivatives of 8-quinolinol and dimethylamine. Polyhedron 2020, 184, 114535. [Google Scholar] [CrossRef]
- Litecká, M.; Prachařová, J.; Kašpárková, J.; Brabec, V.; Smolková, R.; Gyepes, R.; Obuch, J.; Kubíček, V.; Potočňák, I. Low-dimensional compounds containing bioactive ligands. Part XV: Antiproliferative activity of tris(5-nitro-8-quinolinolato)gallium(III) complex with noticeable selectivity against the cancerous cells. Polyhedron 2020, 187, 114672. [Google Scholar] [CrossRef]
- Origin; Version 2022B; OriginLab Corporation: Northampton, MA, USA, 2022.
- CrysAlisPRO; Version 1.0.43; Oxford Diffraction/Agilent Technologies UK Ltd.: Yarnton, UK, 2020.
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–885. [Google Scholar] [CrossRef]
- Spek, A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Brandenburg, K. Diamond; Version 3.2K; Crystal Impact GbR: Bonn, Germany, 2014. [Google Scholar]
- Rojas, J.J.; Ochoa, V.J.; Ocampo, S.A.; Muñoz, J.F. Screening for antimicrobial activity of ten medicinal plants used in Colombian folkloric medicine: A possible alternative in the treatment of non-nosocomial infections. BMC Complement. Altern. Med. 2006, 6, 2. [Google Scholar] [CrossRef]
- Andrews, J.M. Determination of minimum inhibitory concentrations. J. Antimicrob. Chemother. 2001, 48 (Suppl. S1), 5–16. [Google Scholar] [CrossRef]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free. Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef] [PubMed]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a free radical method to evaluate antioxidant activity. LWT Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]

| 1a | 1b | 3 | 6 | |
|---|---|---|---|---|
| Cu1–O1 | 1.922(3) | 1.920(2) | 1.922(2) | 1.926(2) |
| Cu1–N1 | 1.964(3) | 1.965(2) | 1.958(2) | 1.953(2) |
| O1–Cu1–N1i | 94.90(11) | 94.76(8) | 94.57(9) | 95.81(9) |
| O1–Cu1–N1 | 85.10(11) | 85.24(8) | 85.43(9) | 84.19(9) |
| Bonds | Angles | ||
|---|---|---|---|
| Cu1–O1 | 1.9508(14) | O1–Cu1–N1 i | 96.82(7) |
| Cu1–N1 | 1.9706(17) | O1–Cu1–N1 | 83.18(7) |
| Cu1–O2 | 2.4920(19) | O1–Cu1–O2 | 94.14(6) |
| N1–Cu1–O2 | 91.91(7) | ||
| O1i–Cu1–O2 | 85.86(6) | ||
| N1i–Cu1–O2 | 88.09(7) |
| Cell Lines (IC50 in µM) | ||||||||
|---|---|---|---|---|---|---|---|---|
| MCF-7 | MDA-MB-231 | HCT116 | Caco-2 | HeLa | A549 | Jurkat | Cos-7 | |
| 1a | NT | NT | NT | NT | NT | NT | NT | NT |
| 1b | ˃200 | ˃200 | ˃200 | 106.8 | ˃200 | ˃200 | ˃200 | ˃200 |
| 2 | NT | NT | NT | NT | NT | NT | NT | NT |
| 3 | 5.8 | 6.0 | 5.3 | 5.4 | 5.4 | 5.6 | 5.7 | 5.8 |
| 4 | 204.8 | ˃200 | ˃200 | 46.4 | ˃200 | ˃200 | ˃200 | ˃200 |
| 5 | NT | NT | NT | NT | NT | NT | NT | NT |
| 6 | ˃200 | ˃200 | ˃200 | ˃200 | ˃200 | ˃200 | ˃200 | ˃200 |
| HClBrQ | 36.4 | 6.2 | 23.1 | 19.7 | 40.2 | 24.4 | 5.7 | 65.3 |
| HBrQ | 9.1 | 6.1 | 5.6 | 6.5 | 20.1 | 6.4 | 5.9 | 6.5 |
| HdNQ | 78.1 | 79.5 | 81.5 | ˃200 | ˃200 | ˃200 | 72.8 | ˃200 |
| HClNQ | 6.8 | 13.0 | 8.1 | 75.8 | 47.1 | 28.0 | 34.2 | 112.2 |
| cisplatin | 29.7 | 7.1 | 7.4 | 23.2 | 35.4 | 13.5 | 6.3 | 18.3 |
| RIZD (%) | 1b | 3 | 4 | 6 | HClBrQ | HBrQ | HdNQ | HClNQ |
|---|---|---|---|---|---|---|---|---|
| E. coli | NA | NA | NA | NA | NA | NA | NA | 123.51 |
| S. aureus | NA | NA | NA | NA | NA | NA | NA | 153.31 |
| 1:1 | 1:2 | 1:4 | 1:8 | |
|---|---|---|---|---|
| E. coli | ||||
| HClNQ | 0.047 ± 0.001 | 0.051 ± 0.001 | 0.105 ± 0.007 | 0.166 ± 0.043 |
| Positive control | 0.363 ± 0.007 | 0.363 ± 0.007 | 0.363 ± 0.007 | 0.363 ± 0.007 |
| S. aureus | ||||
| HClNQ | 0.046 ± 0.001 | 0.063 ± 0.003 | 0.107 ± 0.001 | 0.224 ± 0.012 |
| Positive control | 0.341 ± 0.035 | 0.341 ± 0.035 | 0.341 ± 0.035 | 0.341 ± 0.035 |
| ABTS | DPPH | |||
|---|---|---|---|---|
| IC50 (μM) | SC (%) | IC50 (μM) | SC (%) | |
| 3 | 33.33 ± 0.31 | 100 | - | 6.09 ± 2.08 |
| HClBrQ | 109.29 ± 1.50 | 82.80 ± 1.87 | - | 24.08 ± 0.78 |
| HBrQ a | 8.23 ± 0.52 | 100 | 46.03 ± 1.09 | 100 |
| HdNQ | - | 0 | - | 13.489 ± 0.87 |
| L-ascorbic acid | 21.03 ± 0.30 | 100 | 35.24 ± 1.25 | 100 |
| Compound | 1a | 1b | 3 | 5 | 6 |
|---|---|---|---|---|---|
| Empirical formula | C18H8.30Br1.70Cl2CuN2O2 | C18H8Br2Cl2CuN2O2 | C18H10Br2CuN2O2 | C24H22CuN8O12 | C18H8Cl2CuN4O6 |
| Formula weight [g·mol−1] | 554.85 | 578.52 | 509.64 | 678.03 | 510.72 |
| Temperature [K] | 100(2) | 100(2) | 173(2) | 173(2) | 100(2) |
| Wavelength [Å] | 1.54184 | 1.54184 | 0.71073 | 0.71073 | 1.54184 |
| Crystal system | monoclinic | monoclinic | monoclinic | triclinic | monoclinic |
| Space group | P21/c | P21/c | P21/n | P − 1 | P21/c |
| Unit cell dimensions [Å, °] | a = 4.89650(10) | a = 4.88650(10) | a = 4.9383(2) | a = 6.5539(6) | a = 3.7670(2) |
| b = 10.32300(10) | b = 10.3474(3) | b = 10.1335(4) | b = 10.1953(7) | b = 12.4567(6) | |
| c = 17.5282(2) | c = 17.5221(4) | c = 16.2276(6) | c = 11.1513(7) | c = 18.1614(5) | |
| β = 90.182(1) | β = 90.201(2) | β = 90.581(3) | α = 103.833(5) | β = 93.452(3) | |
| β = 100.445(6) | |||||
| γ = 103.446(7) | |||||
| Volume [Å3] | 885.99(2) | 885.96(4) | 812.02(5) | 681.18(9) | 850.67(7) |
| Z; density (calculated) [g·cm−3] | 2; 2.080 | 2; 2.169 | 2; 2.084 | 1; 1.653 | 2; 1.994 |
| Absorption coefficient [mm−1] | 9.162 | 9.962 | 6.280 | 0.883 | 5.193 |
| F(000) | 538 | 558 | 494 | 347 | 510 |
| Crystal shape, colour | needle, yellow | needle, yellow | needle, yellow | prism, green | needle, green |
| Crystal size [mm] | 0.161 × 0.078 × 0.031 | 0.050 × 0.040 × 0.020 | 0.560 × 0.228 × 0.064 | 0.654 × 0.216 × 0.152 | 0.048 × 0.015 × 0.009 |
| θ range for data collection [°] | 4.972–77.225 | 4.964–76.881 | 3.216–28.707 | 3.295–28.514 | 4.306–77.467 |
| Index ranges | −5 ≤ h ≤ 6, −13 ≤ k ≤ 13, −21 ≤ l ≤ 22 | −5 ≤ h ≤ 6, −12 ≤ k ≤ 11, −21 ≤ l ≤ 21 | −6 ≤ h ≤ 6, −13≤ k ≤ 11, −20 ≤ l ≤ 21 | −8 ≤ h ≤ 8, −13 ≤ k ≤ 13, −11 ≤ l ≤ 14 | −2≤ h ≤ 4, −11≤ k ≤ 15, −22≤ l ≤ 22 |
| Reflections collected/independent | 30,356/1833 [R(int) = 0.0494] | 8341/1769 [R(int) = 0.0323] | 5221/1863 [R(int) = 0.0257] | 7356/3093 [R(int) = 0.0261] | 6520/1709 [R(int) = 0.0335] |
| Data/restrains/parameters | 1833/0/124 | 1769/0/124 | 1863/0/115 | 3093/0/237 | 1709/0/142 |
| Goodness-of-fit on F2 | 1.161 | 1.062 | 1.070 | 1.089 | 1.161 |
| Final R indices [I > 2 σ(I)] | R1 = 0.0347, | R1 = 0.0271, | R1 = 0.0312, | R1 = 0.0428, | R1 = 0.0367, |
| wR2 = 0.0768 | wR2 = 0.0776 | wR2 = 0.0646 | wR2 = 0.0905 | wR2 = 0.1057 | |
| R indices (all data) | R1 = 0.0371, | R1 = 0.0296, | R1 = 0.0441, | R1 = 0.0549, | R1 = 0.0452, |
| wR2 = 0.0777 | wR2 = 0.0788 | wR2 = 0.0718 | wR2 = 0.0975 | wR2 = 0.1093 | |
| Largest diff. peak and hole [e·Å−3] | 0.435 and −0.476 | 0.458 and −0.573 | 0.424 and −0.384 | 0.402 and −0.452 | 0.461 and −0.643 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kepeňová, M.; Kello, M.; Smolková, R.; Goga, M.; Frenák, R.; Tkáčiková, Ľ.; Litecká, M.; Šubrt, J.; Potočňák, I. Low-Dimensional Compounds Containing Bioactive Ligands. Part XIX: Crystal Structures and Biological Properties of Copper Complexes with Halogen and Nitro Derivatives of 8-Hydroxyquinoline. Inorganics 2022, 10, 223. https://doi.org/10.3390/inorganics10120223
Kepeňová M, Kello M, Smolková R, Goga M, Frenák R, Tkáčiková Ľ, Litecká M, Šubrt J, Potočňák I. Low-Dimensional Compounds Containing Bioactive Ligands. Part XIX: Crystal Structures and Biological Properties of Copper Complexes with Halogen and Nitro Derivatives of 8-Hydroxyquinoline. Inorganics. 2022; 10(12):223. https://doi.org/10.3390/inorganics10120223
Chicago/Turabian StyleKepeňová, Martina, Martin Kello, Romana Smolková, Michal Goga, Richard Frenák, Ľudmila Tkáčiková, Miroslava Litecká, Jan Šubrt, and Ivan Potočňák. 2022. "Low-Dimensional Compounds Containing Bioactive Ligands. Part XIX: Crystal Structures and Biological Properties of Copper Complexes with Halogen and Nitro Derivatives of 8-Hydroxyquinoline" Inorganics 10, no. 12: 223. https://doi.org/10.3390/inorganics10120223
APA StyleKepeňová, M., Kello, M., Smolková, R., Goga, M., Frenák, R., Tkáčiková, Ľ., Litecká, M., Šubrt, J., & Potočňák, I. (2022). Low-Dimensional Compounds Containing Bioactive Ligands. Part XIX: Crystal Structures and Biological Properties of Copper Complexes with Halogen and Nitro Derivatives of 8-Hydroxyquinoline. Inorganics, 10(12), 223. https://doi.org/10.3390/inorganics10120223










